Abstract
Interactions between the T cell receptor and cognate peptide-MHC are crucial initiating events in the adaptive immune response. These binding events are highly specific yet occur with micromolar affinity. Even weaker interactions between TCR and self-pMHC complexes play critical regulatory roles in T cell development, maintenance and coagonist activity. Due to their low affinity, the kinetics and thermodynamics of such weak interactions are difficult to study. In this work, we used M15, a high-affinity TCR engineered from the 3.L2 TCR system, to study the binding properties, thermodynamics, and specificity of two altered peptide ligands (APLs). Our affinity measurements of the high-affinity TCR support the view that the wild type TCR binds these APLs in the millimolar affinity range, and hence very low affinities can still elicit biological functions. Finally, single methylene differences among the APLs gave rise to strikingly different binding thermodynamics. These minor changes in the pMHC antigen were associated with significant and unpredictable changes in both the entropy and enthalpy of the reaction. As the identical TCR was analyzed with several structurally similar ligands, the distinct thermodynamic binding profiles provide a mechanistic perspective on how exquisite antigen specificity is achieved by the T cell receptor.
Keywords: T cell receptor, peptide-MHC, kinetics, thermodynamics, SPR
1.) Introduction
The T cell receptor (TCR) specifically recognizes peptide-MHC (pMHC) complexes displayed by antigen presenting cells (Davis et al., 1998). This interaction between TCR and pMHC represents a crucial event in the adaptive immune response. In the thymus, TCR α- and β–chains are generated by somatic recombination of various gene segments, yielding αβ dimers capable of recognizing a vast array of molecular structures. Positive selection preserves cells with TCRs able to bind self peptide-bound MHC, while negative selection eliminates cells that bind and are activated by these complexes (Jameson et al., 1995). These selection events generate a repertoire of mature T cells reactive to foreign but not self-antigens, providing concomitant protection from both infection and autoimmunity.
Peripheral T cell activation and proliferation rely on interactions of TCRs with foreign pMHC that are unique, highly specific but low affinity, with KD values in the micromolar range (van der Merwe and Davis, 2003). The kinetic proofreading model has been invoked to explain this behavior, suggesting that signaling complexity downstream of the TCR temporally separates receptor ligation from T cell activation. Consequently, weak ligands not stably engaging the TCR would dissociate before the activation signaling pathway is initiated (McKeithan, 1995; Rabinowitz et al., 1996). Despite their low-affinity, considerable evidence exists showing the functional importance of these interactions in a variety of processes. First, thymic T cells are positively selected on the basis of interactions with weak pMHC ligands that do not trigger T cell activation on binding (Jameson et al., 1995). Further, studies have shown weak TCR-pMHC interactions to be necessary for survival of peripheral naïve T cells (Brocker, 1997; Ernst et al., 1999; Kirberg et al., 1997; Takeda et al., 1996), as well as for maintenance of CD4+ memory T cell metabolism, gene expression and effector function (De Riva et al., 2007; Kassiotis et al., 2002). Finally, self-pMHC molecules have been found to assemble at the immunological synapse and participate as coagonists to T cell activation (Krogsgaard et al., 2005; Yachi et al., 2005).
The study of altered peptide ligands (APLs) has provided valuable insight in understanding the functional outcomes of TCR-pMHC interactions. APLs are generated by making single amino acid substitutions to an agonist peptide ligand, resulting in altered TCR binding affinity and stimulatory potency (Kersh and Allen, 1996). Due to the already naturally low affinity of the TCR, some single mutations may reduce the affinity of the interaction the point where it now only poorly induces T cell activation. These weak agonist APLs can be useful surrogate ligands in investigating the TCR-self pMHC interactions required for selection, maintenance and coagonism of T cells. We have studied APLs using 3.L2, a well characterized TCR recognizing residues 64-76 from the minor d allele of the β-chain of mouse hemoglobin (Hb) bound to the class II molecule I-Ek (Evavold and Allen, 1991). APLs described in this system have biological activities ranging from full T cell agonists to antagonists. Antagonist or partial agonist pMHC ligands induce activation phenotypes and patterns of TCR zeta chain phosphorylation distinct from agonist ligands (Sloan-Lancaster et al., 1994; Sloan-Lancaster et al., 1996). Furthermore, work from our and other laboratories has shown that ligand potency increases with binding half-life, thus implicating TCR-pMHC binding kinetics as a key contributor to T cell activation (Kersh et al., 1998; Lyons et al., 1996; Matsui et al., 1994; Williams et al., 1999). However, examples of TCR-pMHC interactions exist where half-life and biological activity do not correlate (Baker et al., 2000; Krogsgaard et al., 2003). Thus, the precise contribution of TCR-pMHC binding kinetics to T cell activation remains unclear.
A complication of studying weak agonist APLs is that their interaction with a TCR can be so weak as to preclude analysis by even highly sensitive techniques like surface plasmon resonance (SPR). APLs falling into this category may then be classified as null ligands when they can productively, albeit weakly, engage the TCR. In fact, even the affinities of some weak agonist ligands have been unable to be characterized by SPR because they exhibit KD values that are above 100 mM (Li et al., 2005). As a result, it has been impossible to make kinetic or thermodynamic measurements to better appreciate how a TCR distinguishes its cognate ligand from a subtly different APL to give distinct functional effects. Approaches using pMHC or TCR tetramers have provided estimates of KD values for low affinity interactions (Deng et al., 2007; Huseby et al., 2006; Norris et al., 2006), but such methods cannot be used to obtain binding kinetic parameters. We have therefore employed M15, a high-affinity single-chain TCR (scTCR) engineered by in vitro evolution of 3.L2 to probe these important interactions with APLs. Previous work showed that M15 binds Hb/I-Ek with 800-fold higher affinity than does 3.L2, providing a larger affinity window within which to study TCR-pMHC interactions. T cells that express M15 are functional and retain antigen specificity for Hb/I-Ek, showing an extent of T cell activation equivalent to 3.L2 (Weber et al., 2005). M15 is thus a valid and relevant model of the 3.L2 TCR that can extend our biophysical analyses to ligands too weak to study otherwise.
High-affinity TCRs have a wide range of useful applications (Richman and Kranz, 2007), including blockage of T cell activation by staphylococcal superantigen (Buonpane et al., 2007), quantitation of specific pMHC ligand expression (Zhang et al., 2007), and targeting specific viral attachment and entry as a potential gene therapy strategy (Peng et al., 2004). We have used high-affinity TCRs in our work to show that T cells can display both degeneracy and specificity in recognizing antigen, and to estimate the KD values of weak TCR-APL interactions to be in the millimolar range (Donermeyer et al., 2006). In the current study, we used M15 to study the binding kinetics and thermodynamics of Q72/I-Ek and D73/I-Ek, two APLs differing from Hb by a single methylene group (Hb peptide shown in Figure 1A). Q72 is a positively selecting ligand that has an Asn to Gln substitution at the solvent-accessible P5 position, while D73 is a partial agonist that has a Glu to Asp substitution at the solvent-inaccessible, MHC contact, P6 position. Because single methylene changes give rise to the vastly different functions elicited by Hb and these two APLs, we used M15 to study the biophysical parameters governing how a TCR distinguishes a cognate ligand from an APL. The results provide a mechanistic view of how exquisite antigen specificity is achieved by the TCR.
