Abstract
Genome-wide association studies (GWAS) have emerged as an important tool for discovering regions of the genome that harbor genetic variants that confer risk for different types of cancers. The success of GWAS in the last 3 years is due to the convergence of new technologies that can genotype hundreds of thousands of single-nucleotide polymorphism markers together with comprehensive annotation of genetic variation. This approach has provided the opportunity to scan across the genome in a sufficiently large set of cases and controls without a set of prior hypotheses in search of susceptibility alleles with low effect sizes. Generally, the susceptibility alleles discovered thus far are common, namely, with a frequency in one or more population of >10% and each allele confers a small contribution to the overall risk for the disease. For nearly all regions conclusively identified by GWAS, the per allele effect sizes estimated are <1.3. Consequently, the findings of GWAS underscore the complex nature of cancer and have focused attention on a subset of the genetic variants that comprise the genomic architecture of each type of cancer, which already can differ substantially by the number of regions associated with specific types of cancer. For instance, in prostate cancer, there could be >30 distinct regions harboring common susceptibility alleles identified by GWAS, whereas in lung cancer, a disease strongly driven by exposure to tobacco products, so far, only three regions have been conclusively established. To date, >85 regions have been conclusively associated in over a dozen different cancers, yet no more than five regions have been associated with more than one distinct cancer type. GWAS are an important discovery tool that require extensive follow-up to map each region, investigate the biological mechanism underpinning the association and eventually test the optimal markers for assessing risk for a disease or its outcome, such as in pharmacogenomics, the study of the effect of genetic variation on pharmacological interventions. The success of GWAS has opened new horizons for exploration and highlighted the complex genomic architecture of disease susceptibility.
Introduction
The history of human genetics has focused on mapping regions of the genome that can explain part or all of a disease or human trait. With the generation of a draft of the human genome in 2001, geneticists quickly set out to comprehensively annotate the genome and apply the evolving knowledge of the pattern of genetic variation to investigate both monogenic, Mendelian disorders and complex diseases, the latter of which by nature are polygenic (1–4). Until recently, the scope and breath of human variation was certainly underappreciated until the advent of early maps of common variants, such as the single-nucleotide polymorphism (SNP), the most common variant in the genome (1,5–7). It is notable that a comprehensive set of genetic variation has shifted the analysis paradigm to finding genetic contributions to complex disease, whereas the capacity to capture environmental exposures and lifestyle decisions is far more rudimentary, even though these factors are essential for understanding complex diseases and traits.
For many years, human genetics has successfully mapped uncommon mutations with large effect sizes in studies conducted in families or special populations, such as the BRCA1/BRCA2 mutations in Ashkenazi women with breast cancer and ovarian cancer (8). The search for highly penetrant mutations in familial aggregation has been based on genetic linkage analysis, an approach that has used microsatellite markers across the genome to scan for markers that segregate within a family (9,10). Based on the identification of linkage peaks using rigorous statistical approaches, follow-up of regions was pursued based on strong signals. Because of the wide spacing of markers across the genome, signals often pointed to regions over multiple megabases that in turn required sequencing large regions of the genome in search of the causative mutations, a daunting task in scope and until recently hampered by technical limitations. Nonetheless, successes in families loaded with melanoma, breast cancer and sets of cancers (Li-Fraumeni Syndrome) (8,11–14) are notable and provided an important substantiation of the approach of using markers indirectly. In retrospect, the use of markers to conclusively identify regions for detailed analysis has been an important lesson for mapping germ line genetic variants associated with risk for cancer, but the approach yielded only mutations with very strong effects.
Over the past 20 years, a parallel approach has been pursued to discover common genetic variants that confer susceptibility to different types of cancers. Initially, association studies were conducted using a handful of annotated genetic variants for which a strong hypothesis could be formulated. In a genetic association study, the analysis consists of a comparison of the distribution of a marker allele between cases and controls, in search of a statistical difference that can be reflected in an estimated effect size—usually quite small compared with mapped linkage signals due to highly penetrant mutations. Naively, at first, investigators searched for alleles with high estimated effect sizes (e.g. per allele odds ratios > 2.0), but with time, it has become apparent that common alleles confer small risk overall in sufficiently large case–control studies of unrelated subjects, the primary study design for association analyses (15).
Nominally, investigators focused on SNPs that altered the coding sequence and resulted in a non-synonymous change, namely a shift in the amino acid sequence of the protein. The approach was predicated on a more simplistic model: changes in the amino acid content would lead to a pronounced (e.g. measurable) change in function and thus influence the disease or trait of interest. Due to the inadequately sized studies, issues of study design and the overestimation of effect size, nearly all published candidate gene association studies, probably represent false positives. In this regard, the candidate gene approach has yielded very few notable findings, namely those that are conclusive and do not represent false positives. To date, perhaps a handful have been adequately replicated and confirmed in follow-up studies. For example, GSTM1 null and NAT2 slow acetylator genotypes have been associated with increased overall risk of bladder cancer and could account for up to 31% of the disease because of their high prevalence (16). Similarly, candidate genes have shown robust findings for a promoter SNP in TNF in non-Hodgkin’s lymphoma and a coding variant in CASP8 in breast cancer (17,18). But overall, very few candidate studies have yielded convincing results worthy of the enormous investment of time to pursue the biological basis of the association.
In the early part of the new millennium, candidate gene studies expanded in scope, looking at sets of genetic markers across a gene of interest. This transition adopted the use of sets of markers defined on the basis of genetic correlation, known as linkage disequilibrium (LD) discussed below. Often, markers are located in introns or intergenic regions, raising the possibility that genetic variants could alter expression or regulation of a gene, thus not only widening the spectrum of variants to be examined but also increasing the scope of underlying mechanisms. As this approach began to find variants associated with cancer risk, the focus was on markers for risk. For examples, Garcia-Closas et al. (19) identified a promising marker near the VCAM1 gene in association with bladder cancer as part of an exploration of genes in several pathways related to cancer biology. Again, the approach was hypothesis driven, in that specific genes were chosen for the best markers but the scope was enlarging and increasing the number and types of variants explored (20).
