Abstract
The prevalence of obesity in children and adults in the United States has increased dramatically over the past decade. Besides environmental factors, genetic factors are known to play an important role in the pathogenesis of obesity. A number of genetic determinants of adult BMI have already been established through genome wide association studies. In this study, we examined 25 single nucleotide polymorphisms (SNPs) corresponding to thirteen previously reported genomic loci in 6,078 children with measures of BMI. Fifteen of these SNPs yielded at least nominally significant association to BMI, representing nine different loci including INSIG2, FTO, MC4R, TMEM18, GNPDA2, NEGR1, BDNF, KCTD15 and 1q25. Other loci revealed no evidence for association, namely at MTCH2, SH2B1, 12q13 and 3q27. For the 15 associated variants, the genotype score explained 1.12% of the total variation for BMI z-score. We conclude that among thirteen loci that have been reported to associate with adult BMI, at least nine also contribute to the determination of BMI in childhood as demonstrated by their associations in our pediatric cohort.
In the past three years, thirteen genetic loci have been implicated for BMI from the outcomes of genome wide association studies (GWA) studies primarily in adults. Insulin-induced gene 2 (INSIG2) was the first locus to be reported by this method to have a role in obesity1 but replication attempts have yielded inconsistent outcomes2–6. The second reported locus, the fat mass- and obesity-associated gene (FTO)7 has been more robustly observed by others8–11. Subsequent larger studies have uncovered eleven additional genes12–14, firstly melanocortin 4 receptor (MC4R) from a multi-center meta-analysis12, then the GIANT consortium revealed six more genes [transmembrane protein 18 (TMEM18), potassium channel tetramerisation domain containing 15 (KCTD15), glucosamine-6-phosphate deaminase 2 (GNPDA2), SH2B adaptor protein 1 (SH2B1), mitochondrial carrier 2 (MTCH2) and neuronal growth regulator 1 (NEGR1)]14 , five of which were confirmed in the GWA study reported from Iceland (but not GNPDA2 due to an unavailable proxy SNP), who also uncovered and reported loci on 1q25, 3q27 and 12q1313 and verified association with the brain-derived neurotrophic factor (BDNF) gene15.
In this study we aimed at examining these finding in a large pediatric cohort with BMI measures and to determine the relative impact of these variants in childhood. For this purpose, we leveraged genotyping data from our ongoing GWA study of BMI variation in children. The twenty five SNPs corresponding to the thirteen previously reported obesity loci were investigated with respect to their association to normalized pediatric BMI (Table 1; also Supplementary Table 1 for analyses by age categories).
Table 1.
Quantitative association results for the candidate loci in the European American BMI cohort (n=6,078), sorted by chromosomal location.
| Chr | SNP | Position (Build 36) |
Nearby genes(s) | NMISS | BETA | SE | R2 | T | P |
|---|---|---|---|---|---|---|---|---|---|
| 1p31 | rs3101336 | 72523773 | NEGR1 | 6077 | −0.04721 | 0.01637 | 0.001367 | −2.884 | 0.0039 |
| 1p31 | rs2568958 | 72537704 | NEGR1 | 6078 | −0.04654 | 0.01636 | 0.00133 | −2.844 | 0.0045 |
| 1q25 | rs10913469 | 176180142 | SEC16B/RASAL2 | 6076 | 0.04859 | 0.02037 | 0.000936 | 2.385 | 0.017 |