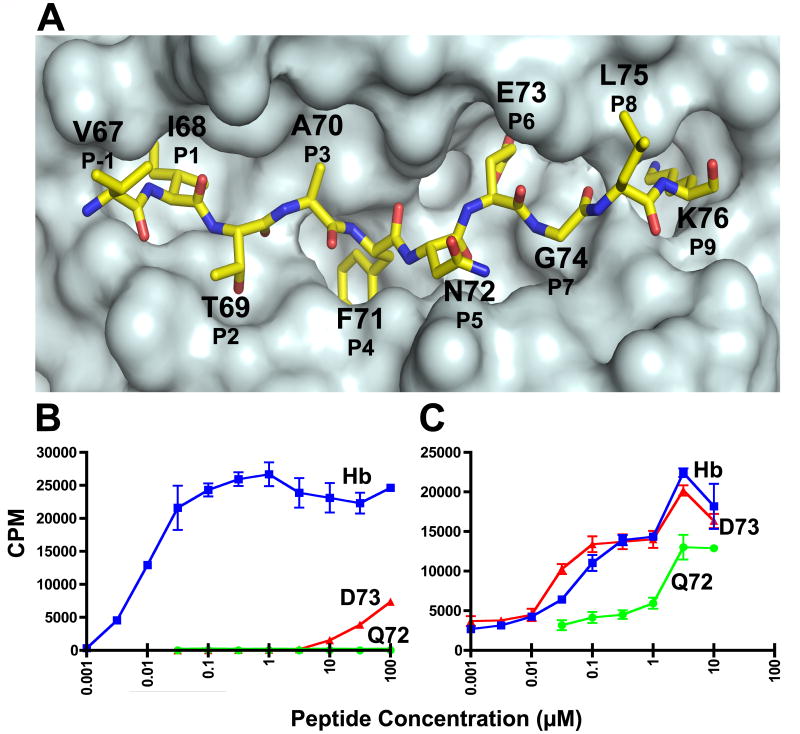
Fig. 1.
T cell response of the 3.L2 T cell and the high-affinity M15. A. A top view of the surface of the crystal structure of Hb/I-Ek showing the anatomy of the Hb peptide. A surface was generated for the I-Ek molecule without the Hb peptide, and then a representation of the Hb peptide was superimposed onto the surface. Each amino acid position of the peptide is designated by the amino acid using the one letter code, the residue number found in the Hb protein chain, and standard peptide register number. There are 4 MHC anchor residues (P1, P4, P6, and P9) and 4 TCR contact residues (P2, P3, P5, and P8). B. The response of the 3.L2 T cell to Hb, D73, and Q72, and C. the response of M15 to Hb, D73, and Q72. T cell hybridoma cells were stimulated by indicated peptide concentrations for 24 hours in the presence of CH27 antigen presenting cells. The level of simulation was assayed by 3H-TdR incorporation into the IL-2 dependent cell line, CTLL-2. The values represent the mean ± triplicate values. Representative experiments of > 3 are shown. Please note, we have published identical findings, but different experiments, of these same T cells and peptides (Donermeyer et al., 2006; Weber et al., 2005). These data are provided to establish the biological responses that are investigated in this study.
2.) Experimental Procedures
2.1) Proteins and Peptides
T cell receptors analyzed include the single-chain version of the 3.L2 TCR and M15, a Hb-specific, high-affinity TCR engineered from 3.L2 by in vitro evolution (Weber et al., 2005). The peptides used in this study and their sequences are as follows: Hb, residues 64-76, GKKVITAFNEGLK; Q72, Asn to Gln substitution of Hb at the P5 position, GKKVITAFQEGLK; D73, Glu to Asp substitution of Hb at P6, GKKVITAFNDGLK; MCC, moth cytochrome C, residues 88-103, ANERADLIAYLKQATK. Boldfaced residues are those occupying the I-Ek peptide binding groove, P1 to P9.
2.2) Protein Production
3.L2 and M15 scTCR proteins were expressed in E. coli and refolded from inclusion bodies as described (Garcia et al., 2001). Briefly, 200 mg of purified inclusion bodies were solubilized in 2 mL Tris EDTA buffer containing 7 M guanidine HCl and 10 mM beta-mercaptoethanol, microfuged to remove undissolved material and added to a 400 mL solution of 3M urea, 10 mM Tris pH 8.0, 2 mM reduced glutathione and 0.2 mM oxidized glutathione for 4 hours with stirring at 4°C. The fold was then diluted to 2 L with 200 mM NaCl, 50 mM Tris pH 8.0 over 16-18 hours with stirring at 4°C. After another 24 hrs, the mixture was filtered through a 0.22 μm filter, returned to 4°C with stirring and 1 mL of Ni agarose beads (Qiagen, Valencia, CA) added. After 24 hours, the agarose beads were recovered by filtration, washed with 10 mM HEPES, 150 mM NaCl pH 7.4, and then eluted with 500 mM imidazole in HEPES buffer. The eluted protein was immediately purified by S200 FPLC chromatography. Purified protein was concentrated with an Amicon Ultra 15 centrifugal filter (Millipore, Bedford, MA) and stored at 4°C. The concentration of the scTCR was determined by A280 using an extinction coefficient of 1.264. Monomeric pMHC complexes were generated by refolding I-Ek from E. coli inclusion bodies in the presence of either Hb, D73, Q72 or MCC peptides as described in a published protocol (Altman et al., 1993). Ek-Ig dimers were produced and purified as described previously (Masteller, 2003). Dimers were loaded with either Hb, D73, Q72, or MCC peptides for 24h at 37°C at pH 5.0, then neutralized prior to use (Masteller, 2003).
2.3) T cell hybridoma assay
Stimulation of the 3.L2 and M15 T cell hybridomas was performed as described (Donermeyer et al., 2006). Briefly, 105 T hybridoma cells in a 96 well tissue culture plate were incubated with 5 × 104 CH27 antigen presenting B cells, along with the Hb, Q72, or D73 peptides (0.001 μM to 100 μM) for 24 hours. The level of T cell stimulation was determined by a bioassay for IL-2 involving 3H-TdR incorporation of the IL-2 dependent cell line, CTLL-2.
2.4) Structural Figures
The molecular graphics program MacPyMol (DeLano Scientific LLC, San Carlos, CA) was used to make the structural figures, using the PDB deposited coordinates of Hb/I-Ek (1FNG) and D73/I-Ek (1FNE) (Kersh et al., 2001).
2.5) Surface Plasmon Resonance
scTCR-pMHC binding kinetics and thermodynamics were analyzed using a Biacore 2000 surface plasmon resonance instrument. For experiments using monomeric pMHC as the ligand, CM5 sensor chips (Biacore AB, Uppsala, Sweden) were activated with a 1:1 mixture of 100 mM N-hydroxysuccinimide and 75 mg/mL 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (Biacore AB, Uppsala, Sweden). Amine coupling of activated chips was accomplished using 1 mg/mL Neutravidin (Pierce, Rockford, IL) diluted 1:15 in 20 mM sodium citrate pH 4.5. Activated but non-Neutravidin bound moieties on the chip surface were blocked with 1 M ethanolamine pH 8.5 (Biacore AB, Uppsala, Sweden). Biotinylated pMHC monomer was immobilized to the chip surfaces to a total response level of 250-500 resonance units (RU) as described (Wu et al., 2002). For experiments with pMHC-Ig dimer as the ligand, CM5 sensor chips were activated as described above and amine coupled directly to dimers to a total response level of 1500-2500 RU, followed by blocking by ethanolamine. For all experiments, injection of sample entailed flowing 80 μL scTCR in HEPES-buffered saline (10 mM HEPES, 3 mM EDTA, 150 mM NaCl and 0.005% Tween 20) over the chip surface with the KINJECT command at a flow rate of 30 μL/min. The flow rate and ligand coupling levels have shown no indication of mass transport or rebinding artifacts. All sensorgrams were corrected for bulk flow effects and nonspecific binding by subtracting the response from a surface bound with MCC/I-Ek monomer, MCC-loaded Ek-Ig dimer or from a blocked, Neutravidin-bound surface with no immobilized pMHC. All subtraction methods produced consistently equivalent results.
