Summary
Here we report on a functional gene-mining method developed to isolate stress tolerance genes without any prior knowledge of the genome or genetic mapping of the source germplasms. The feasibility of this approach was demonstrated by isolating novel salt stress tolerance genes from salt cress (Thellungiella halophila), an extremophile that is adapted to a harsh saline environment and a close relative of the model plant Arabidopsis thaliana. This gene-mining method is based on the expression of salt cress cDNA libraries in Arabidopsis. A cDNA expression library of the source germplasm, salt cress, was constructed and used to transform Arabidopsis via Agrobacterium-mediated gene transfer. A transgenic seed library consisting of >125 000 independent lines was generated and screened for salt-tolerant lines via a high-throughput genetic screen. A number of salt-tolerant lines were isolated, and the salt cress cDNAs were identified by PCR amplification and sequencing. Among the genes isolated, several novel small protein-encoding genes were discovered. The homologs of these genes in Arabidopsis have not been experimentally analyzed, and their functions remain unknown. The function of two genes isolated by this method, ST6-66 and ST225, and their Arabidopsis homologs, were investigated in Arabidopsis using gain- and loss-of-function analyses, and their importance in salt tolerance was demonstrated. Thus, our functional gene-mining method was validated by these results. Our method should be applicable for the functional mining of stress tolerance genes from various germplasms. Future improvements of the method are also discussed.
Keywords: gene-mining, salt tolerance, Arabidopsis, salt cress, cDNA library, high-throughput
Introduction
Isolation of genes with agronomic importance has been one of the focal points in plant biotechnology. With the genomic sequencing of model plants, comparative genomics and the fine mapping and cloning of quantitative trait loci (QTLs) have become important means to isolate these genes from crops. However, the approaches are not easily applicable to wild extremophile germplasms that are rich sources of stress tolerance genes. Therefore, a fast, economical, gene function-based approach is needed for isolating genes with specific functions from wild germplasms, without prior knowledge of their the genome or without the need for genetic mapping. Similar approaches have been exploited to isolate Arabidopsis loss-of-function (LeClere and Bartel, 2001; Mou et al., 2002) and gain-of-function mutants with morphological changes (Ichikawa et al., 2006). Despite the power of this functional approach, it has not been applied for mining stress tolerance genes. Here, we report a proof of concept for this functional gene-mining method by isolating salt-tolerance genes from salt cress (Thellugiella halophila). The procedure and time frame of this method is illustrated in Figure 1, in which the highly efficient transformation (Bent, 2000; Clough and Bent, 1998) makes it possible to transform an entire cDNA library into Arabidopsis, and to generate a large collection of transgenic lines in a relatively short period of time. The transgenic seed library can then be screened for salt-tolerant transformants, and the genes conferring salt tolerance are easily isolated. This approach emphasizes the functionality of the expressed genes, and therefore the cloned genes are directly linked to their function in salt tolerance. In addition, this approach is cost-effective and saves time.
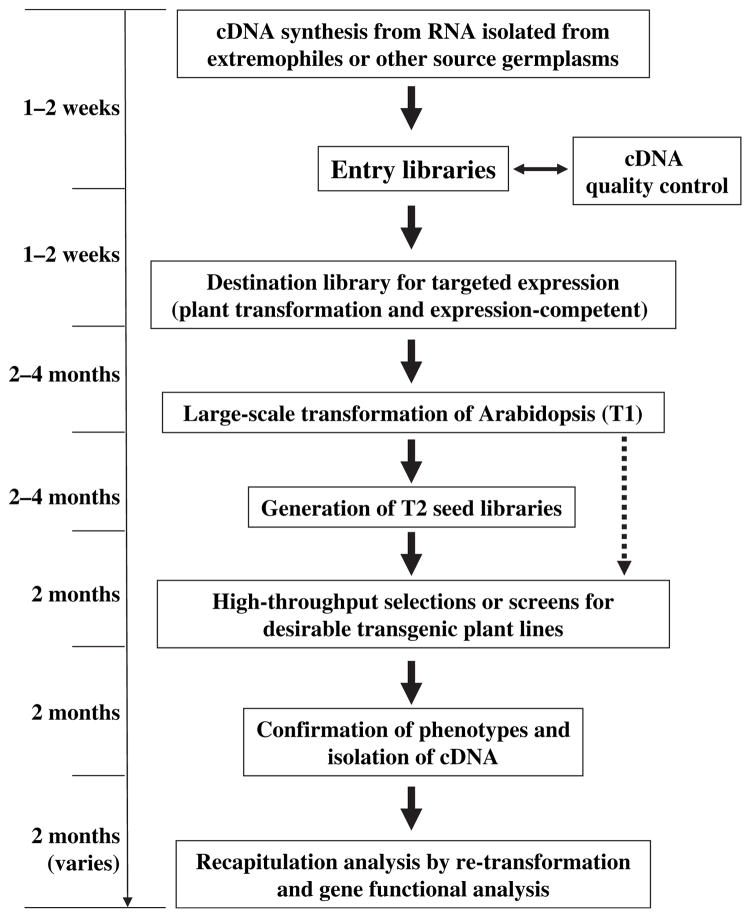
Figure 1.
Flow chart illustrating the functional gene-mining method with the power of Arabidopsis, and the minimal time required for each step.
Starting from cDNA synthesis for the source germplasms (for example, salt cress), through entry and destination expression library construction, large-scale transformation of Arabidopsis to generate the T2 seed library, primary high-throughput screen of the T2 seed library (T1 seeds can also be screened if the T2 library is not maintained) for salt tolerance, for example, secondary screen to confirm the phenotype and isolation of cDNA, to the final recapitulation and gene function analysis: the whole procedure requires a minimal time of 1 year. The broken arrow indicates an alternative route.
Thellungiella halophila, a halophyte endemic to the highly saline coastal areas in eastern China, is ideal for demonstrating the feasibility of this functional gene-mining method, because it is closely related to Arabidopsis thaliana, and has nearly all of the same model plant features of A. thaliana (Amtmann et al., 2005; Inan et al., 2004; Zhu, 2001). Microarray analysis showed that a large number of known abiotic and biotic stress-inducible genes were expressed in salt cress at high levels, even in the absence of stress (Gong et al., 2005; Inan et al., 2004; Oh et al., 2007; Taji et al., 2004). Analysis of the expressed sequence tags for salt cress indicated that the salt cress genome contains genes missing from the Arabidopsis genome, implying that salt cress might have evolved novel salt-tolerance genes and mechanisms (Inan et al., 2004; Wong et al., 2005). It is expected that the expression of the salt-tolerance genes from salt cress in Arabidopsis may render the transgenic plants more tolerant to salt stress.