In 1996, Risch and Merikangas argued that for complex diseases, such as most cancers, large scale linkage studies will be both difficult and not as well powered to detect susceptibility alleles with low estimated effect sizes, of the type that are probably to contribute in a polygenic model (15,21,22). Instead, they suggested that large-scale association testing could be more efficient and more effective (15,21) in the discovery phase. Moreover, the practicality of collecting large sets of family pedigrees was identified as a daunting, and perhaps overwhelming challenge. Indeed, the age of genome-wide association studies (GWAS) has established the association study as an integral tool for discovering the contribution of common genetic susceptibility alleles to different types of cancer.
The value of conducting statistically sound studies that are well powered has become a central tenet of the GWAS era because of the enormous risk for false-positive discovery. The threshold for discovery has been established at a high level, known as genome-wide significance, which serves two dual purposes (23,24). First, it necessitates careful consideration of the power to detect the effect sizes expected to be observed in the study. Second, the high bar of genome-wide significance protects against the probability of a false-positive finding (25,26). The latter is critical because GWAS are discovery tools that point investigators toward long arduous follow-up studies for unraveling the underlying biology and the pursuit of markers for risk assessment (27).
Background
The scope of genetic variation
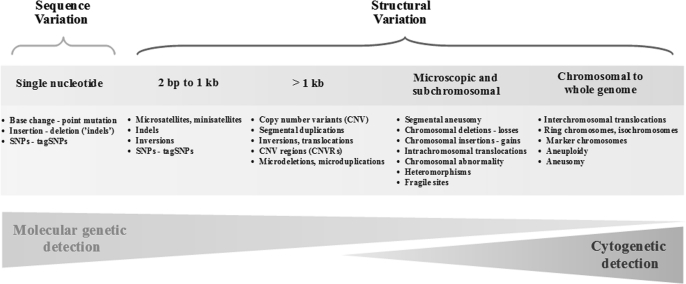
Based on the international annotation projects and the sequencing of nearly a dozen full human genomes, the spectrum of human genetic variation is enormous with respect to the types of genetic variation and the magnitude of variants in any given genome (28–34). Although two genomes are estimated to differ by <0.5%, there are at least several million differences, only a small subset of which contributes to disease risk while the majority is probably vestigial. The most common type of variation is a single-nucleotide base substitution, known as the SNP. Next generation sequence analysis has begun to identify the large set of small insertions or deletions in sequence (30,35,36). Progressively, larger structural alterations and copy number variants are fewer in absolute number but impact more bases across the genome (Figure 1).
Fig. 1.
Types of genetic variations in the human genome. Common types of genetic variations can be categorized into two major groups—those that involve single base changes (e.g. SNPs) and those that alter more than one base (e.g. microsatellites or structural variants).
Most common variants namely those with a minor allele frequency (MAF) >5% are common to all populations, although the distribution of allele frequencies can vary greatly across the globe (37). Ascertainment estimates for lower frequency variants depend on both the number of subjects as well as the population genetic history of those examined. With next generation sequencing applied to high-profile regions in large numbers, greater complexity in different human populations is emerging, particularly with variants of lower frequency (36,38,39). Interestingly, the scope of structural variants is much greater than previously recognized, though the majority of large-scale polymorphisms appear to be less common, namely <1–5% in unrelated populations, unlike SNPs and insertions and deletions, of which there are millions with frequencies >5%. Accordingly, the GWAS approach in unrelated subjects has been most successfully applied to SNPs and it has been far less successful applied to structural variants, also known as copy number variations (CNVs).
The most common sequence variation in the germ line genome is SNP, which, by definition, is observed in at least 1% of a population. By definition, the MAF is a relative term and applies to the allele with the lower frequency at a locus in a reference population. In many instances, there can be major differences in MAFs between populations with distinct histories. For the common SNPs (MAF >5%), <10% of SNPs are specific to a given population (28,37). This observation suggests the common ancestry of common SNPs. The literature suggests that there are at least 10 million SNPs with a MAF >1% (40–42) and 5 million SNPs with a MAF >10% (3,4,40) but recent large-scale sequencing efforts, such as the 1000 Genome project, indicate that these estimates are low (www.1000genomes.org/) (43). In fact, there could be double or triple the earlier estimates. Lastly, there is a small subset of SNPs that are tri-allelic; at a given base on the reference genome, there can be three different bases, though these are rare, they can be formidable technical challenges for quality control metrics.
It is estimated that between 50 000 and 250 000 common SNPs could be biologically active, as non-synonymous coding variants or regulators of gene expression or splicing (7,15). For candidate gene studies, there was a premium assigned to SNPs in coding regions, usually based on in silico predictions. These coding SNPs, known as cSNPs, can be divided into non-synonymous variety (which alters the predicted amino acid codon) and synonymous SNPs (which do not alter the codon sequence). The latter are far more common and less probably alter function. Though intense interest has been directed at non-synonymous SNPs, few have been conclusively associated with human diseases and even fewer have corroborative biological data to provide plausibility for the association (7,15). There has been considerable effort to predict the effect of a non-synonymous cSNP and putative conformational protein changes, but the biological significance is based on laboratory evidence only. Recently, it has emerged that there are subset of SNPs that alter regulation or expression of a gene. These regulatory SNPs are difficult to identify using informatic tools and thus have to be defined on the basis of laboratory data (44).
More than 5 million human SNPs of the international public repository for SNPs, known as dbSNP (www.ncbi.nih.gov/SNP/), have been validated to date with genotyping assays by the SNP Consortium and the International HapMap Project (1,28). Until recently, sequence validation was applied to a small subset but this is about to shift with the completion of the 1000 Genome Project, so that the majority of entries will be sequence based (45,46). Historically, many variants in dbSNP are monoallelic, due to either genotyping error or, more probably, sequencing errors (47,48). It is notable that the reported SNPs have been biased toward high-frequency variants in populations of European ancestry. The catalog of uncommon variation, namely SNPs with MAF under 1%, is incomplete but the 1000 Genome Project is expected to generate a catalog of variants between 0.5 and 5% frequency, which will complement the International HapMap of common variants above 5–10%. Already, the latest build of dbSNP has >20 million variants, mainly less common ones. In addition, dbSNP contains downloads from many disease-specific mutation databases, which will make the curation and utility of less common variants even more daunting for analytical approaches toward prioritization of variants for study. Still, the contribution of uncommon variants represents an untapped portion of the genomic architecture and will necessitate new approaches toward mining these variants for cancer susceptibility. Highly penetrant disease mutations are cataloged in a public database, the Online Mendelian Inheritance in Man or OMIM (www.ncbi.nlm.nih.gov/sites/entrez?db=OMIM/).