2.6) Kinetic analysis
Concentration series of scTCR covering at least two orders of magnitude were injected in duplicate or triplicate at 25°C over surfaces coupled with either monomeric peptide-I-Ek or peptide-Ek-Ig dimers. Sensorgrams from the concentration series were fitted to a 1:1 Langmuir binding model using BiaEvaluation version 4.1 (Biacore AB, Uppsala, Sweden) to calculate the koff, kon, and KD (=koff/kon). KD and maximum response (RMax) values for equilibrium binding analysis were obtained by plotting the equilibrium response (Req) at each concentration and fitting these data to a one-step binding model using GraphPad Prism version 4.0c for Macintosh (GraphPad Software, San Diego, CA). Scatchard plots were generated to confirm 1:1 binding stoichiometry by graphing Req/[scTCR] versus Req. The lines drawn for each Scatchard plot have an x-intercept of RMax and a y-intercept of RMax/KD. The Gibbs free energy change of binding was calculated from the KD using Equation 1 as follows:
| (1) |
where ΔG is the binding free energy change in kcal/mol, R is the universal gas constant, 1.987 cal/mol-K, and T is temperature in Kelvin.
2.7) Van’t Hoff Analysis
scTCR was injected at temperatures in the range of 10°C to 30°C in 2°C increments with 2 to 4 replicates taken per temperature. All thermodynamic data were obtained with SPR surfaces coupled with monomeric peptide-Ek molecules. Sensorgrams were individually fit to a 1:1 Langmuir model using BiaEvaluation version 4.1 (Biacore AB, Uppsala, Sweden) to obtain multiple KA (=kon/koff) values at each temperature. The natural logarithm of each KA was then plotted against inverse absolute temperature. Data were fitted to a nonlinear van’t Hoff model, as shown in Equation 2, and weighted by 1/SD2 as described (Anikeeva et al., 2003; Davis-Harrison et al., 2005) using GraphPad Prism version 4.0c for Macintosh (GraphPad Software, San Diego, CA):
| (2) |
where ΔH is the enthalpy change in kcal/mol, ΔS is the entropy change in kcal/mol-K, ΔCP is the heat capacity change in kcal/mol-K, and T and To are temperatures in Kelvins. All calculations were done using T = 298.15 K. The nonlinear van’t Hoff model was fitted to the data using Equation 3, and ΔH and ΔS were calculated from the fit using Equations 4 and 5 (Huecas and Andreu, 2003):
| (3) |
| (4) |
| (5) |
where a, b and c are constants to be determined. Equation 2 assumes a constant ΔCP that may be calculated using Equation 6:
| (6) |
3.) Results
3.1) Functional and kinetic analysis of scTCR binding to Q72 Hb/I-Ek, a positively selecting APL mutated at a TCR-contact position
As a positively selecting APL for T cells that express the 3.L2 TCR (Williams et al., 1999), the 3.L2-Q72/I-Ek interaction is an ideal candidate for studying the kinetic and thermodynamic parameters of weak TCR-self pMHC interactions. As shown in our previous work, (Donermeyer et al., 2006), wild type Hb peptide, but not Q72 peptide, could induce proliferation in 3.L2 expressing T cells at the indicated peptide concentrations (Figure 1B). With M15-expressing T cells, both Hb and Q72 elicited responses, with Hb inducing a response similar in strength as with the 3.L2 TCR and Q72 inducing a less effective and potent response compared to Hb (Figure 1C). Thus, M15 retains the ability to recognize its cognate ligand and can also respond to a 3.L2 positively selecting ligand.
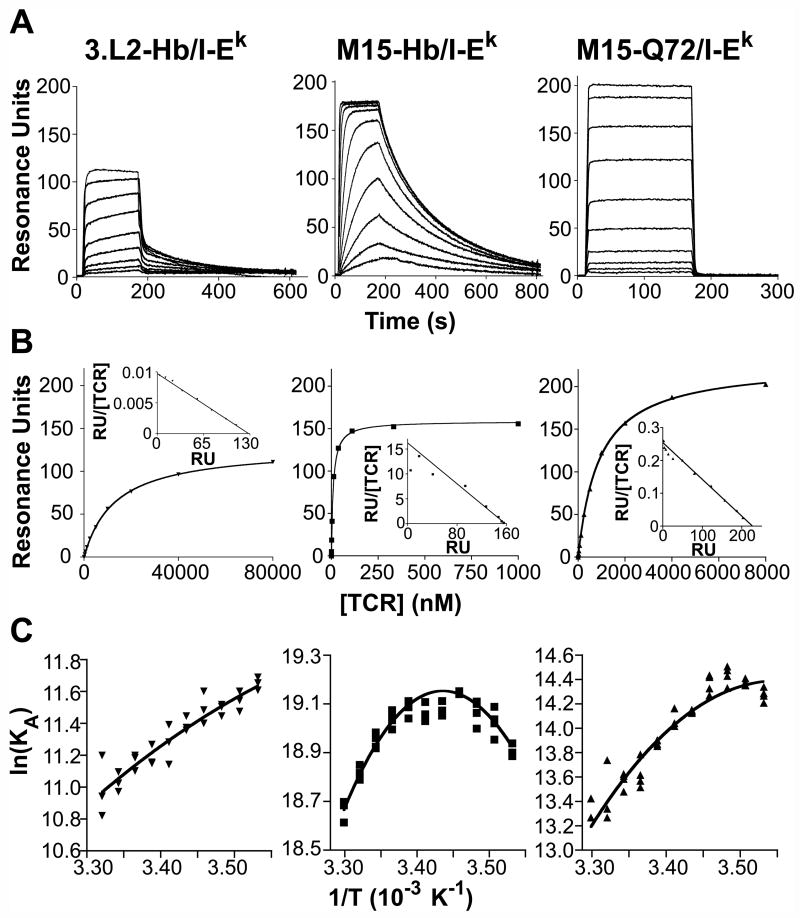
We next used surface plasmon resonance to kinetically explain why M15 can respond to Q72/I-Ek. Consistent with our previous work (Weber et al., 2005), we found that M15 interacts with Hb/I-Ek with a KD of 4.94 nM based on kinetic constants (kon = 1.05 × 106 M-1 s-1, koff = 0.00512 s-1), approximately 5-fold lower than the published value due to faster association kinetics (Figure 2A; Table 1). The 3.L2-Hb/I-Ek interaction had a KD of 17.25 μM (kon = 5.6 × 103 M-1 s-1, koff = 0.0954 s-1), agreeing with our published data. Equilibrium analysis confirmed these values, yielding a KD of 7.27 nM for the M15-Hb/I-Ek interaction and 21.3 μM for the 3.L2-Hb/I-Ek interaction (Figure 2B; Table 1). Comparison of the kinetic values for 3.L2 and M15 interactions with Hb/I-Ek in this work showed that M15 achieved its ~3492-fold higher affinity with a 187.5-fold faster kon and a 18.6-fold slower koff (Table 1). SPR performed with immobilized Q72/I-Ek showed a KD value of 1.2 μM for the interaction based on kinetic constants (kon = 3.33 × 105 M-1 s-1, koff = 0.405 s-1) and 0.68 μM based on the equilibrium binding analysis (Figure 2B; Table 1). Thus, Q72’s agonist activity with M15 correlates with a binding affinity within the expected range for TCR-agonist pMHC interactions.
Fig. 2.