Using the plant transformation-ready expression library of salt cress that we previously constructed (Lei et al., 2007; Ni et al., 2007), a large-scale floral-dip transformation of Arabidopsis was carried out (Bechtold et al., 1993; Clough and Bent, 1998), and a large collection of 125 000 independent transgenic lines was generated. Using a high-throughput genetic screen for salt-tolerant seedlings, the transgenic seed library was screened, and hundreds of putative salt-tolerance lines were isolated in the primary screen. A secondary screen was used to confirm the salt tolerance and a number of salt-tolerance lines were isolated. Analysis of the cDNAs from the salt-tolerance lines revealed novel genes that confer salt tolerance when overexpressed in Arabidopsis. To validate this approach, ST6-66 and ST225, two of the unknown genes identified by the screening, were functionally analyzed and their importance in salt tolerance demonstrated. This functional gene-mining method may be applicable to the isolation of other abiotic stress-tolerance genes, as well as to biotic stress-resistance genes such as disease-resistance genes.
Results
Generation of a large collection of Arabidopsis transgenic lines with the salt cress destination cDNA expression library
The destination cDNA library we had constructed from salt cress (Ni et al., 2007) was introduced into Agrobacterium tumefaciens strain GV3101 (pMP90) (Koncz and Schell, 1986) in order to transform wild-type Columbia Arabidopsis plants using the floral-dip method (Bechtold et al., 1993; Clough and Bent, 1998) on a large scale, with the efficient Basta herbicide resistance selection on soil. A collection of 125 000 independent herbicide resistance transgenic lines was generated. The T1 seeds were harvested in pools of 200 individual lines. The transgenic Arabidopsis seed library consisted of 625 pools that could be screened for salt-tolerant lines.
Isolation of salt-tolerance transgenic lines from the library via a high-throughput genetic screen
To isolate transgenic lines that are tolerant to salt stress, a high-throughput genetic screen was set up by germinating seeds in the library on half-strength MS medium containing 220 mM NaCl, as described by Saleki et al. (1993). Unlike other salt-tolerant germination screens where the emergence of the radicle was defined as tolerance in germination (Quesada et al., 2002), the emergence of green cotyledons and continued growth of the seedling under salt stress were used as the criteria of salt-tolerant germination for our genetic screen (Figure S1). We routinely screened up to 3000 seeds on a single 15-cm-diameter plate. The primary screen of the whole library yielded >800 putative salt-tolerant seedlings. These plants were rescued and grown in soil to maturity, and T2 seeds were harvested from individual plants.
The T2 seeds from the individual putative salt-tolerant lines were subjected to a secondary screen, as described in the Experimental procedures. Approximately 10% of the lines showed salt tolerance in the next generation. Some of the lines showed salt tolerance only during germination, whereas others also showed salt tolerance during later developmental stages.
Novel genes discovered with the functional gene-mining method
After confirming salt tolerance by the secondary screen, the salt-tolerant lines were subjected to molecular analysis. The salt cress cDNA transferred to Arabidopsis was isolated by PCR amplification, as described in the Experimental procedures, and sequenced. Full-length cDNA sequences are listed in Table 1. Truncated cDNA sequences missing the 5′ end are listed in Table 2. Most of the cDNAs were isolated more than once from independent salt-tolerant lines. Among the isolated genes, most of the sequences show high similarity to their homologs in Arabidopsis, although there are also a few that do not have high similarity to any Arabidopsis sequences. The predicted functions of the cDNAs include transcription factors, photosynthesis-related proteins, chaperones and damage-protection proteins. About 30% of the isolated genes encode unknown proteins that do not have a known function or known domain. The fact that truncated cDNA sequences apparently led to increased salt tolerance suggests that these sequences might have caused the silencing of homologous Arabidopsis sequences, which may play negative roles in salt tolerance. Among the truncated cDNAs, five sequences (ST2-4, ST15-18-1, ST10, ST27-4 and ST37-9) have a complete open reading frame, and encode small peptides of around 100 amino acids. The possibility that the truncated peptides function in salt tolerance cannot be ruled out. The salt tolerance associated with full-length cDNAs is likely to be caused by the overexpression of the sequences in the transgenic lines. These results demonstrate that salt-tolerance genes can be isolated by using this functional gene-mining approach.
Table 1.
Full-length salt cress cDNAs isolated from the salt-tolerant transgenic Arabidopsis lines, and their Arabidopsis homologs
| Salt-tolerant lines | Accession number | cDNA size (bp/aa) | No. independent lines | Predicted function | Arabidopsis homolog (bp/aa) | % Similarity (cDNA/aa) |
|---|---|---|---|---|---|---|
| ST5-18-3 | EU714066 | 708/132 | 2 | SPT4 HOMOLOG 2; positive transcription elongation factor | At5G63670 (563/116) | 81/98 |
| ST6-40 | EU714067 | 658/169 | 1 | Unknown | At4G01150 (821/164) | 86/89 |
| ST6-66 | EU714068 | 679/177 | 2 | Unknown | At1G13930 (684/155) | 79/67 |
| ST7-5-2 | EU714069 | 973/234 | 1 | Auxin/aluminum-responsive protein | At5G19140 (1215/234) | 89/94 |
| ST19 | EU714070 | 644/210 | 1 | DNAJ heat shock N-terminal domain-containing protein | At5G59610 (924/268) | 89/87 |
| ST20 | EU714071 | 655/118 | 2 | LTP1 (nonspecific lipid transfer protein 1) | At2G38540 (670/118) | 78/87 |
| ST45-2 | EU714072 | 518/92 | 2 | Unknown | At4G33640 (583/95) | 74/79 |
| ST45-109 | EU714073 | 543/143 | 1 | 60S ribosomal protein L28 | At2G19730 (722/143) | 90/96 |
| ST49-6 | EU714074 | 646/215 | 2 | LHCA2 (Photosystem-I light harvesting complex gene 2) | At3G61470 (1107/257) | 89/89 |
| ST63-2 | EU714075 | 634/90 | 1 | Unknown | At4G33467 (546/101) | 74/82 |
| ST103-2 | EU714076 | 630/85 | 2 | Unknown, ozone-induced protein | At1G01170 (515/83) | 80/97 |
| ST112 | EU714077 | 448/92 | 6 | Glycine-rich protein | At2G05530 (603/115) | 87/70 |
| ST120 | EU714078 | 633/107 | 1 | Unknown, proline-rich family protein | At3G20850 (405/134) | 50/42 |
| ST140 | EU714079 | 497/141 | 1 | CAB1 (CHLOROPHYLL A/B BINDING PROTEIN 1) | At1G29930 (1044/267) | 87/99 |
| ST225 | EU714080 | 509/116 | 1 | Unknown | At1G19530 (700/117) | 81/75 |
| ST240 | EU714081 | 490/125 | 1 | J8; heat-shock protein binding/unfolded protein binding | At1G80920 (848/163) | 78/71 |
| ST244 | EU714082 | 643/146 | 3 | PSBR (photosystem-II subunit R) | At1G79040 (720/140) | 86/83 |
Table 2.