The spectrum of genetic variation in the genome can range from single base substitutions to small insertions/deletions to structural variations that can be cytologically observed. The short tandem repeat, also known as the microsatellite, represents a class of polymorphisms used in linkage analysis that are defined by repeats of two or more nucleotides but display notable differences in the frequencies of the repeat units. Typically, they are located in non-coding regions. However, most large-scale structural variation is submicroscopic and ranges in size from a few base pairs to thousands of base pairs (49,50). Collectively, the submicroscopic variants are known as CNVs, a focus of intense interest in large-scale association studies. Estimates of segmental duplications in the genome have been suggested to approach 10% of the genome, but most are not common enough to be effectively analyzed using current GWAS (51–53). Current surveys suggest that CNVs are less common than previously reported (54,55) and in fact, perhaps, three-quarters of common CNVs are in LD with common SNPs (55).
Correlation of common genetic variants
It has been observed that the majority of SNPs are not inherited independently but segments on a chromosome, inherited from generation to generation (41,56,57). A central concept in germ line genetics is the inheritance of correlated markers on the same chromosome, known as LD. It is defined as the non-random association between allelic markers on a chromosome and is classically measured using one of two estimators, D′ or r2 (58). Individual SNPs that are strongly correlated with each other are said to be in LD, but with time and geographic distribution, LD can erode by recombination events (e.g. exchange of genetic material) during meiosis (59).
Haplotypes are defined as sets of SNPs or polymorphisms (e.g. insertions, deletions or large copy events) in strong LD, in which one or more can serve as surrogates for the other markers on the haplotype. A haplotype can be determined in most cases with family trios but in GWAS or large association studies, family structure is usually not available. Still, the offspring haplotype phase can be determined if the parental genotypes are known or established by biochemical methods and then applied to study to best estimate the common haplotypes (58). However, the phasing of haplotypes is more challenging in unrelated subjects but accurate estimates based by well-developed statistical methods that can account for the ambiguity of unobserved haplotypes can provide haplotypes with assigned probabilities (58). Some have argued that haplotypes are preferable for candidate gene studies but for GWAS, the approach is laborious and less nimble in analyzing the thousands of markers genotyped. The methods are not as robust for conducting analysis across thousands of variants.
The appreciation of applying LD to the millions of SNPs observed in human populations that has given rise to the fundamental principle of GWAS, testing across the genome with well-chosen markers that serve as surrogates for untested markers (60–62). The ‘indirect approach’ represents the first step in identifying regions with strong association with cancer or a human trait and relegates the investigation of the optimal variants to study for understanding the biological basis of the association signal (59). The commonly used approach to select optimal SNPs is the ‘greedy algorithm’, which estimates highly correlated SNPs, on the basis of MAFs and creates heuristic bins of ‘tagged’ SNPs. It is the set of tags that function as proxies for the highly correlated untested variants (60).
Practical issues in GWAS
GWAS have emerged as a powerful tool to identify susceptibility loci with low effect sizes in unrelated subjects with specific cancers and related outcomes. Though epidemiologic design is important, in the discovery phase, there has been a relaxation of epidemiologic rigor in order to discover novel regions, mainly because of the need to gather a sufficiently large enough data set to detect low effect sizes. Often, groups have used convenient or publicly available controls for the discovery analysis in GWAS (23), of which the Wellcome Trust Case Control Consortium has been a notable example. These steps could come at a cost, such as a slightly higher rate of false positives, or in related manner, the apparent contradiction of regions or loci that do not robustly replicate in separate scans, suggesting subtle, but real differences related to selection and exposure criteria. Consequently, the estimates are slightly unstable and maybe refined as better studies if analyzed with high quality epidemiologic and environmental exposure data. In order to meet the requirements of a sufficiently large enough data set to observe significant differences between cases and controls, many scans, particularly for rarer cancers, have had to amalgamate data sets.
Replication of results is critical in a separate comparable set of studies (63). The value of replication is to guard against the blizzard of false positives observed with common alleles with low effect sizes. By scaling the studies, GWAS can effectively shed the majority of false positives. The industry standard that has emerged has targeted genome-wide statistical significance for a GWAS with a P value less than between 5 × 10−7 and 1 × 10−8 using either a trend or genotype test, adjusted for minimal cofactors/covariates (23,64–66).
Because GWAS are conducted in unrelated subjects, there has been intense interest in the background population substructure of cases and controls. The capacity to examine thousands of markers with minimal or no LD can be used to effectively discriminate differences in population substructure (67–69). Population stratification is present when there is a measurable difference in the distribution of alleles between subgroups that have different population histories, which can certainly alter association analyses, providing false-positive findings, such as in early case–control studies, in which the cases and controls were drawn from individuals of different populations. Stratification between cases and controls based on differences in exposures can also be problematic, but less so in GWAS. The ability to detect stratification with sets of markers depends on the allele frequencies in each subgroup (70). Subjects with admixture coefficients >15–20% can be removed from association analyses (71) based on attempt to separate subjects into groups and determining the distribution of shared alleles. Further, detection of population stratification is conducted on the GWAS data set to adjust simultaneously for a fixed number of top-ranked principal components resulting from a principal component analysis (67). The search for underlying subgroups in stratified samples can be investigated with genetic markers not linked to the phenotype, using a principal component analysis that yields eigenvectors, used to adjust for possible inflation of test statistics due to stratification (67,72,73).
One of the fundamental reasons for the success of GWAS has been the foresight to collect biospecimens in case–control and cohort studies over the past decades, each of which affords advantages for studying exposures or avoiding survivorship bias. Since the high throughput genotype platforms that analyze thousands of commercially determined SNPs and now CNVs demand high performance DNA, most investigators have used native DNA—either from blood or buccal cells. The latter works quite well when optimally collected and extracted (74). Neither whole genome amplified DNA can be effectively used in GWAS or can materials from tumor tissue (or its adjacent region) due to problems with allelic imbalance. High-quality genotypes are generated using widely accepted quality control metrics for SNP completion, sample completion, heterozygosity scores, testing for fitness for proportion of Hardy–Weinberg equilibrium (70) and assay verification with a second technology (75).