Kinetic and thermodynamic features of 3.L2 and M15 scTCR binding to pMHC. Concentration series of the 3.L2 and M15 scTCRs were injected over a Biacore CM5 SPR chip surface with immobilized pMHC molecules to determine binding kinetics and KD values. A. Sample SPR sensorgrams used to determine binding kinetic parameters of the 3.L2-Hb/I-Ek interaction (left, [3.L2] are 80, 40, 20, 10, 5, 2.5, 1.25, 0.625, and 0.313 μM from top to bottom), M15-Hb/I-Ek interaction (middle, [M15] are 500, 250, 125, 62.5, 31.3, 15.6, 7.8, 3.9, 1.95 and 0.98 nM from top to bottom) and the M15-Q72/I-Ek interaction (right, [M15] are 8, 4, 2, 1, 0.5, 0.25, 0.125, 0.063, 0.032 and 0.016 μM from top to bottom). Sensorgrams shown are representative of three to six experiments. B. Equilibrium binding analysis of 3.L2-Hb/I-Ek (left), M15-Hb/I-Ek (middle) and M15-Q72/I-Ek (right) interactions. Scatchard plots (insets) are linear, confirming 1:1 TCR-pMHC binding stoichiometry. Data are presented as the mean ± standard deviation, and plots are representative of three or four experiments. C. Van’t Hoff analysis of 3.L2-Hb/I-Ek (left), M15-Hb/I-Ek (center) and M15-Q72/I-Ek (right), conducted by plotting the natural logarithm of KA against inverse absolute temperature, to calculate the enthalpy, entropy and heat capacity changes. Two to four KA values were determined per temperature in each experiment, with each replicate displayed on the plots. Each plot has a different scale to better illustrate subtle differences between plots. Plots shown are representative of three to five experiments.
Table 1.
Kinetic and thermodynamic binding parameters for the 3.L2 and M15 TCR interactions with pMHC ligands. Kinetic values were determined using surface plasmon resonance and equilibrium binding analysis, and thermodynamic parameters were calculated with van’t Hoff analysis of KD values obtained with SPR over the temperature range of 10-30°C. Values are presented as mean ± standard deviation of two to six experiments.
| 3.L2-Hb/I-Ek | M15-Hb/I-Ek | M15-Q72/I-Ek | M15-D73/I-Ek | 3.L2-D73/Ek dimer | |
|---|---|---|---|---|---|
| kon (M-1 s-1) | 5600 ± 756 | 1050000 ± 155957 | 333000 ± 30380 | 572000 ± 26986 | 659 ± 145.8 |
| koff (s-1) | 0.0954 ± 0.0098 | 0.00512 ± 0.0000469 | 0.405 ± 0.023 | 0.0402 ± 0.0027 | 0.164 ± 0.029 |
| t1/2a (s) | 6.90 ± 0.29 | 134.77 ± 1.25 | 1.71 ± 0.10 | 17.23 ± 0.97 | 4.21 ± 0.815 |
| KD (koff/kon) (nM) | 17250 ± 2722 | 4.94 ± 0.80 | 1220 ± 77.0 | 70.4 ± 0.591 | 259000 ± 79728 |
| Equilibrium KD (nM) | 21255 ± 7889 | 7.27 ± 1.74 | 682.2 ± 282.1 | 74.1 ± 24.5 | 304858 ± 150365 |
| ΔG (kcal mol-1) | -6.50 ± 0.09 | -11.33 ± 0.09 | -8.07 ± 0.037 | -9.76 ± 0.005 | Not tested |
| ΔH (kcal mol-1) | -7.86 ± 0.79 | -7.98 ± 0.32 | -14.46 ± 0.28 | -15.04 ± 0.63 | Not tested |
| TΔS (kcal mol-1) | -1.36 ± 0.79 | +3.35 ± 0.32 | -6.39 ± 0.28 | -5.28 ± 0.63 | Not tested |
| ΔS (cal mol-1 K-1) | -4.55 ± 2.66 | +11.25 ± 1.08 | -21.48 ± 0.83 | -17.71 ± 2.09 | Not tested |
| ΔCp (kcal mol-1 K-1) | -0.29 ± 0.17 | -1.12 ± 0.14 | -0.91 ± 0.16 | -0.60 ± 0.17 | Not tested |
t1/2 = 0.69/koff
Why Q72 stimulates M15-expressing cells but only positively selects 3.L2 expressing cells would be more ideally investigated by direct kinetic analysis of the 3.L2-Q72/I-Ek interaction by SPR. However, even at 200 μM injected 3.L2 scTCR, we have observed poor binding to immobilized monomeric Q72/I-Ek or Q72/I-Ek-Ig dimer, as expected for a positively selecting ligand (data not shown). To estimate the unknown affinity of 3.L2 for Q72/I-Ek, we assumed that the difference in affinity between the M15-Hb/I-Ek and M15-Q72/I-Ek interactions could be directly extrapolated to the difference between 3.L2-Hb/I-Ek and 3.L2-Q72/I-Ek. Using the known value for the 3.L2-Hb/I-Ek interaction (17.3 μM) and applying to it the 247-fold lower affinity of M15 interacting with Q72/I-Ek compared to Hb/I-Ek, we find a predicted KD value for 3.L2-Q72/I-Ek of 4.3 mM (17.3 μM × 247). This estimated value is consistent both with the lack of 3.L2 binding Q72/I-Ek by SPR and the lack of 3.L2 T cell activation by Q72/I-Ek.
3.2) Thermodynamic studies of scTCR-Q72/I-Ek interactions
The dramatic effect of a single additional methylene group in Q72/I-Ek on the kinetics of binding M15 led us to consider the underlying thermodynamics of Q72/I-Ek and Hb/I-Ek binding to M15. We therefore conducted van’t Hoff analysis in order to calculate ΔH, ΔS, and ΔCP. This method examines the dependence of the binding affinity on temperature to extract thermodynamic information about a protein-protein interaction. The use of van’t Hoff analysis to study the thermodynamics of TCR-pMHC interactions has been reported in the past (Boniface et al., 1999; Garcia et al., 2001; Lee et al., 2004) and shown in several studies to give similar results to microcalorimetry (Davis-Harrison et al., 2005; Ely et al., 2006; Krogsgaard et al., 2003; Willcox et al., 1999).
The thermodynamics of 3.L2 and M15 binding with Hb/I-Ek were first studied to determine how the M15 TCR achieves its increased affinity (Figure 2C; Table 1). The 3.L2-Hb/I-Ek interaction was driven by a negative ΔH (-7.86 kcal/mol) offset by a small negative TΔS (-1.36 kcal/mol) at 25°C. The same analysis of M15 showed that it achieves higher affinity binding not through an increase in enthalpy (ΔH = -7.98 kcal/mol) but by a more favorable entropy change (TΔS = +3.35 kcal/mol).
Despite only a single methylene difference at the interface, the M15-Q72/I-Ek interaction showed a very different thermodynamic binding mechanism than either the 3.L2-Hb/I-Ek or M15-Hb/I-Ek interactions. M15-Q72/I-Ek binding was associated with a much more negative ΔH (-14.46 kcal/mol) but the interaction exhibited considerably more unfavorable entropy (TΔS = -6.39 kcal/mol) (Figure 2C; Table 1). Similar to most other TCR-pMHC interactions that have been examined, each of the TCR- pMHC pairs examined showed a negative ΔCP (ranging from -0.29 to -1.12 kcal/mol-K), consistent with the burial of hydrophobic surface area upon binding (Ely et al., 2006; Krogsgaard et al., 2003).