Truncated salt cress cDNAs isolated from the salt-tolerant transgenic Arabidopsis lines, and their Arabidopsis homologs
| Salt-tolerant lines | Accession number | cDNA size (bp/aa) | Number of independent lines | Predicted function | Arabidopsis homolog (bp/aa) | % Similarity (cDNA/aa) |
|---|---|---|---|---|---|---|
| ST2-4 | EU714083 | 494/62 | 1 | Unknown | At5G02390 (2508/836) | 83/87 |
| ST5-18-1 | EU714084 | 462/82 | 3 | beta-carotene hydroxylase | At4G25700 (1210/311) | 83/97 |
| ST6-32 | EU714085 | 668/136 | 2 | eIF-5A | At1G26630 (897160) | 90/89 |
| ST7-5-1 | EU714086 | 471/140 | 6 | NAC domain-containing protein, BTF3b-like transcription factor | At1G73230 (898/166) | 91/88 |
| ST10 | EU714087 | 613/136 | 1 | Ribosomal protein L19 family protein | At4G17560 (1173/225) | 87/84 |
| ST27-4 | EU714088 | 663/141 | 3 | DRT100 (DNA-DAMAGE REPAIR/TOLERATION 100) | At3G12610 (1409/373) | 80/73 |
| ST37-9 | EU714089 | 424/64 | 1 | SCPL49 (serine carboxypeptidase-like 49) | At3G10410 (1828/517) | 71/72 |
| ST54-6 | EU714090 | 369/64 | 1 | Peptidyl-prolyl cis-trans isomerase, putative | At3G55920 (798/229) | 79/90 |
| ST63-4 | EU714091 | 419/37 | 2 | AIM1 (ABNORMAL INFLORESCENCE MERISTEM); enoyl-CoA hydratase | At4G29010 (2553/722) | 64/94 |
| ST73-2 | EU714092 | 397/54 | 1 | GRF3 (GENERAL REGULATORY FACTOR 3) | At5G38480 (1141/256) | 88/96 |
| ST236 | EU714093 | 442/68 | 1 | Phosphoethanolamine N-methyltransferase 2 (NMT2) | At1G48600 (1770/476) | 80/78 |
Functional analysis of the isolated ST6-66 cDNA
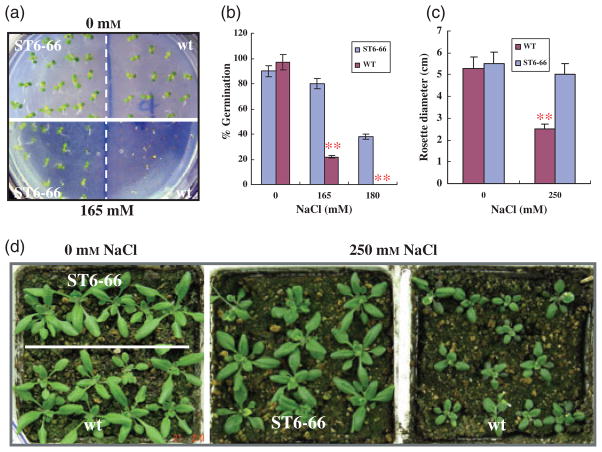
To further validate our functional gene-mining method, we characterized the salt cress cDNA isolated from the salt-tolerant line ST6-66, which displayed salt tolerance in germination, as shown in Figure 2a, b. The transgenic line showed a substantially higher percentage of germination than the wild type (80% versus 20% under 165 mM NaCl stress, and about 40% versus 0% under 180 mM NaCl stress). Soil-grown transgenic plants also showed substantially higher tolerance to salt stress. Figure 2c, d show that the growth of the wild-type plants was significantly more suppressed than that of the transgenic plants under 250 mM NaCl stress, as reflected in the sizes of the rosette plants.
Figure 2.
The Arabidopsis transgenic line expressing the salt cress cDNA ST6-66 shows improved salt tolerance.
(a) Salt-tolerant germination assay. Seeds of the wild type (WT) and the transgenic line ST6-66 were germinated on half-strength MS medium containing 0, 165 or 180 mM NaCl. Photographs were taken for 0 and 165 mM NaCl treatment, when seedlings were 1-week old. About 20 seeds each of the wild type and ST6-66 were sowed on each plate. Three replication plates were used.
(b) Comparison of germination in the experiments shown in (a). Values are the mean of the three replications. Error bars represents SEM. **Significant differences between WT and ST6-66 plants at P < 0.001.
(c) Salt-tolerance assay in soil. ST6-66 transgenic and wild-type plants were grown under identical soil and short day conditions, and were treated with 0 or 250 mM NaCl solutions when seedlings were 2-weeks old. Growth continued for one more week under salt stress before the rosette diameters were measured, as described in Figure 2c. Values are the mean of three replication pots, each containing between nine and 12 plants. Error bars represents SEM. **P < 0.001.
(d) Photograph of representative pots for control (0 mM NaCl) and 250 mM NaCl treatment at the time of rosette-diameter measurement.
Genetic analysis of ST6-66
To study the inheritance of salt tolerance in the ST6-66 line, a cross was made between the mutant as the male parent and the wild-type plants as the female parent. F1 seeds were assayed for salt tolerance during germination. A segregation ratio of 1:1 (salt-tolerant:salt-sensitive) was observed (Table S1). The progeny of selfed F1 plants showed a segregation of 3:1 (salt-tolerant:salt-sensitive). Salt tolerance co-segregated with the herbicide resistance bar gene on the same T-DNA (Table S1 and Figure S2a). The genetic analysis demonstrates that the salt tolerance of ST6-66 was conferred by a single dominant nuclear gene that co-segregated with the bar gene on the T-DNA.
Confirmation of ST6-66 cDNA-conferred salt tolerance by recapitulation
The cDNA of ST6-66 is 679 bp in length and encodes a protein of 177 amino acids. The nucleotide sequence shares 79% similarity with its Arabidopsis homolog At1g13930, encoding an expressed protein. At the amino acid level, ST6-66 was 59% identical and 67% similar to At1g13930. The Arabidopsis gene has not been analyzed experimentally so far.