| 2p25 | rs2867125 | 612827 | TMEM18 | 6076 | −0.07964 | 0.02042 | 0.002498 | −3.9 | 9.72×10−5 |
| 2p25 | rs4854344 | 628144 | TMEM18 | 6046 | −0.07734 | 0.0204 | 0.002372 | −3.791 | 1.52×10−4 |
| 2p25 | rs7561317 | 634953 | TMEM18 | 6065 | −0.07905 | 0.02033 | 0.002488 | −3.889 | 1.02×10−4 |
| 2q14 | rs17047697* | 118544280 | INSIG2 | 6072 | 0.03516 | 0.01672 | 0.000728 | 2.103 | 0.036 |
| 3q27 | rs7647305 | 187316984 | SFRS10/ETV5/DGKG | 6075 | 0.01377 | 0.01932 | 0.00008368 | 0.7129 | 0.48 |
| 4p13 | rs13130484** | 44870448 | GNPDA2 | 6078 | 0.06016 | 0.01572 | 0.002404 | −3.826 | 1.32×10−4 |
| 11p12 | rs10838738 | 47619625 | MTCH2 | 6070 | −0.01098 | 0.01659 | 0.00007224 | −0.6621 | 0.51 |
| 11p14 | rs4074134 | 27603861 | BDNF | 6078 | −0.03607 | 0.0191 | 0.0005866 | −1.889 | 0.059 |
| 11p14 | rs4923461 | 27613486 | BDNF | 6076 | −0.03339 | 0.0191 | 0.0005027 | −1.748 | 0.081 |
| 11p14 | rs925946 | 27623778 | BDNF | 6078 | 0.01886 | 0.01758 | 0.0001895 | 1.073 | 0.28 |
| 11p14 | rs10501087 | 27626684 | BDNF | 6077 | −0.02955 | 0.0191 | 0.0003937 | −1.547 | 0.12 |
| 11p14 | rs6265 | 27636492 | BDNF | 6078 | −0.04517 | 0.01997 | 0.0008411 | −2.262 | 0.024 |
| 12q13 | rs7138803 | 48533735 | BCDIN3D/FAIM2 | 6078 | 0.02931 | 0.01623 | 0.0005365 | 1.806 | 0.071 |
| 16p11 | rs8049439 | 28745016 | SH2B1 | 6077 | −0.003809 | 0.01622 | 0.000009073 | −0.2348 | 0.81 |
| 16p11 | rs4788102 | 28780899 | SH2B1 | 6075 | −0.003331 | 0.01626 | 0.00000691 | −0.2049 | 0.84 |
| 16q12 | rs6499640 | 52327178 | FTO | 6077 | −0.01216 | 0.01624 | 0.00009223 | −0.7485 | 0.45 |
| 16q12 | rs8050136 | 52373776 | FTO | 6078 | 0.06086 | 0.01597 | 0.002385 | 3.811 | 1.40×10−4 |
| 16q12 | rs3751812 | 52375961 | FTO | 6071 | 0.06232 | 0.01598 | 0.002501 | 3.901 | 9.68×10−5 |
| 16q12 | rs7190492 | 52386253 | FTO | 6024 | −0.03824 | 0.01656 | 0.0008843 | −2.309 | 0.021 |
| 16q12 | rs8044769 | 52396636 | FTO | 6077 | −0.06233 | 0.0157 | 0.002588 | −3.97 | 7.26×10−5 |
| 18q21 | rs12970134 | 56035730 | MC4R | 6078 | 0.05519 | 0.01806 | 0.001534 | 3.056 | 0.0023 |
| 19q13 | rs29941 | 39001372 | KCTD15 | 6057 | −0.03519 | 0.01715 | 0.0006952 | −2.052 | 0.040 |
NMISS: number of individuals tested; BETA: regression coefficient for the test SNP; SE: standard error of the regression coefficient; R2: r2 value in linear regression; T: test statistic; P: two-sided trend test P-value. The direction of effect is shown for the minor allele in each case. NEGR1 r2 between rs3101336 and rs2568958 = 1; TMEM18: r2 between rs2867125 rs4854344 and rs7561317 = 1; BDNF r2 between rs4074134, rs4923461 and rs10501087 =1 and r2 between rs4074134 and rs925946, rs6265 = 0.14 and 0.85 respectively; SH2B1: r2 between rs8049439 AND rs4788102 = 0.965; FTO r2 between rs8050136 and rs3751812, rs7190492, rs8044769, rs6499640 = 1, 0.38, 0.61 and 0.18 respectively.
perfect surrogate for rs7566605
perfect surrogate for rs10938397
In summary, fifteen of these SNPs yielded at least nominally significant association to BMI (P < 0.05), representing nine different loci with the same direction of effect as previously reported. Of these nine loci, variants at the FTO locus yielded the strongest association with P<10−4, namely rs8044769 and rs3751812 (P = 7.26×10−5 and 9.68×10−5, respectively); in addition, this locus also yielded association with rs8050136 and rs7190492 (P = 1.40×10−4 and 0.021, respectively) but not with rs6499640.