3.3) Functional and thermodynamic analysis of M15 binding to D73/I-Ek, a peptide variant at a MHC contact residue
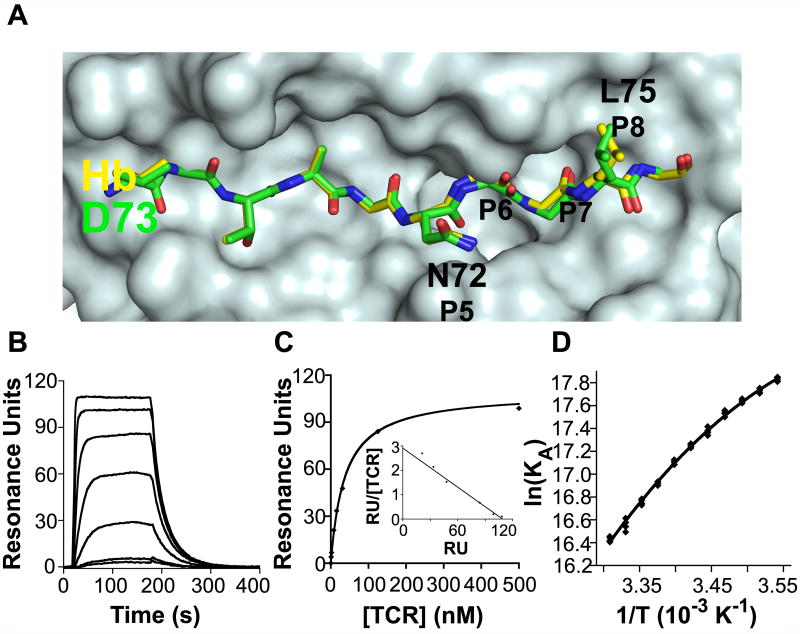
Our kinetic and thermodynamic characterization of M15-Q72/I-Ek demonstrates the remarkable differences between a TCR interaction with cognate pMHC and an APL differing only by a methylene group at a solvent-accessible position. We therefore set out to determine the ramifications of TCR binding to a potentially even more subtle modification of the Hb peptide, D73/I-Ek, which differs from Hb/I-Ek by one fewer methylene group at the solvent inaccessible P6 side chain. The crystal structure for D73/I-Ek has been solved (Figure 3A), revealing that the D73 peptide main chain occupying the P5-P8 positions is slightly displaced compared to Hb and the leucine side chain at the P8 TCR contact position adopts a different rotamer configuration (Kersh et al., 2001). We have shown that the 3.L2 TCR is sensitive to even this relatively minor structural change caused by subtle modification of a solvent inaccessible residue, as D73 is a weak agonist for 3.L2 T cells (Figure 1B). Cells expressing M15 appear functionally insensitive to the methylene change at P6, as they respond similarly to D73 as to Hb (Figure 1C).
Fig. 3.
Structure, kinetics and thermodynamics of D73/I-Ek, a weak agonist ligand. A. A top view of the pMHC surface comparing the crystal structures of Hb and D73 (Kersh et al., 2001). A surface was generated from the I-Ek molecule including the 4 Hb anchor residues side chains, onto which the stick representations of Hb (yellow) and D73 (green) were superimposed. As previously reported, the structures are identical except in the P6 to P8 regions of the peptides, where there is a shift in the P6 carbonyl oxygen, a shift in the P7 main chain, and a different P8 leucine rotamer. B. Sample SPR sensorgram used to determine kinetic binding parameters for the M15-D73/I-Ek interaction ([M15] are 500, 250, 125, 62.5, 31.3, 15.6 and 7.8 nM from top to bottom). Sensorgram shown is representative of three experiments. C. Equilibrium binding analysis of the M15-D73/I-Ek interaction to determine the equilibrium KD. Data are presented as the mean ± standard deviation, and plots are representative of three separate experiments. D. Van’t Hoff analysis to calculate thermodynamic binding properties of the M15- D73/I-Ek interaction. Data are representative of three experiments.
The M15-D73/I-Ek interaction had a KD value of 70.4 nM based on kinetic constants (kon = 5.72 × 105 M-1 s-1, koff = 0.0402 s-1) and 74.1 nM based on equilibrium analysis (Figures 3B and 3C; Table 1). Again, we observed approximately 5-fold stronger affinity than our previous measurements, due to a faster kon. M15-D73/I-Ek had approximately 14-fold weaker binding affinity compared to M15-Hb/I-Ek due to a 1.8-fold slower kon and a 7.9-fold faster koff. Extrapolation of these differences to the 3.L2 TCR predicts a KD value of 246 μM for the 3.L2-D73/I-Ek interaction. This value is near the lower range of TCR affinities that have been proposed to yield weak agonist activity (Li et al., 2005), consistent with the weak activity observed for 3.L2 with D73 (Figure 1B).
Van’t Hoff analysis of the M15-D73/I-Ek interaction (Figure 3D) revealed that the thermodynamic strategy used by M15 to bind D73/I-Ek resembles that used to bind Q72/I-Ek, with very favorable enthalpy (ΔH = -15.04 kcal/mol) driving an entropically unfavorable (ΔS = -5.28 kcal/mol) interaction (Table 1). A negative ΔCP (-0.60 kcal/mol-K) was again observed, although this value was lower than that observed for M15 binding to Q72/I-Ek or Hb/I-Ek.
3.4) Validation of binding affinity for the 3.L2-D73/I-Ek interaction
The estimated binding affinity for the 3.L2-D73/I-Ek interaction is near, but still within, the limit of sensitivity for SPR. We therefore attempted to validate our affinity extrapolations of 3.L2-D73/I-Ek with direct measurement by SPR. However, injections of scTCR over monomeric D73/I-Ek routinely gave low quality sensorgrams that could not be interpreted by curve-fitting or equilibrium analysis. To circumvent what appeared to be an issue specific to a monomeric D73/I-Ek-bound surface, we prepared dimeric D73-loaded I-Ek-Ig fusion protein for use as an SPR ligand. The kinetic constants obtained for the TCR interactions with the monomeric pMHC used in this study agreed with those obtained with Ig dimers, confirming that the system can substitute for monomeric pMHC.
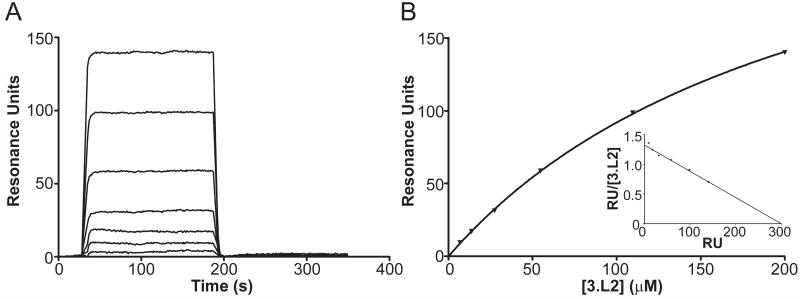
With Ig dimers coupled to our SPR surfaces, we were able to obtain readily analyzable sensorgrams, yielding a KD of 259 μM (koff = 0.164s-1, kon= 659 M-1 s-1) from kinetic constants and 304.9 μM by equilibrium analysis (Figure 4A and 4B, Table 1), consistent with our estimation of 246 μM. Scatchard plots for Ig-dimers interactions are linear, demonstrating the appropriateness of a 1:1 binding model and the independence of the two TCR binding sites (Figure 4B). Compared to 3.L2-Hb/I-Ek, 3.L2- D73/I-Ek has a ~9-fold slower kon and 1.6-fold faster koff. Thus, the factor most clearly distinguishing an activating from a non-activating TCR-pMHC interaction is kon, contrasting with the association with koff seen with many of the interactions in this and other studies.
Fig. 4.
Validation of the kinetics and binding affinity of the 3.L2-D73/I-Ek interaction. A. Sample SPR sensorgram used to determine kinetic binding parameters for the 3.L2-D73/I-Ek interaction ([3.L2] are 200, 109.5, 54.8, 27.4, 13.7, 6.9, and 3.5 μM from top to bottom). Sensorgram shown is representative of four experiments. B. Equilibrium binding analysis of the 3.L2-D73/I-Ek interaction to determine the equilibrium KD. Data are presented as the mean ± standard deviation, and plots are representative of four experiments.