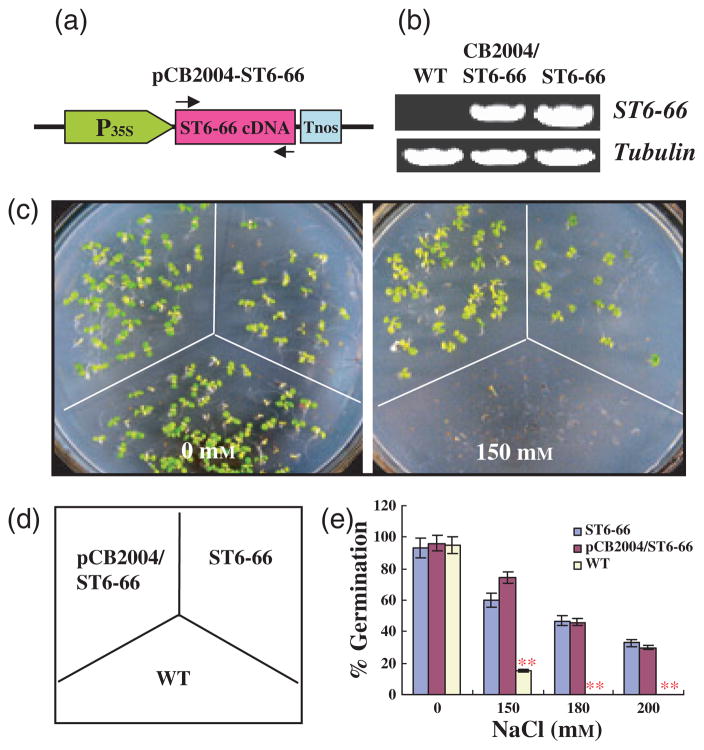
To confirm that the salt tolerance was conferred by the ST6-66 cDNA, the cDNA was cloned into the binary expression vector pCB2004 for generating transgenic plants. The cDNA is under the control of the 35S promoter, and the construct pCB2004-ST6-66 is illustrated in Figure 3a. Homozygous lines (T2 generation) were assayed for the overexpression of the transgene (Figure 3b) and for salt tolerance. Typical results from the experiments are shown in Figures 3c–e. Under normal conditions, there was no significant difference in germination between the pCB2004-ST6-66 transgenic line, the ST6-66 line and the wild type. However, under the three different salt-stress conditions, the germination of the transgenic seeds (both pCB2004-ST6-66 and ST6-66) was substantially higher than that of the wild type (Figure 3c, e). Soil-grown transgenic plants expressing pCB2004-ST6-66 showed similar salt tolerance to the original line ST6-66 (data not shown). These results confirm that ectopic overexpression of the ST6-66 cDNA of salt cress confers improved salt tolerance in Arabidopsis.
Figure 3.
Confirmation of salt tolerance conferred by ST6-66 overexpression.
(a) The overexpression construct of ST6-66 cDNA, pCB2004-ST6-66, is illustrated (not drawn to scale). This construct was used to generate transgenic plants, and homozygous lines were used for the following salt-tolerance assay.
(b) Overexpression of ST6-66 in the wild type (WT) and transgenic lines by RT-PCR analysis. RNA was isolated from WT and transgenic seedling (CB204/ST6-66 and ST6-66), and ST6-66 transcript levels were analyzed using ST6-66-specific primers, as described in the Experimental procedures.
(c) Salt-tolerant germination assay. Seeds of the WT and pCB2004-ST6-66, as well as ST6-66, were germinated on the half-strength MS medium containing 0, 150, 180 or 200 mM NaCl. Photographs were taken for 0 and 150 mM NaCl treatment when seedlings were 1-week old. About 30 seeds each of the WT, pCB2004-ST6-66 and ST6-66 were sown on each plate. Three replication plates were used.
(d) The key for the plates in (c).
(e) Comparison of germination for the experiments in (b). Values are the mean of three replications. Error bars represent SEM. **Significant differences between WT and transgenic plants at P < 0.001.
Loss-of-function analysis of the ST6-66 homolog At1g13930, and complementation with the ST6-66 cDNA
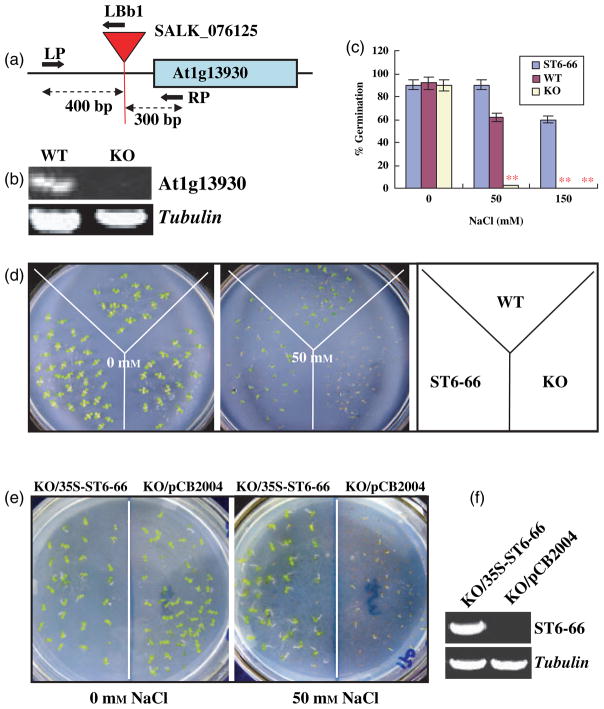
To further confirm the importance of this gene in salt tolerance, a loss-of-function analysis was carried out in Arabidopsis. Since the Arabidopsis homolog with the highest similarity is At1g13930, the T-DNA insertion line SALK_076125 was obtained. The T-DNA insertion is in the 5′ regulatory sequence of At1g13930, as illustrated in Figure 4a. The homozygous plants were screened by genomic PCR, and its null expression was confirmed by RT-PCR analysis, as shown in Figure 4b. The knock-out line was subjected to the salt-tolerance assay. The results in Figure 4c, d show that the germination of the knock-out mutant was <5% under 50 mM NaCl treatment, in contrast with a more than 60% germination for the wild type, demonstrating that knock-out of At1g13930 renders the mutant hypersensitive to salt stress. These results indicate that At1g13930 plays an important role in salt tolerance.
Figure 4.
At1g13930 mutant and complementation analysis.
(a) Illustration of the At1g13930 locus with the T-DNA insertion in SALK_076125, and primers used for identifying homozygous progeny.
(b) Confirmation of the knock-out of At1g13930 by RT-PCR. The transcript level of the SALK-076125 homozygous line (KO) was not detectable, in contrast with that of the wild type (WT). Tubulin was used for equal loading.
(c) Knock-out of At1g13930 renders the mutant more sensitive to salt stress. Seeds of the WT and KO, as well as of ST6-66, were germinated on half-strength MS medium containing 0, 50 or 150 mM NaCl. Germination was counted when the seedlings were 1-week old. Seeds (15–30 each) of WT, KO and ST6-66 were sown on each plate. Three replication plates were used. Values are the mean of three replications. Error bars represent SEM. **Significant differences between ST6-66 and WT and KO lines at P < 0.001.
(d) Photographs were taken for 0 and 50 mM NaCl treatment, and germination was counted when seedlings were 1-week old. The key for the plates is shown on the right.