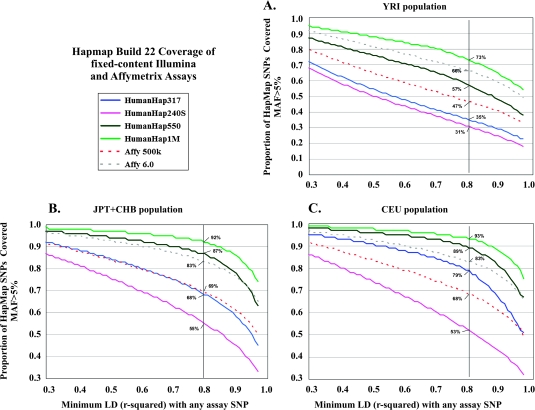
Scanning the genome with SNPs can be performed with commercially available fixed products that provide hundreds of thousands of SNPs, chosen either on the basis of the tag strategy, spacing across the genome or inclusion of obligate SNPs either known or predicted to be functionally important. Great importance has been attached to the extent of ‘coverage’ afforded by the fixed content chips, which for each commercial product has translated into higher cost for greater coverage (24). The bias of the chips has been to select SNPs that most efficiently tag common SNPs in individuals of European background based on the successive builds of the International HapMap Project (Figure 2). Specifically, the level of coverage is generally measured by determining the percentage of ‘bins’ tagged by SNPs (with MAF > 5 or 10%) for each of the three HapMap II populations, individuals of European background (known as CEU), Yoruban of West Africa (YRI) and East Asians (CHN and JPN) (24,59,60). Over 500 regions of the genome have now been conclusively associated (e.g. report signals with P value <5 × 10−7) in >100 human diseases or traits (76–78).
Fig. 2.
Coverage of various genotyping platforms on HapMap II SNPs. The coverage of commercially available genotyping platforms in HapMap populations are plotted based on estimates of linkage disequilibrium using r2, the correlation coefficient. A vertical bar depicts the cut off of an r2 = 0.8, which is commonly used as a threshold to effectively tag monitored SNPs. The three HapMap populations of Phase II are labeled and the percentage estimated at the threshold is provided. (A): Coverage plot in Yoruban population (Ibadan, Nigeria), (B): coverage plot in Japanese (Tokyo, Japan) and Han Chinese (Bejing, China) and (C): coverage plot of US residents with northern and western European ancestry by the Centre d'Etude du Polymorphisme Humain (CEPH).
The analysis of dense genotyping data can be carried out with publicly available tools in either Genotype Library and Utilities (GLU) or PLINK (79), each of which permits archiving, manipulation and basic analyses of data sets, including assessment of population substructure and association testing for SNPs. CNVs are more challenging because the primary image files have to be analyzed and quality control metrics applied to predict CNVs with varying degrees of probability. It is this latter issue, together with the evolving annotation of CNVs, which has hampered the widespread application of this type of analysis to yield association results comparable to those from common SNPs. Consequently, only a handful of common CNVs have been conclusively associated with complex diseases. In cancer GWAS, only one conclusive finding has been reported, the association of a region on chromosome 1 with the rare pediatric cancer neuroblastoma (80).
The first look at GWAS findings in cancer
Theme and variations
The age of GWAS and cancer have quickly ushered in a new era of discovery of regions that harbor germ line genetic variants (common and uncommon) associated with susceptibility to specific cancers. Currently, >75 regions of the genome (some harboring multiple independent signals) have been conclusively associated with susceptibility to specific cancers. Notably, in a handful of few circumstances, more than one type of cancer maps to the same set of genetic variants but overall, it appears that the contribution of common germ line variation has a strong component of tissue specificity. It is also notable that no single locus identified by the current crop of etiologically driven GWAS has also been shown to influence outcome, as measured by progression, disease stage, metastases or survivorship. This latter observation suggests that the germ line factors responsible for development of a cancer could differ from those genetic factors that sustain carcinogenesis or lead to progression. It is interesting to note that for the 29 independent loci identified in prostate cancer GWAS, so far, not a single locus exclusively associates with the more aggressive form of the disease (65,66,81–84). In the Cancer Genetic Markers of Susceptibility Initiative of a GWAS in prostate cancer, the analysis plan specifically addressed the early and advanced forms of prostate cancer, yet did not identify a locus specific to disease state (65,66,84). Consequently, it will be necessary to conduct distinct GWAS in studies designed to address these important outcomes, but it will most probably require new collections and collaborative networks to achieve the required numbers to discover the low to moderate effect alleles influencing cancer outcomes.
It was unanticipated that GWAS studies in certain cancers would yield many novel regions (e.g. prostate cancer with perhaps 29, breast cancer with 13 and colon with 10) (64,66,75,81–93), whereas other cancers strongly associated with environmental exposures have yielded so few regions: three for lung cancer in primarily smokers and three in bladder cancer despite analysis of sufficiently large data sets. Thus, it is plausible that the effect of tobacco use is substantially stronger than any single region with low estimated effect sizes (below 1.3 in GWAS). The lung cancer findings are also notable in that the strongest signal on chromosome 15q25 maps to a region that has also been identified in GWAS of smoking phenotypes (94–97). Prior to GWAS, it was also considered on the list of candidate genes because it contains nicotine receptors (e.g. CHNRA3 and CHRNA5) (98,99). Further studies are urgently needed in non-smoking cases and controls to discriminate between signals that could be driven by tobacco exposure versus primary carcinogenesis (94). Fine-mapping studies in different populations may accelerate the pinpointing of the set of variants in this region requiring further study to understand the biology underlying the association study.