4.) Discussion
The goal of this study was to understand the functional, kinetic and thermodynamic properties governing how the 3.L2 TCR binds to Q72/I-Ek and D73/I-Ek, two APLs of Hb/I-Ek generated by addition or removal of single methylene groups, respectively. The major barrier to characterizing the interaction of these APLs with the wild-type 3.L2 TCR is that the mutations introduced weaken the affinities to levels difficult to measure by SPR. In general, many normal TCR-pMHC interactions share this limitation, making kinetic and thermodynamic evaluation virtually impossible in such cases. This is compounded by the fact that many TCRs cannot be produced in sufficient quantity to conduct such studies (Rudolph et al., 2006). Even with sufficient TCR for study, low quality sensorgrams arising from aggregation or other solubility issues becomes problematic with high injected concentrations of scTCR. The enhanced affinity of the M15 TCR gave us a unique opportunity to biophysically study how a TCR distinguishes subtly different ligands that would be too weak to probe using the wild type TCR. We have shown that the peptide specificity and function of M15 are the same as the 3.L2 TCR, confirming M15 as a relevant model for studying kinetics and thermodynamics of weak 3.L2 ligands. Thus, we have presented a novel system for differentiating interactions that lead to T cell activation from those that play critical roles in T cell effector function, development and maintenance.
The principal finding in the present study is that a single TCR uses a variety of thermodynamic mechanisms to bind to ligands differing by a single methylene group. The gain or loss of single methylene groups from the Hb/I-Ek molecules, as seen in the Q72/I-Ek and D73/I-Ek ligands, respectively, had profound effects on binding thermodynamics with the M15 TCR. In both cases, binding to M15 was more favorable enthalpically with a compensating entropy penalty, compared to Hb/I-Ek. One might anticipate such dramatic enthalpy-entropy compensation with recognition of structurally very different pMHC ligands (Mazza et al., 2007), or with structurally very different TCRs (Anikeeva et al., 2003; Davis-Harrison et al., 2005; Ely et al., 2006).
For ligands like Q72/I-Ek differing at a residue exposed to the TCR, it is perhaps not surprising that a methylene change affected binding thermodynamics. Indeed, the loss of a methyl group from MCC peptide at its P8 position due to a Thr to Ser mutation had such an effect (Krogsgaard et al., 2003). The thermodynamic differences we observed in the M15-Q72/I-Ek interaction could result from perturbation of the TCR-pMHC interface in a region of close packing by an extended side chain at P5. It could also be that desolvating P5 upon binding is better compensated when it is Asn rather than Gln, due possibly to formation of a stable network of hydrogen bonds or interactions with buried water molecules not possible when P5 is Gln. The difference in thermodynamics of M15-D73/I-Ek compared to M15-Hb/I-Ek is quite noteworthy, as this difference originates from a change at a residue that itself contacts only the MHC. However, this perturbation alters the peptide backbone orientation from P5 to P8 and the rotamer configuration at P8. Thus, the dramatic differences in thermodynamic mechanisms between M15 binding to Hb and D73 are most likely due to indirect conformational effects on the ligand, which are sensed by the TCR and thus contribute to its exquisite specificity. Crystal structures will be necessary, however, to rigorously interpret the basis of these thermodynamic differences
Our thermodynamic analyses also showed that the M15’s ~3500-fold higher affinity than 3.L2 for Hb/I-Ek, is accounted for solely by more favorable entropy. This contrasts with a recent study of another high-affinity TCR called m6, which exhibited an improvement in enthalpy but an unfavorable entropy, compared to the wild type 2C TCR, for binding to the class I ligand QL9/Ld (Colf et al., 2007). Thus, high-affinity interactions can be achieved by different thermodynamic means. While initial studies with wild type TCRs suggested that they have conformational flexibility that resulted in an entropy penalty upon pMHC binding (Boniface et al., 1999; Garcia et al., 2001; Willcox et al., 1999), recent studies have shown that this is not universally the case (Anikeeva et al., 2003; Davis-Harrison et al., 2005; Ely et al., 2006; Krogsgaard et al., 2003; Mazza et al., 2007). Thus, while the M15 TCR has been engineered in vitro for higher affinity, it exhibits contributions from both enthalpic factors (as seen with the wild type 3.L2 TCR) and entropic factors (as seen only with M15). The favorable entropy change seen for M15 binding to Hb/I-Ek could be related to the greatly increased kon of M15, compared to 3.L2. The slow kon of a typical TCR-pMHC interaction can be explained by conformational flexibility of the TCR, as this would lower the chance that a TCR will have the correct conformation to recognize its ligand (van der Merwe and Davis, 2003). The loss of this flexibility upon ligand engagement would represent a loss of entropy and would thus be unfavorable. The directed evolution process used to generate M15 may have selected TCRs that exhibited a “lock and key” improved fit for the pMHC, as has been seen with affinity-matured antibodies (Yin et al., 2001). Alternatively, there may be significant differences between the two interfaces (M15-Hb/I-Ek and 3.L2-Hb/I-Ek) in terms of hydrophobic effects.
The significantly more negative heat capacity seen in the M15-Hb/I-Ek interaction compared to 3.L2-Hb/I-Ek is also consistent with more entropically favorable binding, as it suggests more extensive burial of nonpolar protein surface area than seen with 3.L2. As these hydrophobic surfaces are buried, the ordered shell of water surrounding them would disperse, resulting in greater entropy. However, assuming that a heat capacity change of -0.2 to -0.3 kcal mol-1 K-1 predicts a burial of ~1000 Å2 of protein surface area (Murphy and Freire, 1992; Murphy, 1993), the M15-Hb/I-Ek interaction would bury approximately between 3700 and 5600 Å2 of surface area, much larger than the 1000-2500 Å2 expected for a TCR-pMHC interaction. M15 interactions with Q72/I-Ek and D73/I-Ek pMHC molecules also showed more negative ΔCP values than the 3.L2-Hb/I-Ek interaction. We would not expect the differences between the pMHC ligands studied and between the M15 and 3.L2 TCRs to significantly increase the hydrophobicity of these interacting surfaces, suggesting that hydrophobicity alone is not responsible for the observed heat capacity changes. As previously discussed (Krogsgaard et al., 2003), one possible explanation for large ΔCP values in TCR-pMHC interactions is that these values indicate conformational change and/or change in TCR flexibility that may contribute to T cell activation. Support of such a mechanism in our system, however, necessitates structural data and more extensive thermodynamic analysis of 3.L2 and M15 interactions with pMHC than are presented here.
Our use of M15 also gave us an opportunity to estimate KD values for the 3.L2-Q72/I-Ek and 3.L2-D73/I-Ek interactions to lie in the millimolar range (4.30 mM and 0.246 mM for Q72/I-Ek and D73/I-Ek, respectively), as suggested in previous work (Donermeyer et al., 2006). To validate these estimates, we attempted direct binding studies of 3.L2 to D73/I-Ek and Q72/I-Ek. As expected, scant binding to Q72/I-Ek was observed even with 3.L2 scTCR concentrations up to 200 μM, strongly suggesting that the concentration at which we would theoretically observe half-maximal binding (ie, the KD) indeed lies well into the millimolar range. Using a D73/I-Ek-Ig dimer as an SPR ligand, we were able to obtain KD values (259 μM and 304 μM by kinetics and equilibrium analysis, respectively) that agree with our extrapolation (246 μM).
Though useful in evaluating the affinity range of interactions too weak to study conventionally, we recognize that there is likely not a general ability to extrapolate KD values obtained with TCR binding one ligand to another ligand. However, examples exist demonstrating correlations to this effect. For example, a high-affinity version of a Vβ8 TCR binds the ligand SEC-1A4 with 10-fold greater affinity and the ligand SEC3 with 1000-fold greater affinity than the wild type Vβ8 TCR (Kieke et al., 2001). Further, a high-affinity version of the 2C TCR binds the ligand SIY/Kb with 2000-fold greater affinity and the ligand dEV8/Kb with 12-fold greater affinity than the wild type 2C receptor (Holler et al., 2003). Given the myriad complication with studying weak TCR-pMHC interactions as already discussed, we believe our method, though based upon estimations, provides information not reliably obtainable by alternative methods.