(e) Complementation analysis. The same pCB2004-ST6-66 line as described in Figure 4a was used to transform SALK-076125 homozygous plants (KO), and the empty vector pCB2004 was used to generate control plants. T1 seeds were assayed for salt sensitivity by germinating seeds on MS medium containing 50 mg L−1 herbicide glufosinate and 0 or 50 mM NaCl. When seedlings were 1-week old, germination was counted and photographs were taken. Between 30 and 50 seeds each of the control (KO/pCB2004) and the complemented line (KO/pCB2004-ST6-66) were sown on each plate. Three replica plates were used. Photos of representative treatments are shown.
(f) Overexpression of ST6-66 in the transgenic line by RT-PCR analysis. RNA was isolated from KO control (KO/pCB2004), and transgenic seedling (KO/pCB2004-ST6-66) and ST6-66 transcript levels were analyzed using the ST6-66-specific primers, as described in the Experimental procedures.
To functionally confirm that ST6-66 is a homolog of At1g13930, a complementation experiment was conducted by transferring the pCB2004-ST6-66 construct into the At1g13930 knock-out mutant (SALK_076125). The results in Figure 4e demonstrate that ST6-66 was indeed able to restore the salt tolerance of SALK_076125 to the wild-type level when ectopically overexpressed (Figure 4f). The germination of the complemented line under 50 mM NaCl treatment was 72%, in contrast to 2% for the knock-out mutant.
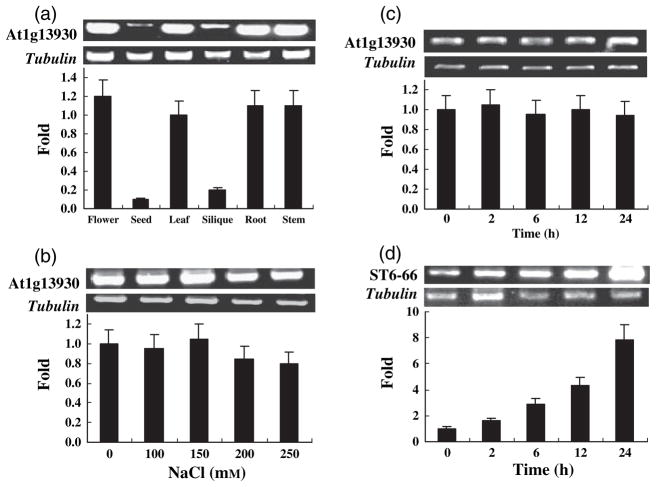
Expression pattern of At1g13930 and ST6-66
RT-PCR and real-time RT-PCR were used to analyze the expression pattern of At1g13930 and ST6-66. It was found that At1g13930 was constitutively expressed in Arabidopsis, but that its transcript level was much lower in seeds and siliques than in other organs, as shown in Figure 5a. This gene was not responsive to salt stress, as demonstrated in Figures 5b, c. In contrast, ST6-66 was responsive to salt stress in salt cress, as shown in Figure 5d. These results indicate that the response of this gene to salt stress is different in Arabidopsis and salt cress. It is interesting to speculate how the ability to induce ST6-66 might contribute to increased salt tolerance in salt cress, as compared with Arabidopsis.
Figure 5.
Expression analysis of ST6-66 and its Arabidopsis homolog At1g13930 with RT-PCR and real-time RT-PCR.
(a) Analysis of the expression pattern of At1g13930 by RT-PCR (top) and real-time RT-PCR (bottom). RNA was isolated from flower, seed, leaf, silique, root and inflorescence stem, and was analyzed as described in the Experimental procedures. Tubulin was used as an equal loading control. Real-time RT-PCR was performed using the same RNA samples and primers. The relative abundance of the At1g13930 transcript was first normalized to the internal tubulin level, and then to the At1g13930 transcript level in the leaves. Values are the average of three repeats after normalization. Error bars represent SEM.
(b) Response of At1g13930 to salt stress. Columbia wild-type seedlings (10-days old) were treated with NaCl solutions of the indicated concentrations for 5 h before RNA was isolated. RT-PCR and analysis was conducted as described in (a). The relative abundance of the At1g13930 transcript was first normalized to the internal tubulin level, and then to the At1g13930 transcript level in the 0 mM NaCl control sample.
(c) Response of At1g13930 to salt stress. Columbia wild-type seedlings (10-days old) were treated with 100 mM NaCl solution for the indicated time before RNA was isolated. RT-PCR analysis was conducted as described in (a). The relative abundance of the At1g13930 transcript was first normalized to the internal tubulin level, and then to the At1g13930 transcript level in the 0-h control sample.
(d) Response of ST6-66 to salt stress in salt cress. Salt cress wild-type seedlings (10-days old) were treated with 100 mM NaCl solution for the indicated times before RNA was isolated. RT-PCR analysis was conducted as described in (a), except that ST6-66-specific primers were used. The relative abundance of the At1g13930 transcript was first normalized to the internal tubulin level, and then to the At1g13930 transcript level in the 0-h control sample.
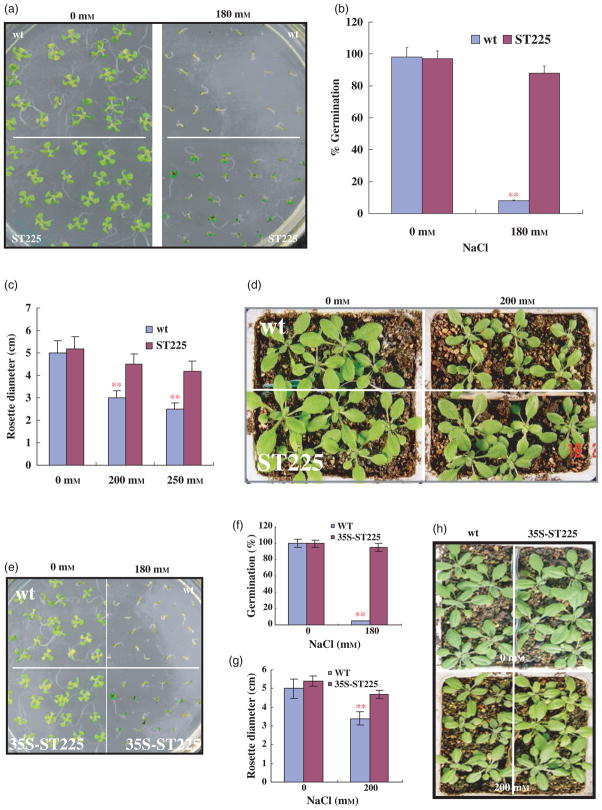
ST225 cDNA-conferred salt tolerance
The salt-tolerant line ST225 displayed salt tolerance during germination and later developmental stages, as shown in Figure 6a. This line showed nearly 90% germination on medium containing 180 mM NaCl, in contrast with 10% for the wild type, whereas no significant difference was found between them under normal conditions without salt stress (Figure 6a, b). When soil-grown plants were stressed with 200 and 250 mM NaCl, respectively, the growth of the wild-type plants was significantly decreased, as reflected in the rosette diameter, whereas the transgenic plants showed improved tolerance to salt stress, as shown in Figure 6c, d.