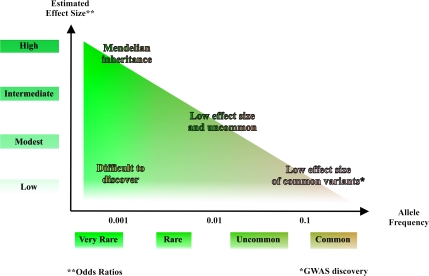
There are few notable exceptions to the observation that the per allele estimated effect is <1.5 for alleles discovered in cancer GWAS (100). In fact, most are <1.3, and it is anticipated that more will be discovered in the vicinity of 1.1–1.2 as consortial activities permit meta-analyses with larger sets of scanned subjects (Figure 3). Still, it was notable that two recent testicular cancer scans each identified two regions with effect sizes considerably greater than what had been observed previously in cancer GWAS. The loci mapped to regions on chromosomes 5 and 12 that harbored candidate genes previously implicated in testicular development, the ligand for the receptor tyrosine kinase (KITLG) and sprouty 4 (SPRY4). Moreover, the studies were notable for the high effect sizes detected for chromosome 5, namely >2.5, as well as the biological plausibility of the candidate genes (101,102). This was not surprising in light of the marked increase risk for family members (103,104). Another cancer with a familial aggregation, thyroid cancer, also yielded alleles with relatively high estimated effect sizes, and interestingly, they were detected in a small primary scan (105).
Fig. 3.
The relationship between the estimated effect size and the allele frequency of disease susceptibility locus. The majority of disease susceptibility loci identified by GWAS in different cancers have low effect size (per allele estimated effect size of 1.1–1.3).
In select GWAS, the findings have pointed to genes previously investigated in that cancer. Pancreatic cancer is a highly lethal disease with a 5-year relative survival of <5% (106), with known risk factors of family history of pancreatic cancer, type 2 diabetes mellitus and cigarette smoking. Interestingly, the first reported GWAS in pancreatic cancer identified a variant in an intron of the ABO blood group antigen, which confirmed a finding suggested 50 years ago (107,108). This is a striking example of how a GWAS hit points to a finding previously described in the epidemiology literature and has been confirmed with a recent study, in which comparable effect sizes have been observed by known blood type (109).
In prostate cancer, the signal on chromosome 10q13 points to a variant in the promoter of the MSMB gene, which encodes a protein, PSP94, under intense investigation as a biomarker for prostate cancer (65,89). The T allele of rs10993994, 57 bp centromeric to the first exon of the MSMB gene, showed significant association with prostate cancer in two independent studies (65,89), and it is known to have influence in the MSMB gene expression (prostate secretory protein 94, PSP94) in tumor (110,111). Now that the region has been extensively resequenced, further investigation of additional variants in strong LD with rs10993994 is warranted and it is possible that a neighboring gene, NCOA4, could also be a candidate gene for analysis because it is an androgen receptor coactivator.
A GWAS of neuroblastoma, a rare pediatric cancer, has implicated three different chromosomal regions, one of which is a copy number variation at chromosome 1q21.1 (80,112,113). The first region is at 6p22 and it is plausible that the risk alleles have dosage effect on the severity of disease by subgrouping patients into patients of metastatic stage 4, patients with somatic MYCN amplification and patients with relapse. The second region is at 2q35 within the BARD1 gene (112).
Despite the enormous effort focused on choosing candidate genes or pathways, based on current models, so far, the results of cancer GWAS have pointed to primarily new or unknown regions and genes. However, there are a few notable exceptions, such as two GWAS of pediatric lymphoblastic leukemia, which have uncovered three sets of markers pointing to genes involved in B-cell development (114,115), but the clustering of related genes has not been observed. Moreover, for a disease such as breast cancer, which has been epidemiologically linked to hormones, surprisingly, none of the major signals map to regions harboring estrogen/progesterone genes in women of European background. However, in a scan of Asian women, a GWAS convincingly discovered markers near the estrogen receptor alpha (known as ESR1) (93).
Discovering more complexity
GWAS have uncovered a series of possible interesting and unexpected relationships between different diseases. For example, three of the regions identified in prostate cancer GWAS also map to type two diabetes susceptibility regions. For some time, there has been a controversial literature reporting an inverse relationship between type two diabetes and prostate cancer; it is further speculated that the protection against prostate cancer is more apparent several years after the diagnosis of diabetes. For two of regions, the markers appear to be inversely related, namely the apparent risk allele for prostate cancer is protective for diabetes for HNF1B on chromosome 17q24 and for THADA on chromosome 2p21. The signal on chromosome 7p15 localizes to intron 2 of JAZF1, a very large gene, whereas the diabetes signal, as well SNPs for height, body stature and systemic lupus erythematosus are localized to a distinct region >200 kb away in intron 1 with no residual LD, suggesting different variants.
Differences in study design can lead to important observations related to both the genetic and environmental contributions to cancer etiology. In one notable instance, two distinct GWAS efforts in prostate cancer have yielded different results for a region of chromosome, 19q13.33, that harbors the gene responsible for the prostate serum antigen (PSA), used by many, but not all for screening for prostate cancer (116,117). In one study, that used clinically advanced cases with controls that had low PSA levels, a strong signal for a SNP in KLK3 was observed, replicating with a substantially lower degree of statistical significance in the follow-up studies, whereas in Cancer Genetic Markers of Susceptibility Initiative, comprised of mainly cohort studies, there was little effect for prostate cancer risk (39,89,118,119). In fact, the Cancer Genetic Markers of Susceptibility Initiative analysis reported that the SNP in the region of KLK3 was associated with PSA levels, raising the possibility that the locus could be related to PSA levels instead of prostate carcinogenesis, though it is possible it could be a both but further studies are needed. Indeed, now that the KLK3 region has been resequenced, it will be possible to investigate this issue with the optimal markers (36).
Most studies have relied on combining data from different designs and often combining histologic or molecular subtypes of a classically defined cancer. The result has been to identify regions that appear to be associated with biological processes common to the development of a tissue-specific type of cancer. For example, the follow-up analysis of the initial set of signals identified in breast cancer GWAS suggests that there could be a differential effect for some regions based on estrogen receptor status for some regions (120). The preponderance of estrogen receptor-positive cases in the discovery studies certainly could have contributed to this observation, but additional reports have identified regions with stronger effects in estrogen receptor-positive subjects (92). In other GWAS, subtype GWAS have yielded convincing findings for a histologic subtype, such as the chromosome 5p15.33 locus in lung cancer (in predominately smokers), which is significantly associated in the adenocarcinoma subtype but not in squamous cell carcinoma (121,122). Similarly, in non-Hodgkin’s lymphoma, distinct regions have been identified in the chronic lymphocytic leukemia (114) and follicular subtypes (123). On the other hand, for the associations with high effect sizes in testicular cancer, there was no appreciable difference by subtype analysis for seminoma and non-seminoma cancers, suggesting the common contribution of the two regions to testicular carcinogenesis (101,102,124).