Protein interactions with millimolar-range affinities are not unprecedented, though few studies have focused on their biological importance (Nooren and Thornton, 2003). One such study involves the identification of an interaction between the focal adhesion proteins Nck-2 and PINCH-1, which bind with a KD of approximately 3.0 mM (Vaynberg et al., 2005). This interaction, although extremely weak, was shown to be an important regulator of cell shape and migration via the integrin signaling pathway. Another such study found that a pMHC ligand that bound the 2C TCR with millimolar-range affinity exhibited weak agonist activity (Sykulev et al., 1994). Such low affinities presumably are capable of eliciting biological functions, including signaling, by virtue of the multivalent interactions that occur at cell membranes (e.g. with TCRs in an immunological synapse). It is important to note that while the affinities of such protein interactions measured by SPR may be low, this may not be the case when these interactions occur at adjacent cell membranes. Interactions found to be low affinity according to solution measurements may be sufficiently strong when both binding partners are constrained to two-dimensional movement in their respective plasma membranes (Qi et al., 2006).
Finally, as previously reported, the koff appears to correlate with biological activity for many of the interactions studied here. M15-Hb/I-Ek (koff = 0.00512 s-1), M15-D73/I-Ek (koff = 0.0402 s-1), and 3.L2-Hb/I-Ek (koff = 0.0954 s-1) trigger a similarly high level of T cell activation exceeding that seen with Q72/I-Ek (koff = 0.405 s-1), with no apparent association with the kon. However, 3.L2-D73/I-Ek, a poorly stimulatory interaction, has a koff that is only ~1.6-fold faster than 3.L2-Hb/I-Ek, a fully stimulatory interaction. 3.L2-D73/I-Ek is also less stimulatory than M15-Q72/I-Ek, despite the fact that its koff is 2.5-fold slower than M15-Q72/I-Ek. Dissociation kinetics alone, therefore, do not distinguish productive from nonproductive interactions in this case. The kon for 3.L2-D73/I-Ek is ~9-fold lower than for 3.L2-Hb/I-Ek, and could be an important distinguishing factor for these two interactions. It is possible that even with similar half-lives in the bound state, the relative infrequency of 3.L2 binding to D73/I-Ek upon collision at a cell-cell contact is insufficient to trigger productive signaling. However, since both kon and koff are different between these two interactions, we cannot attribute the difference in stimulatory behavior solely to one or the other. Even so, it is remarkable that over such a narrow range of association and/or dissociation rate constants, a distinction between an activating and non-activating stimulus can be made.
Our data have allowed us to kinetically and thermodynamically analyze how a TCR differentiates subtly different pMHC to give rise to different functional effect. Our use of a high-affinity TCR has allowed us to probe ligands whose affinities with the wild-type receptor would have precluded analysis by conventional molecular techniques. Such studies are informative of the molecular and biophysical basis of antigen specificity, using surrogate probes such as M15, for the surface recognized by normal TCRs.
Acknowledgments
We thank Steve Horvath for peptide and pMHC synthesis. This work was supported by grants from The National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman JD, Reay PA, Davis MM. Formation of functional peptide complexes of class II major histocompatibility complex proteins from subunits produced in Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:10330–10334. doi: 10.1073/pnas.90.21.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anikeeva N, Lebedeva T, Krogsgaard M, Tetin SY, Martinez-Hackert E, Kalams SA, Davis MM, Sykulev Y. Distinct molecular mechanisms account for the specificity of two different T-cell receptors. Biochemistry. 2003;42:4709–16. doi: 10.1021/bi026864+. [DOI] [PubMed] [Google Scholar]
- Baker BM, Gagnon SJ, Biddison WE, Wiley DC. Conversion of a T cell antagonist into an agonist by repairing a defect in the TCR/peptide/MHC interface: implications for TCR signaling. Immunity. 2000;13:475–84. doi: 10.1016/s1074-7613(00)00047-9. [DOI] [PubMed] [Google Scholar]
- Boniface JJ, Reich Z, Lyons DS, Davis MM. Thermodynamics of T cell receptor binding to peptide-MHC: evidence for a general mechanism of molecular scanning. Proc Natl Acad Sci U S A. 1999;96:11446–51. doi: 10.1073/pnas.96.20.11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker T. Survival of mature CD4 T lymphocytes is dependent on major histocompatibility complex class II-expressing dendritic cells. J Exp Med. 1997;186:1223–32. doi: 10.1084/jem.186.8.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonpane RA, Churchill HR, Moza B, Sundberg EJ, Peterson ML, Schlievert PM, Kranz DM. Neutralization of staphylococcal enterotoxin B by soluble, high-affinity receptor antagonists. Nat Med. 2007;13:725–729. doi: 10.1038/nm1584. [DOI] [PubMed] [Google Scholar]
- Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–44. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- Davis-Harrison RL, Armstrong KM, Baker BM. Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J Mol Biol. 2005;346:533–50. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- De Riva A, Bourgeois C, Kassiotis G, Stockinger B. Noncognate interaction with MHC class II molecules is essential for maintenance of T cell metabolism to establish optimal memory CD4 T cell function. J Immunol. 2007;178:5488–95. doi: 10.4049/jimmunol.178.9.5488. [DOI] [PubMed] [Google Scholar]
- Deng L, Langley RJ, Brown PH, Xu G, Teng L, Wang Q, Gonzales MI, Callender GG, Nishimura MI, Topalian SL, Mariuzza RA. Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor. Nat Immunol. 2007;8:398–408. doi: 10.1038/ni1447. [DOI] [PubMed] [Google Scholar]
- Donermeyer DL, Weber KS, Kranz DM, Allen PM. The study of high-affinity TCRs reveals duality in T cell recognition of antigen: specificity and degeneracy. J Immunol. 2006;177:6911–9. doi: 10.4049/jimmunol.177.10.6911. [DOI] [PubMed] [Google Scholar]
- Ely LK, Beddoe T, Clements CS, Matthews JM, Purcell AW, Kjer-Nielsen L, McCluskey J, Rossjohn J. Disparate thermodynamics governing T cell receptor-MHC-I interactions implicate extrinsic factors in guiding MHC restriction. Proc Natl Acad Sci U S A. 2006;103:6641–6. doi: 10.1073/pnas.0600743103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst B, Lee DS, Chang JM, Sprent J, Surh CD. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 1999;11:173–81. doi: 10.1016/s1074-7613(00)80092-8. [DOI] [PubMed] [Google Scholar]
- Evavold BD, Allen PM. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 1991;252:1308–1310. doi: 10.1126/science.1833816. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Radu CG, Ho J, Ober RJ, Ward ES. Kinetics and thermodynamics of T cell receptor- autoantigen interactions in murine experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2001;98:6818–23. doi: 10.1073/pnas.111161198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- Huecas S, Andreu JM. Energetics of the cooperative assembly of cell division protein FtsZ and the nucleotide hydrolysis switch. J Biol Chem. 2003;278:46146–54. doi: 10.1074/jbc.M307128200. [DOI] [PubMed] [Google Scholar]
- Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- Kassiotis G, Garcia S, Simpson E, Stockinger B. Impairment of immunological memory in the absence of MHC despite survival of memory T cells. Nat Immunol. 2002;3:244–50. doi: 10.1038/ni766. [DOI] [PubMed] [Google Scholar]