With a similar magnitude of association to FTO was TMEM18, with rs2867125 yielding a P = 9.72×10−5, together with almost as strongly associated SNPs, rs7561317 and rs4854344 (P = 1.02×10−4 and 1.52×10−4, respectively). Indeed, it is interesting to note that the two adult cohorts that uncovered TMEM18 in obesity also showed it to be second only in significance to FTO13, 14. The third most significantly associated locus was at GNPDA2, (rs13130484; P = 1.32×10−4).
Overall, in addition to FTO, TMEM18 and GNPDA2, we found evidence for association at the INSIG2, MC4R, NEGR1, 1q25, BDNF and KCTD15 loci. One could argue that we have carried out multiple testing in our BMI cohort for these previously reported SNPs, albeit at a number of magnitudes less than for a full GWA study. If we were to apply the strictest correction, i.e. the Bonferroni correction based on twenty five SNPs, then FTO, TMEM18, GNPDA2 and MC4R would still be considered significant. This is very much in line with the observations made with the pediatric cohort utilized by Willer et al14; however the one exception is that we do not observe strong association with KCTD15.
It was also observed that SNPs residing at the 12q13, 3q27, MTCH2 and SH2B1 loci did not reveal any evidence of association with BMI in our pediatric cohort.
The positive results for FTO and MC4R come as no surprise as we have previously reported their association with the CDC-defined 95th percentile of BMI, i.e. obesity, in our pediatric cohort, but limited to ages 2–18 years old8, 16. In this current study, where we utilized BMI z-score on all children 0–18 years old, it is satisfying that they continue to show association at a similar magnitude.
One of the more notable results is the positive association with INSIG2. This association with pediatric BMI, albeit at just the nominal level, will further add to the debate on the relevance of INSIG2 in BMI determination.
For the loci we did not observe any evidence for association for at all may be due to power issues, but could also indicate that they have a less pronounced role in a pediatric setting. Indeed, many of the newly uncovered genes have been implicated in neurological functions, genes which may be more important in BMI determination in adults rather than in children, where other more direct metabolic genes could play a more important role.