Based on follow-up fine mapping of the regions, often using HapMap chosen SNPs or those defined by comprehensive resequence analysis (36,38,39), intense effort has focused on the investigation of the genomic architecture of each GWAS region. It is plausible that more than one common variant, each with small effect sizes, could contribute to cancer susceptibility and in fact, this has been demonstrated in three regions identified in prostate cancer susceptibility. For 8q24, there are at least four distinct prostate cancer susceptibility loci in men of European background (66,82,84,85,90,125). In men of other backgrounds (e.g. African, East Asian or Latino/admixed), it is possible that even more population-specific loci could be important and perhaps partially explain some of the disease disparity among different ethnic groups (85,90). For the HNF1B locus on chromosome 17q24, further mapping identified a second independent signal (126). Similarly, the gene desert of 11q13 harbors at least two independent signals and perhaps more (127).
Cancer GWAS Nexus regions
8q24, a cancer susceptibility region for many unrelated cancers
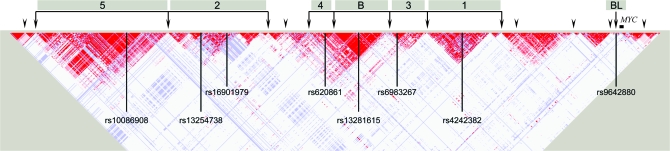
A region of ∼600 kb, centromeric to the well studied, MYC oncogene, is a region that has been repeatedly discovered to harbor distinct independent markers associated with cancer risk (Figure 4). MYC encodes for nuclear phosphoprotein that involves in growth regulation, cell differentiation and apoptosis, and its amplification/overexpression is a frequent event in bladder tumors (128,129). The findings have unexpectedly found that prostate, breast, colorectal, bladder and perhaps ovarian cancers are associated with common genetic variants in this region (66,75,82,88,90,130–134). The region is also notable because it is frequently amplified in epithelial cancers and does not harbor candidate genes, but instead several pseudogenes, whose function and presence are not well established. In this regard, the findings of 8q24 attest to the complexity of the region and the likelihood that regulatory elements of both MYC and other regions could underlie the cancer susceptibility.
Fig. 4.
Linkage disequilibrium pattern and cancer susceptibility loci indentified in 8q24 region. The 8q24 region harbors multiple cancer susceptibility loci identified by GWAS. The linkage disequilibrium heat map was drawn using HapMap I + II release 22 CEU data from 127 948 to 128 950 kb genomic region (reference build 36.3). The arrowheads indicate probable recombination hotspots according to the HapMap I + II. Five distinct regions have been associated with prostate cancer risk (regions 1–5). Region 3 is also conclusively associated with colorectal cancer and precancerous colorectal adenomas. Region B harbors a breast cancer susceptibility locus rs13281615, and BL indicate a bladder cancer susceptibility locus rs9642880, which is telomeric to the region 1, and ∼30 kb centromeric to the MYC oncogene.
The 8q24 region was first implicated as a prostate cancer risk locus by a genome-wide linkage scan in Icelandic men, followed by identification of an allele of the microsatellite marker, DG8S737, and A allele of rs1447295 from replication association studies in three case–control samples of European ancestry from Iceland, Sweden and USA (125). The region was also discovered by an admixture mapping in African-Americans (135). The SNP, rs1447295, was reconfirmed by a large nested case–control study using 6637 cases and 7361 matched controls (91). Independent of the rs1447295, which marked as ‘region 1’, two independent loci, rs16901979 and rs6983267, marked as region 2 and region 3, respectively, centromeric to the region 1 were identified by three independent studies (66,82,90). Notably, the rs16901979 showed clear association in African-Americans with higher risk allele frequency than Europeans. In two recent studies, another independent prostate cancer susceptibility locus rs620861 was identified, located in between region 2 and region 3 and overlapping with a region previously identified in a breast cancer GWAS (81,84,136).
For colorectal cancer, four different studies reported the same variant, rs6983267 (in region 3 of prostate cancer), as the strongest signal by GWAS (88,90,132,137). Recently, published work has begun to generate insights in the functional nature of the rs6983267 variant, which has only two other variants in strong LD compared with rsw1447295 with 49 variants in strong LD (36,138,139). The two studies suggest that in colorectal cancer, rs6983267 shows long-range interaction with MYC as well as possible enhancement of the Wnt-signaling pathway. Interestingly, the prostate specific effect is more complex and as of now, not well explained except for the presence of multiple regions across the 600 kb of 8q24.
Kiemeney et al. (130) reported that the T allele of rs9642880 located ∼30 kb upstream of MYC oncogene showed significant association with bladder cancer (odds ratio = 1.22, P = 9.34 × 10−12). Wu et al. (140) reported that rs2294008 located in exon 1 of PSCA on the other side of MYC is significantly associated with bladder cancer risk. Since the SNP, rs2294008, is located in the exon 1 of PSCA and yields a missense variant that alters the start codon, Wu et al. further performed an in vitro reporter assay using the four most frequent haplotypes of the PSCA 5′ upstream region including rs2294008 and showed significantly lower promoter activity of the T allele-containing haplotypes.
5p15.33
Common variants in the TERT-CLPTM1L locus on 5p15.33 have been identified by GWAS to harbor susceptibility alleles for cancer of the brain and lung (96,97,122,141,142). For lung cancer, it appears that the signal is strongly associated with the adenocarcinoma subtype and not squamous or other subtypes (122). In the region, there is an attractive candidate gene, TERT, the reverse transcriptase component of the telomerase a gene that is critical for telomere replication and stabilization by controlling telomere length. TERT promotes epithelial proliferation and telomere maintenance has been implicated in the progression from KRAS-activated adenoma to adenocarcinoma in a murine model (143,144). There is additional evidence for associations with cancer of the bladder, prostate, uterine cervix and skin including basal cell carcinoma and melanoma based on candidate studies in follow-up of GWAS hits (145).