- Kersh GJ, Allen PM. Essential flexibility in the T-cell recognition of antigen. Nature. 1996;380:495–8. doi: 10.1038/380495a0. [DOI] [PubMed] [Google Scholar]
- Kersh GJ, Kersh EN, Fremont DH, Allen PM. High- and low-potency ligands with similar affinities for the TCR: the importance of kinetics in TCR signaling. Immunity. 1998;9:817–826. doi: 10.1016/s1074-7613(00)80647-0. [DOI] [PubMed] [Google Scholar]
- Kersh GJ, Miley MJ, Nelson CA, Grakoui A, Horvath S, Donermeyer DL, Kappler J, Allen PM, Fremont DH. Structural and functional consequences of altering a peptide MHC anchor residue. J Immunol. 2001;166:3345–54. doi: 10.4049/jimmunol.166.5.3345. [DOI] [PubMed] [Google Scholar]
- Kieke MC, Sundberg E, Shusta EV, Mariuzza RA, Wittrup KD, Kranz DM. High affinity T cell receptors from yeast display libraries block T cell activation by superantigens. J Mol Biol. 2001;307:1305–1315. doi: 10.1006/jmbi.2001.4560. [DOI] [PubMed] [Google Scholar]
- Kirberg J, Berns A, von Boehmer H. Peripheral T cell survival requires continual ligation of the T cell receptor to major histocompatibility complex-encoded molecules. J Exp Med. 1997;186:1269–75. doi: 10.1084/jem.186.8.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–43. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- Krogsgaard M, Prado N, Adams EJ, He XL, Chow DC, Wilson DB, Garcia KC, Davis MM. Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation. Mol Cell. 2003;12:1367–78. doi: 10.1016/s1097-2765(03)00474-x. [DOI] [PubMed] [Google Scholar]
- Lee JK, Stewart-Jones G, Dong T, Harlos K, Di Gleria K, Dorrell L, Douek DC, van der Merwe PA, Jones EY, McMichael AJ. T cell cross-reactivity and conformational changes during TCR engagement. J Exp Med. 2004;200:1455–66. doi: 10.1084/jem.20041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bbound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DS, Lieberman SA, Hampl J, Boniface JJ, Chien Y, Berg LJ, Davis MM. A TCR binds to antagonist ligands with lower affinities and faster dissociation rates than to agonists. Immunity. 1996;5:53–61. doi: 10.1016/s1074-7613(00)80309-x. [DOI] [PubMed] [Google Scholar]
- Masteller EL, Warner MR, Ferlin W, Judkowski V, Wilson D, Glaichenhaus N, Bluestone JA. Peptide-MHC Class II Dimers as Therapeutics to Modulate Antigen-Specific T Cell Responses in Autoimmune Diabetes. J Immunol. 2003;171:5587–5595. doi: 10.4049/jimmunol.171.10.5587. [DOI] [PubMed] [Google Scholar]
- Matsui K, Boniface JJ, Steffner P, Reay PA, Davis MM. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc Natl Acad Sci U S A. 1994;91:12862–6. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza C, Auphan-Anezin N, Gregoire C, Guimezanes A, Kellenberger C, Roussel A, Kearney A, van der Merwe PA, Schmitt-Verhulst A, Malissen B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci U S A. 1995;92:5042–6. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP, Freire E. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- Murphy KP, Xie D, Garcia KC, Amzel LM, Freire E. Structural energetics of peptide recognition: angiotensin II/antibody binding. Proteins. 1993;15:113–120. doi: 10.1002/prot.340150203. [DOI] [PubMed] [Google Scholar]
- Nooren IM, Thornton JM. Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- Norris PJ, Stone JD, Anikeeva N, Heitman JW, Wilson IC, Hirschkorn DF, Clark MJ, Moffett HF, Cameron TO, Sykulev Y, Stern LJ, Walker BD. Antagonism of HIV-specific CD4+ T cells by C-terminal truncation of a minimum epitope. Mol Immunol. 2006;43:1349–1357. doi: 10.1016/j.molimm.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng KW, Holler PD, Orr BA, Kranz DM, Russell SJ. Targeting virus entry and membrane fusion through specific peptide/MHC complexes using a high-affinity T cell receptor. Gene Ther. 2004;11:1234–1239. doi: 10.1038/sj.gt.3302286. [DOI] [PubMed] [Google Scholar]
- Qi S, Krogsgaard M, Davis MM, Chakraborty AK. Molecular flexibility can influence the stimulatory ability of receptor-ligand interactions at cell-cell junctions. Proc Natl Acad Sci U S A. 2006;103:4416–4421. doi: 10.1073/pnas.0510991103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Beeson C, Lyons DS, Davis MM, McConnell HM. Kinetic discrimination in T-cell activation. Proc Natl Acad Sci U S A. 1996;93:1401–5. doi: 10.1073/pnas.93.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman SA, Kranz DM. Display, engineering, and applications of antigen-specific T cell receptors. Biomol Eng. 2007 doi: 10.1016/j.bioeng.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Sloan-Lancaster J, Shaw AS, Rothbard JB, Allen PM. Partial T cell signaling: altered phospho-zeta and lack of zap70 recruitment in APL-induced T cell anergy. Cell. 1994;79:913–922. doi: 10.1016/0092-8674(94)90080-9. [DOI] [PubMed] [Google Scholar]
- Sloan-Lancaster J, Steinberg TH, Allen PM. Selective activation of the calcium signaling pathway by altered peptide ligands. J Exp Med. 1996;184:1525–1530. doi: 10.1084/jem.184.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykulev Y, Brunmark A, Jackson M, Cohen RJ, Peterson PA, Eisen HN. Kinetics and Affinity of Reactions between an Antigen-Specific T Cell Receptor and Peptide-MHC Complexes. Immunity. 1994;1:15–22. doi: 10.1016/1074-7613(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Takeda S, Rodewald HR, Arakawa H, Bluethmann H, Shimizu T. MHC class II molecules are not required for survival of newly generated CD4+ T cells, but affect their long-term life span. Immunity. 1996;5:217–28. doi: 10.1016/s1074-7613(00)80317-9. [DOI] [PubMed] [Google Scholar]
- van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–84. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- Vaynberg J, Fukuda T, Chen K, Vinogradova O, Velyvis A, Tu Y, Ng L, Wu C, Qin J. Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell. 2005;17:513–23. doi: 10.1016/j.molcel.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Weber KS, Donermeyer DL, Allen PM, Kranz DM. Class II-restricted T cell receptor engineered in vitro for higher affinity retains peptide specificity and function. Proc Natl Acad Sci U S A. 2005;102:19033–8. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BE, Gao GF, Wyer JR, Ladbury JE, Bell JI, Jakobsen BK, van der Merwe PA. TCR binding to peptide-MHC stabilizes a flexible recognition interface. Immunity. 1999;10:357–65. doi: 10.1016/s1074-7613(00)80035-7. [DOI] [PubMed] [Google Scholar]
- Williams CB, Engle DL, Kersh GJ, White JM, Allen PM. A kinetic threshold between negative and positive selection based on the longevity of the T cell receptor-ligand complex. J Exp Med. 1999;189:1531–44. doi: 10.1084/jem.189.10.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–6. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- Yachi PP, Ampudia J, Gascoigne NR, Zal T. Nonstimulatory peptides contribute to antigen-induced CD8-T cell receptor interaction at the immunological synapse. Nat Immunol. 2005;6:785–792. doi: 10.1038/ni1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Mundorff EC, Yang PL, Wendt KU, Hanway D, Stevens RC, Schultz PG. A comparative analysis of the immunological evolution of antibody 28B4. Biochemistry. 2001;40:10764–10773. doi: 10.1021/bi010536c. [DOI] [PubMed] [Google Scholar]
- Zhang B, Bowerman NA, Salama JK, Schmidt H, Spiotto MT, Schietinger A, Yu P, Fu YX, Weichselbaum RR, Rowley DA, Kranz DM, Schreiber H. Induced sensitization of tumor stroma leadds to eradication of established cancer by T cells. J Exp Med. 2007;204:49–55. doi: 10.1084/jem.20062056. [DOI] [PMC free article] [PubMed] [Google Scholar]