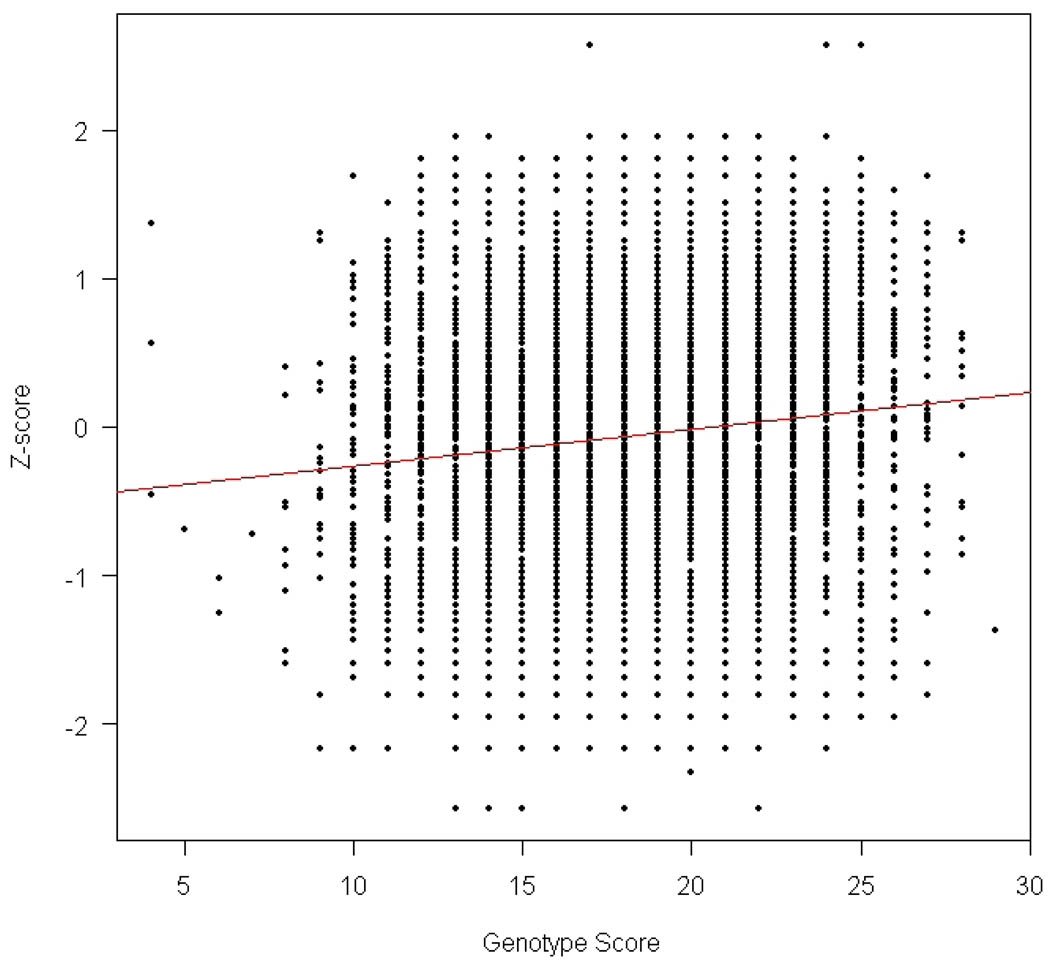
Finally, we investigated the fifteen significant SNPs further by testing for association between BMI Z-score and the genotype score by summing the number of BMI increasing alleles across all these SNPs. The resulting P-value for the genotype score was 2.53×10−16 (Figure 1). The genotype score explains 1.12% of the total variation for BMI z-score. We also tested pair-wise interactions between the fifteen significant SNPs, but none of the interaction effects were significant (Supplementary Table 2), suggesting that these fifteen SNPs act additively on the pediatric BMI z-score. As such, we did observe a cumulative effect but not as striking as reported by the GIANT consortium in their adult cohorts14.
Figure 1.
Scatter plot for association between BMI z-score and the genotype score by summing the number of BMI increasing alleles across all fifteen BMI-associated SNPs.
From this analysis, it is clear that a number of loci previously reported from GWA analyses of adult BMI and / or obesity also play a role in our phenotype of interest. While these recently discovered loci unveil several new biomolecular pathways not previously associated with obesity, it is important to note that these well established genetic associations with obesity explain very little of the genetic risk for this pediatric phenotype, suggesting the existence of additional loci whose number and effect size remain unknown. Once our GWA study is complete, we will have the opportunity to look for other variants in the genome that are associated with BMI in childhood.
RESEARCH METHODS AND PROCEDURES
Study Subjects
All subjects were consecutively recruited from the Greater Philadelphia area from 2006 to 2008 at the Children's Hospital of Philadelphia. Our study cohort consisted of 6,078 children of European ancestry with BMI information. All subjects were biologically unrelated and were aged between 0 and 18 years old. This study was approved by the Institutional Review Board of the Children's Hospital of Philadelphia. Parental informed consent was given for each study participant for both the blood collection and subsequent genotyping.