Figure 6.
The Arabidopsis transgenic line expressing the salt cress cDNA ST225 shows improved salt tolerance.
(a) Salt-tolerant germination assay. Seeds of the wildtype (WT) and the transgenic line ST225 were germinated on half-strength MS medium containing 0 or 180 mM NaCl. Photographs were taken when seedlings were 1-week old. About 20 seeds each of the WT and ST225 were sown on each plate. Three replica plates were used.
(b) Comparison of germination for the experiments in (a). Values are the means of three replications. Error bars represent SEM. **Significant differences between WT and ST225 at P < 0.001.
(c) Salt-tolerance assay in soil. ST225 transgenic and WT plants were grown in soil in the same pot under short-day conditions (10-hour photoperiod), and were treated with NaCl solutions of the indicated concentrations when seedlings were 2-weeks old. The plants were grown for one further week under salt stress before rosette diameters were measured, where the widest diameter was recorded. Values are the mean of three replication pots each containing six plants. Error bar represents SEM. **Significant differences between WT and ST225 at P < 0.001.
(d) Photograph of representative pots for control (0 mM NaCl) and 200 mM NaCl treatment at the time of rosette-diameter measurement.
(e) Salt-tolerant germination assay of re-transformants. The ST225 cDNA was cloned into pCB2004 as described in the Experimental procedures. T2 homozygous seeds were germinated on MS medium containing 0 or 180 mM NaCl. Photographs were then taken again when the seedlings were 1-week old.
(f) Comparison of germination for the experiments in (e). Values are the means of three replications. Error bars represent SEM. **Significant differences between the WT and the transgenic plants at P < 0.001.
(g) Salt-tolerance assay in the soil of re-transformants. T2 homozygous plants were grown in soil, and were challenged with salt stress as described in (c). Values are the mean of three replication pots, each containing six plants. Error bars represent SEM. **P < 0.001.
(h) The photographs of representative pots for control and 200 mM NaCl treatment were taken after 1 week of salt stress.
Genetic analysis demonstrated that the salt tolerance of the ST225 line was conferred by a single dominant nuclear gene that co-segregated with the bar gene on the T-DNA (Table S1; Figure S2b). The result indicates that the transferred salt cress cDNA caused the salt tolerance. Subsequently, the salt cress cDNA was isolated from the ST225 line. To confirm that the ST225 cDNA confers salt tolerance, the cDNA was cloned into the expression vector pCB2004 (Lei et al., 2007), and was transformed into wild-type Columbia plants. The transgenic plants expressing the ST225 cDNA showed salt-tolerant salt tolerance in germination (Figure 6e, f), and also improved the salt tolerance of the transgenic plants grown in soil (Figure 2g, h). The germination of the transgenic seeds was 87%, as compared with 11% for the wild type. When grown in soil, transgenic plants showed improved salt tolerance, as reflected by less growth inhibition compared with wild-type plants (Figure 6g, h). These results demonstrate that the salt cress gene in line ST225 confers elevated salt tolerance in Arabidopsis, and further documents the feasibility of this functional gene-mining approach to isolating salt-tolerance genes.
Discussion
We have developed a simple and straightforward method to functionally mine salt-tolerance genes from a source germplasm by isolating genes conferring salt tolerance from salt cress. This functional gene-mining method outlined in Figure 1 offers several advantages. First, it does not require any genome sequence or expressed sequence tag (EST) database information. Neither is genetic mapping required. Second, this functional gene-mining method saves time and is cost effective. Third, it does not require any expensive experimental facilities or services. Fourth, it may be applicable to the isolation of other genes of agronomic importance, such as disease resistance and drought tolerance genes. Theoretically, any gene can be functionally isolated in this way, if such a gene can be faithfully expressed and is functional in Arabidopsis, and if the function can be efficiently screened. As the cDNA libraries are constructed in plant expression vectors, the cDNA expression in Arabidopsis can be under the control of well-defined promoters. Therefore, the source of germplasms is unlimited. This is advantageous over the BiBAC technology (Hamilton et al., 1996), where genomic DNA is used for library construction, and where the expression of the genes relies on their own regulatory sequences.
The inclusion of smaller cDNAs ranging from 500 to 1000 base pairs in size increases the representation of these cDNAs in the library, and consequently increases the possibility of their being isolated in the screen, thus increasing our chance of isolating novel salt-tolerance genes. Usually, the small cDNAs are discriminated against because they are thought to result from an incompletely synthesized cDNA fraction during library construction. As a result, few such small cDNAs have been isolated, and their roles in salt tolerance have been neglected. Using this method, we have isolated several novel small protein-encoding genes involved in salt tolerance. These small genes have never been experimentally isolated or studied before. Further gain-of-function analysis for ST225 and ST6-66, as well as gain- and loss-of-function analyses of the Arabidopsis homolog of ST6-66, demonstrated the importance of two such small genes in salt tolerance.
The trade-off for inclusion of small-sized cDNA is that the frequency of partial cDNA sequences increases in the library. The average size of the cDNA inserts used in the library was <1 kb (Ni et al., 2007). Because the initial library was not normalized, less abundant and larger transcripts were biased against during PCR amplification, and thus they have a much lower representativity in the library. This is perhaps why salt-tolerance genes such as SOS1 (Shi et al., 2000, 2003) were not isolated in this study. Notwithstanding, the isolation and functional analysis of ST6-66 and ST225 demonstrate the feasibility of this gene-mining approach. In addition, the truncated cDNA of ST6-32 codes for eIF-5A, but lacks the initiation codon and a short stretch of amino acids in the N terminus. It is90%identical at the nucleic acid level to its Arabidopsis homolog At1G26630. As a result, overexpression of this truncated cDNA might result in downregulation of the endogenous eIF-5A in Arabidopsis. Hypusine modification by deoxyhypusine synthase (DHS) is required for eIF-5A activity. Antisense downregulation of DHS suppression was shown to increase stress tolerance (Wang et al., 2003). Another truncated cDNA, ST7-5-1, was independently isolated six times. ST7-5-1 codes for an NAC domain-containing protein, but lacks the initiation codon and a few amino acids in the N terminus. It is 91% identical at the nucleotide level to its Arabidopsis homolog At1G73230. The overexpression of this truncated cDNA might downregulate At1G73230. At1G73230 has not been experimentally analyzed thus far. However, compiled microarray data show that this gene is downregulated by salt and osmotic stresses (https://iii.genevestigator.ethz.ch/at/), indicating it may negatively regulate salt tolerance.
One possible reason for the low frequency of confirmation of putative salt-tolerant lines appearing in the secondary screen might be gene silencing, which is known to be associated with the 35S promoter-driven expression of transgenes (Butaye et al., 2004; Mishiba et al., 2005). The approximately 10% of confirmed salt tolerance in the second generation observed in our study is similar to that observed for 35S-gusA (Butaye et al., 2004).