This region is particularly interesting because of the scope and spectrum of allele frequencies associated with diseases. Mutations in the TERT gene have been described in acute myelogenous leukemia and in the inherited bone marrow failure family pedigrees with dyskeratosis congenita, a cancer predisposition syndromes (146,147). Mutations in the TERT gene have also been described in patients with idiopathic pulmonary fibrosis (148,149) and in families with hematologic disorders and serious liver fibrosis (150). Mutations in TERT have also been shown to result in shorter telomeres and explain a subset of those with familial idiopathic pulmonary fibrosis (151).
Conclusions
The age of genome-wide association studies in cancer have ushered in a new era of discovery of regions of the genome harboring common genetic susceptibility alleles that require extensive effort to map the signal to define the optimal variants for investigating the biological basis of the association. For nearly all signals identified, the markers have not immediately uncovered variants that can easily explain the signal and in most cases, appear to be variants not in coding regions that instead of shifting the amino acid sequence, probably alter the regulation of one or more complex genetic processes. In this regard, GWAS are the first step toward identifying novel regions and pathways associated with both primary carcinogenesis and probably gene–environment interactions.
To make sense of the known GWAS signals and to find more signals, some that could explain major disparities in incidence and outcomes by ethnic backgrounds, it will be critical to conduct GWAS in populations with distinct population genetic histories (and different underlying LD structures) as well as to map known hits in other populations. The age of GWAS has not only uncovered new regions but perhaps provided insights in a subset of the regions that require refined analyses, such as the effect of tobaccos usage and lung cancer risk to unravel the complex nature of these types of cancer.
The recent genomic revolution has produced a comprehensive map of genetic variation that has enabled research to scan the genome in search of statistically sound signals worthy of follow-up. However, the ability to survey environmental and lifestyle exposures is not nearly as advanced, thus hampering the opportunity to explore the dynamic relationship between genomic variants and the environment. Lastly, the age of GWAS is actually the beginning of a new age, one characterized by many new regions of the genome worthy of pursuit as candidate genes to explore the common as well as uncommon variants that contribute to the risk of different cancers.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CNV
copy number variation
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- MAF
minor allele frequency
- PSA
prostate serum antigen
- SNP
single-nucleotide polymorphism
References
- 1. doi: 10.1038/nature02168. The International HapMap Consortium. (2003) The International HapMap Project. Nature, 426, 789–796. [DOI] [PubMed] [Google Scholar]
- 2.Collins FS, et al. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 5. doi: 10.1038/nature04226. The International HapMap Consortium. (2005) A haplotype map of the human genome. Nature, 437, 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sachidanandam R, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 7.Chanock S. Candidate genes and single nucleotide polymorphisms (SNPs) in the study of human disease. Dis. Markers. 2001;17:89–98. doi: 10.1155/2001/858760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 9.NIH/CEPH Collaborative Mapping Group. (1992) A comprehensive genetic linkage map of the human genome. NIH/CEPH Collaborative Mapping Group. Science. 1992;258:67–86. [PubMed] [Google Scholar]
- 10.Elston RC, et al. Overview of model-free methods for linkage analysis. Adv. Genet. 2001;42:135–150. doi: 10.1016/s0065-2660(01)42020-7. [DOI] [PubMed] [Google Scholar]
- 11.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 12.Hussussian CJ, et al. Germline p16 mutations in familial melanoma. Nat. Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 13.Kamb A, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat. Genet. 1994;8:23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 14.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 15.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Closas M, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothman N, et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol. 2006;7:27–38. doi: 10.1016/S1470-2045(05)70434-4. [DOI] [PubMed] [Google Scholar]
- 18.Cox A, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Closas M, et al. Large-scale evaluation of candidate genes identifies associations between VEGF polymorphisms and bladder cancer risk. PLoS Genet. 2007;3:e29. doi: 10.1371/journal.pgen.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunning AM, et al. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum. Mol. Genet. 2009;18:1131–1139. doi: 10.1093/hmg/ddn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol. Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- 22.Risch N, et al. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 23. doi: 10.1038/nature05911. The Wellcome Trust Case Control Consortium. (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature, 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, et al. Evaluating coverage of genome-wide association studies. Nat. Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 25.O’Berg MT. Epidemiologic study of workers exposed to acrylonitrile. J. Occup. Med. 1980;22:245–252. [PubMed] [Google Scholar]
- 26.Wolff MS, et al. Blood levels of organochlorine residues and risk of breast cancer. J. Natl. Cancer Inst. 1993;85:648–652. doi: 10.1093/jnci/85.8.648. [DOI] [PubMed] [Google Scholar]
- 27.Erichsen HC, et al. SNPs in cancer research and treatment. Br. J. Cancer. 2004;90:747–751. doi: 10.1038/sj.bjc.6601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 34.Kim JI, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harismendy O, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeager M, et al. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Hum. Genet. 2008;124:161–170. doi: 10.1007/s00439-008-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 38.Yeager M, et al. Comprehensive resequence analysis of a 97 kb region of chromosome 10q11.2 containing the MSMB gene associated with prostate cancer. Hum. Genet. 2009;126:743–750. doi: 10.1007/s00439-009-0723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parikh H, et al. A comprehensive resequence analysis of the KLK15-KLK3-KLK2 locus on chromosome 19q13.33. Hum. Genet. 2009 doi: 10.1007/s00439-009-0751-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruglyak L, et al. Variation is the spice of life. Nat. Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 41.Reich DE, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 42.Reich DE, et al. Quality and completeness of SNP databases. Nat. Genet. 2003;33:457–458. doi: 10.1038/ng1133. [DOI] [PubMed] [Google Scholar]
- 43.Hayden EC. International genome project launched. Nature. 2008;451:378–379. doi: 10.1038/451378b. [DOI] [PubMed] [Google Scholar]
- 44.Hudson TJ. Wanted: regulatory SNPs. Nat. Genet. 2003;33:439–440. doi: 10.1038/ng0403-439. [DOI] [PubMed] [Google Scholar]
- 45.Packer BR, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephens M, et al. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat. Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- 47.Marth G, et al. Sequence variations in the public human genome data reflect a bottlenecked population history. Proc. Natl Acad. Sci. USA. 2003;100:376–381. doi: 10.1073/pnas.222673099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marth GT, et al. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 49.McCarroll SA, et al. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 50.Scherer SW, et al. Challenges and standards in integrating surveys of structural variation. Nat. Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 52.Bailey JA, et al. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 54.Buckley PG, et al. Copy-number polymorphisms: mining the tip of an iceberg. Trends Genet. 2005;21:315–317. doi: 10.1016/j.tig.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 55.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 56.Bonnen PE, et al. Haplotype and linkage disequilibrium architecture for human cancer-associated genes. Genome Res. 2002;12:1846–1853. doi: 10.1101/gr.483802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 58.Slatkin M. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orr N, et al. Common genetic variation and human disease. Adv. Genet. 2008;62:1–32. doi: 10.1016/S0065-2660(08)00601-9. [DOI] [PubMed] [Google Scholar]