In the future, several aspects may be modified that will improve the overall efficiency of this method. First, normalization of cDNA libraries would greatly even out the representation of low- and high-abundance cDNAs (Bonaldo et al., 1996; Fischer et al., 2003; Soares et al., 1994), thereby significantly reducing the number of clones required to transform Arabidopsis. Subsequently, a much-reduced number of transgenic plants would be required to represent the whole cDNA library. This will also benefit cDNA isolation and identification, as highly abundant cDNAs with salt-tolerance functions are over-represented, thus producing more redundant salt-resistant transgenic plants from a cDNA library that is not normalized. Second, the gene-silencing problem associated with the 35S promoter can be minimized by using Arabidopsis mutants, such as sgs2 (Mourrain et al., 2000; Peragine et al., 2004), as recipients of transformation (Butaye et al., 2004), and/or by choosing promoters with fewer gene-silencing problems. Third, salt-sensitive ecotypes or mutants may be used for the transformation, so that the sensitivity of salt-tolerance selection can be improved. Fourth, the development of a high-throughput screen in soil would simplify the screen procedure, and increase the chance of isolating genes important for salt tolerance at the whole-plant level (as opposed to tolerance at germination only).
Experimental procedures
Plant materials and growth conditions
Arabidopsis Columbia ecotype was used throughout the study. Plants were grown in a commercial soil mix of PeatMoss and perlite (9:1), and were kept in a growth room with a 12-h photoperiod with a light intensity of 200 μE m−2 sec−1, and a constant temperature was maintained at 22°C, unless otherwise specified.
Mobilizing the destination library into Agrobacterium
To mobilize the destination library that we created (Ni et al., 2007) into Agrobacterium for plant transformation, the destination library DNA was electroporated into A. tumefaciens GV3101 (pMP90) (Koncz and Schell, 1986). The transformed agrobacteria were selected on LB agar medium containing 100 mg L−1 kanamycin and 100 mg L−1 gentamycin. More than 107 colonies were obtained, and all the agrobacterial colonies were washed off the agar plates and completely resuspended. A fraction of the bacterial suspension was mixed with an equal volume of 80% glycerol, and aliquots were frozen in liquid nitrogen and stored as agrobacterial library stocks at −80°C, whereas the remaining suspension was diluted with agrobacterial infiltration medium for Arabidopsis transformation (Bechtold et al., 1993; Clough and Bent, 1998).
Generation of a large Arabidopsis transgenic seed library
The above prepared agrobacterial suspension was diluted with agrobacterial infiltration medium to an OD600 of 0.7. Batches of flowering wild-type Columbia plants (when plants were at the peak of flowering) were dipped in the agrobacterial solution as described previously (Bechtold et al., 1993; Clough and Bent, 1998). After dipping, plants were covered to maintain a high level of humidity for 24 h. The plants were then transferred to a greenhouse to continue to grow to maturity. T1 seeds were harvested in bulk, dried in the air and were then cleaned for transformant selection.
The bulk seeds were imbibed in water for 2 days at 4°C before being sown at high density in soil, and were then kept in a glasshouse. When they were about 5 days old, seedlings were sprayed with 0.2% of commercial herbicide (glufosinate ammonium; Aventis, http://en.sanofi-aventis.com). The surviving transformants were grown to maturity, and seeds were collected in pools of approximately 200 independent transgenic lines. These pools constitute the transgenic seed library from which stress-tolerant lines were screened.
A high-throughput genetic screen for salt-tolerant transgenic lines
To find the optimal NaCl concentration for the genetic screen, wild-type Columbia seeds were surface sterilized with 15% commercial bleach for 15 min and then rinsed with sterile water four times. The sterilized seeds were imbibed in sterile water for 2 days at 4°C before being spread at high density (~3000 seeds per 15-cm-diameter plate) on half-strength MS agar medium supplemented with 0, 100, 150, 200 and 250 mM NaCl. Seed germination was inhibited at 150 mM NaCl. To screen the transgenic library for salt-tolerant lines, a higher NaCl concentration of 220 mM was used to ensure the selection for the salt-tolerance function. Approximately 800–1000 seeds for each pool (~200 lines) were germinated on half-strength MS medium containing 25 mg L−1 glufosinate and 220 mM NaCl. Theoretically, there should be three salt-tolerant seeds if one heterozygous salt-tolerant line is present in one pool. Thus, 800–1000 seeds gave a high confidence of representation of salt-tolerance lines during the primary screen.
Rescuing the salt-tolerance transgenic lines, and secondary screen for salt tolerance
In about 2 weeks, green seedlings on half-strength MS medium containing 220 mM NaCl were selected as putative salt-tolerance lines, and were transferred to half-strength MS and grown for 1 week before being transferred to soil to grow to maturity. Seeds were harvested from individual plants and kept as salt-tolerance lines. To confirm the salt tolerance, the secondary screen was conducted with T2 seeds. Briefly, the seeds were re-examined for salt-tolerance germination on half-strength MS medium containing 220 mM NaCl, in comparison with the wild type. In addition, salt-sensitive root elongation was examined on vertically placed half-strength MS medium containing 150 mM NaCl for additional confirmation of salt tolerance. Seeds were germinated on the herbicide-containing half-strength MS medium to select transgenic seedlings, and to verify the segregation ratio. When about 1-week old, herbicide-resistance seedlings were transferred to soil to grow for about 2 weeks before salt stress was imposed.
PCR amplification of salt cress cDNA, and identification of the cDNAs by sequencing
To isolate the cDNA from the salt-tolerance transgenic lines, total DNA was prepared from the leaves of the transgenic lines and then used as the template of PCR amplification with the primers Omega, 5′-TTTTTACAACAATTACCAACAACAACAA-3′, and attB2T12, 5′-TACAAGAAAGCTGGGTTTTTTTTTTTT-3′, that flank the inserted cDNA. The amplified DNA products were gel purified and sequenced.
Genetic analysis of the salt-tolerant lines ST225 and ST6-66
Crosses were performed by transferring the pollen from mature ST225 and ST6-66 anthers to the stigmas of previously emasculated wild-type flowers. F1 and F2 seeds were assayed for salt tolerance and herbicide resistance, as described above in the secondary screen. The germination frequency was recorded and the segregation was analyzed by a χ2 test. For co-segregation of the transgene, DNA was isolated from individual plants of the F2 population, and was then subjected to PCR analysis using ST225-specific (forward primer, 5′-ATGGGTTCTCTAATGTCAGGAT-3′; reverse primer, 5′-TTACTGGCCCACGCTTTTGGCGA-3′) or ST6-66-specific primers (forward primer, 5′-ATGAACTTCATCTCTGATCAGGAA-3′; reverse primer, 5′-TTATTTAATCTTATTATTACGCGCAT-3′).
Validation of the salt-tolerance function of the isolated salt cress cDNAs
To validate the salt-tolerance function conferred by a particular cDNA isolated from a salt-tolerance transgenic line, the cDNA was inserted in the expression binary vector pCB2004 (Lei et al., 2007). The resultant construct was used to transform the wild-type Arabidopsis Columbia. The transgenic lines were examined for salt tolerance, as described for the secondary screen.
Genomic PCR screen for the T-DNA insertion mutant, and RT-PCR verification of the homozygous mutant for At1G13930
The SALK_076125 line was obtained from ABRC (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/abrchome.htm) and was screened with genomic PCR using three primers: primer LP, 5′-TCCCCATAAACATAAACGAAGC-3′; primer RP, 5′-TCCATAAACTAAAGTGGTGCTGTC-3′; and primer LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′. The homozygous mutant was verified by RT-PCR using primer pairs P1, 5′-CCGGAATTCATGAATTTCATCTCCGATCAG-3′, and P2, 5′-ATATACTCGAGTTCAAGAAACCTTGAGCCATCT-3′.
RT-PCR and real-time RT-PCR analysis of the expression pattern of At1G13930 and response to salt stress
For reverse-transcription PCR (RT-PCR) analysis, total RNA was extracted using Trizol reagent (Invitrogen, http://www.invitrogen.com) from various organs of the wild-type Columbia plants, and 10-day-old Arabidopsis and salt cress wild-type seedlings, under normal growth conditions or after salt stress. RT-PCR was carried out with the ThermoScript RT system (Invitrogen) using 1000 ng of RNA as the template and oligo dT25. The RT reaction mixture (1 μl) was used as the template for RT-PCR analysis using At1G13930- and ST6-66-specific primers. The PCR conditions were 1 min of denaturation at 95°C, 1 min of primer annealing at 58°C and 1 min primer extension at 72°C for 30 cycles, or for 25 cycles for tubulin. Real-time RT-PCR was performed and statistically analyzed as described by Livak and Schmittgen (2001). As an internal control, the tubulin transcript was used to quantify the relative transcript abundance of each target gene in each tissue type.
Supplementary Material
Figure S1. NaCl concentrations inhibiting seed germination, primary screen and confirmation of salt tolerance.
Figure S2. Co-segregation analysis of the transgene, with salt-tolerance and herbicide-resistance phenotypes.
Genetic analysis of the salt-tolerant lines ST225 and ST6-66.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (‘863’ program 2002AA224071 and ‘973’ program 2003CB114305) and the Chinese Academy of Science (KSCX2-SW-3). The authors thank the ABRC for providing mutant seeds.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Amtmann A, Bohnert HJ, Bressan RA. Abiotic stress and plant genome evolution. Search for new models. Plant Physiol. 2005;138:127–130. doi: 10.1104/pp.105.059972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Bent AF. Arabidopsis in planta transformation. Uses, mechanisms, and prospects for transformation of other species. Plant Physiol. 2000;124:1540–1547. doi: 10.1104/pp.124.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Butaye KM, Goderis IJ, Wouters PF, Pues JM, Delaure SL, Broekaert WF, Depicker A, Cammue BP, De Bolle MF. Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 2004;39:440–449. doi: 10.1111/j.1365-313X.2004.02144.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Fischer K, Holt DC, Wilson P, Davis J, Hewitt V, Johnson M, McGrath A, Currie BJ, Walton SF, Kemp DJ. Normalization of a cDNA library cloned in lambda ZAP by a long PCR and cDNA reassociation procedure. Biotechniques. 2003;34:250–252. 254. doi: 10.2144/03342bm03. [DOI] [PubMed] [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005;44:826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Frary A, Lewis C, Tanksley SD. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc Natl Acad Sci USA. 1996;93:9975–9979. doi: 10.1073/pnas.93.18.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, et al. The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 2006;48:974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- Inan G, Zhang Q, Li P, et al. Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 2004;135:1718–1737. doi: 10.1104/pp.104.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- LeClere S, Bartel B. A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol. 2001;46:695–703. doi: 10.1023/a:1011699722052. [DOI] [PubMed] [Google Scholar]
- Lei ZY, Zhao P, Cao MJ, Cui R, Chen X, Xiong LZ, Zhang QF, Oliver DJ, Xiang CB. High-throughput binary vectors for plant gene function analysis. J Integr Plant Biol. 2007;49:556–567. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mishiba K, Nishihara M, Nakatsuka T, Abe Y, Hirano H, Yokoi T, Kikuchi A, Yamamura S. Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 2005;44:541–556. doi: 10.1111/j.1365-313X.2005.02556.x. [DOI] [PubMed] [Google Scholar]
- Mou Z, Wang X, Fu Z, Dai Y, Han C, Ouyang J, Bao F, Hu Y, Li J. Silencing of phosphoethanolamine N-methyl-transferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell. 2002;14:2031–2043. doi: 10.1105/tpc.001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Ni WS, Lei ZY, Chen X, Oliver DJ, Xiang CB. Construction of a plant transformation-ready expression cDNA library for Thellungiella halophila using recombination cloning. J Integr Plant Biol. 2007;49:1313–1319. [Google Scholar]
- Oh DH, Gong Q, Ulanov A, Zhang Q, Li Y, Ma W, Yun DJ, Bressan RA, Bohnert HJ. Sodium Stress in the halophyte Thellungiella halophila and transcriptional changes in a thsos1-RNA interference line. J Integr Plant Biol. 2007;49:1484–1496. [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Garcia-Martinez S, Piqueras P, Ponce MR, Micol JL. Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol. 2002;130:951–963. doi: 10.1104/pp.006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleki R, Young PG, Lefebvre DD. Mutants of Arabidopsis thaliana capable of germination under saline conditions. Plant Physiol. 1993;101:839–845. doi: 10.1104/pp.101.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol. 2003;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- Soares MB, Bonaldo MF, Jelene P, Su L, Lawton L, Efstratiadis A. Construction and characterization of a normalized cDNA library. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K. Comparative genomics in salt tolerance between Arabidopsis and aRabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TW, Lu L, Zhang CG, Taylor C, Thompson JE. Pleiotropic effects of suppressing deoxyhypusine synthase expression in Arabidopsis thaliana. Plant Mol Biol. 2003;52:1223–1235. doi: 10.1023/b:plan.0000004332.80792.4d. [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Whitty BR, Diaz-Camino C, Akhter SR, Brandle JE, Golding GB, Weretilnyk EA, Moffatt BA, Griffith M. Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Mol Biol. 2005;58:561–574. doi: 10.1007/s11103-005-6163-6. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. NaCl concentrations inhibiting seed germination, primary screen and confirmation of salt tolerance.
Figure S2. Co-segregation analysis of the transgene, with salt-tolerance and herbicide-resistance phenotypes.
Genetic analysis of the salt-tolerant lines ST225 and ST6-66.