- 60.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardon LR, et al. Using haplotype blocks to map human complex trait loci. Trends Genet. 2003;19:135–140. doi: 10.1016/S0168-9525(03)00022-2. [DOI] [PubMed] [Google Scholar]
- 62.Johnson GC, et al. Haplotype tagging for the identification of common disease genes. Nat. Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 63.Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 64.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 66.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 67.Yu K, et al. Population substructure and control selection in genome-wide association studies. PLoS One. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patterson N, et al. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 70.Ryckman K, et al. Calculation and use of the Hardy-Weinberg model in association studies. Curr. Protoc. Hum. Genet. 2008;57:1.18.1–1.18.11. doi: 10.1002/0471142905.hg0118s57. [DOI] [PubMed] [Google Scholar]
- 71.Falush D, et al. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Devlin B, et al. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 73.Pritchard JK, et al. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feigelson HS, et al. Successful genome-wide scan in paired blood and buccal samples. Cancer Epidemiol. Biomarkers Prev. 2007;16:1023–1025. doi: 10.1158/1055-9965.EPI-06-0859. [DOI] [PubMed] [Google Scholar]
- 75.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manolio TA, et al. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diskin SJ, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 83.Gudmundsson J, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat. Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yeager M, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eeles RA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Houlston RS, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 89.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 90.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schumacher FR, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 92.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 93.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chanock SJ, et al. Genomics: when the smoke clears. Nature. 2008;452:537–538. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- 95.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 96.McKay JD, et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bierut LJ, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Caporaso N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Easton DF, et al. Genome-wide association studies in cancer. Hum. Mol. Genet. 2008;17:R109–R115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 101.Kanetsky PA, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat. Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rapley EA, et al. A genome-wide association study of testicular germ cell tumor. Nat. Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Skinner DG. Urological Cancer. New York: Grune & Stratton; 1983. [Google Scholar]
- 104.Swerdlow AJ, et al. Risks of breast and testicular cancers in young adult twins in England and Wales: evidence on prenatal and genetic aetiology. Lancet. 1997;350:1723–1728. doi: 10.1016/s0140-6736(97)05526-8. [DOI] [PubMed] [Google Scholar]
- 105.Gudmundsson J, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat. Genet. 2009;41:460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jemal A, et al. Cancer statistics, 2008. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 107.Amundadottir L, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bodmer W, et al. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolpin BM, et al. ABO blood group and the risk of pancreatic cancer. J. Natl Cancer Inst. 2009;101:424–431. doi: 10.1093/jnci/djp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chang BL, et al. Fine mapping association study and functional analysis implicate a SNP in MSMB at 10q11 as a causal variant for prostate cancer risk. Hum. Mol. Genet. 2009;18:1368–1375. doi: 10.1093/hmg/ddp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu P, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J. Natl Cancer Inst. 2008;100:1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Capasso M, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat. Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maris JM, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N. Engl. J. Med. 2008;358:2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Papaemmanuil E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Trevino LR, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Andriole GL, et al. Mortality results from a randomized prostate-cancer screening trial. N. Engl. J. Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schroder FH, et al. Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 118.Ahn J, et al. Variation in KLK genes, prostate-specific antigen and risk of prostate cancer. Nat. Genet. 2008;40:1032–1034. doi: 10.1038/ng0908-1032. ; author reply 1035–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eeles R, et al. Reply to “Variation in KLK genes, prostate-specific antigen and risk of prostate cancer”. Nat. Genet. 2008;40:1035–1036. doi: 10.1038/ng0908-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garcia-Closas M, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Broderick P, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Skibola CF, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat. Genet. 2009;41:873–875. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chanock S. High marks for GWAS. Nat. Genet. 2009;41:765–766. doi: 10.1038/ng0709-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 126.Sun J, et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat. Genet. 2008;40:1153–1155. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zheng SL, et al. Two independent prostate cancer risk-associated loci at 11q13. Cancer Epidemiol. Biomarkers Prev. 2009;18:1815–1820. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.DePinho RA, et al. myc family oncogenes in the development of normal and neoplastic cells. Adv. Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- 129.Mhawech-Fauceglia P, et al. Genetic alterations in urothelial bladder carcinoma: an updated review. Cancer. 2006;106:1205–1216. doi: 10.1002/cncr.21743. [DOI] [PubMed] [Google Scholar]
- 130.Kiemeney LA, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 133.Tomlinson IP, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 134.Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Al Olama AA, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 137.Gruber SB, et al. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol. Ther. 2007;6:1143–1147. doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 138.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 139.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wu X, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wrensch M, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shete S, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Choi J, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sweet-Cordero A, et al. Comparison of gene expression and DNA copy number changes in a murine model of lung cancer. Genes Chromosomes Cancer. 2006;45:338–348. doi: 10.1002/gcc.20296. [DOI] [PubMed] [Google Scholar]
- 145.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Calado RT, et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc. Natl Acad. Sci. USA. 2009;106:1187–1192. doi: 10.1073/pnas.0807057106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yamaguchi H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 148.Tsakiri KD, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc. Natl Acad. Sci. USA. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mushiroda T, et al. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J. Med. Genet. 2008;45:654–656. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]
- 150.Calado RT, et al. A spectrum of severe familial liver disorders associate with telomerase mutations. PLoS ONE. 2009;4:e7926. doi: 10.1371/journal.pone.0007926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Armanios MY, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]