Genotyping
We performed high throughput genome-wide SNP genotyping using either the Illumina Infinium™ II HumanHap550 or Human 610 BeadChip technology in the same manner as our center has reported previously17. The SNPs analyzed survived the filtering of the genome wide dataset for SNPs with call rates <95%, minor allele frequency <1%, missing rate per person <2% and Hardy-Weinberg equilibrium P < 10−5.
Loci described from GWA studies published to date have been found using either the Affymetrix or Illumina platform. In the event a locus was reported using both the Illumina and Affymetrix arrays, we used the SNPs present on the Illumina array. In the event of a signal only being described on the Affymetrix array, we either already had that SNP on our Illumina array or we identified and used the best surrogate SNP available. As such, as rs7566605 at INSIG2 and rs10938397 at GNPDA2 were not available on the BeadChip, we employed perfect surrogates for the association analysis i.e. rs17047697 and rs13130484 respectively (r2=1). With rs7498665 at the SH2B1 locus, rs8049439 and rs4788102 are already in strong LD with this SNP so no additional surrogate was selected.
Analysis
From our database of heights and weights for our multi-dimensional scaling (MDS) determined Caucasians, we eliminated BMI outliers using 2% cutoff for each age category in order to remove measurement error and syndromic forms of obesity. As BMI values vary widely across pediatric age groups, each BMI value was adjusted for age and sex then expressed as a z-score.
We queried the data for the indicated SNPs in our pediatric samples. All statistical analyses were carried out using the software package plink18. By treating BMI as a quantitative trait, association analysis for each SNP was carried out using linear regression with the SNP included as an independent variable (coded as 0, 1, and 2).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all participating subjects and families. Hope Thomas, Kisha Harden, Andrew Hill, Kenya Fain, Crystal Johnson-Honesty, Cynthia Drummond, Shanell Harrison and Sarah Wildrick provided expert assistance with genotyping or data collection and management. We would also like to thank Smari Kristinsson, Larus Arni Hermannsson and Asbjörn Krisbjörnsson of Raförninn ehf for their extensive software design and contribution. This research was financially supported by the Children’s Hospital of Philadelphia.
The study is supported in part by a Research Development Award from the Cotswold Foundation (H.H. & S.F.A.G.) and NIH grant 1R01HD056465-01A1.
REFERENCES
- 1.Herbert A, Gerry NP, McQueen MB, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006 Apr 14;312(5771):279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 2.Loos RJ, Barroso I, O'Rahilly S, Wareham NJ. Comment on "A common genetic variant is associated with adult and childhood obesity". Science. 2007 Jan 12;315(5809):187. doi: 10.1126/science.1130012. author reply 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dina C, Meyre D, Samson C, et al. Comment on "A common genetic variant is associated with adult and childhood obesity". Science. 2007 Jan 12;315(5809):187. doi: 10.1126/science.1129402. author reply 187. [DOI] [PubMed] [Google Scholar]
- 4.Rosskopf D, Bornhorst A, Rimmbach C, et al. Comment on "A common genetic variant is associated with adult and childhood obesity". Science. 2007 Jan 12;315(5809):187. doi: 10.1126/science.1130571. author reply 187. [DOI] [PubMed] [Google Scholar]
- 5.Lyon HN, Emilsson V, Hinney A, et al. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet. 2007 Apr 27;3(4):e61. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotta K, Nakamura M, Nakata Y, et al. INSIG2 gene rs7566605 polymorphism is associated with severe obesity in Japanese. J Hum Genet. 2008;53(9):857–862. doi: 10.1007/s10038-008-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007 May 11;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant SF, Li M, Bradfield JP, et al. Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS ONE. 2008;3(3):e1746. doi: 10.1371/journal.pone.0001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinney A, Nguyen TT, Scherag A, et al. Genome Wide Association (GWA) Study for Early Onset Extreme Obesity Supports the Role of Fat Mass and Obesity Associated Gene (FTO) Variants. PLoS ONE. 2007;2(12):e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007 Jun;39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 11.Scuteri A, Sanna S, Chen WM, et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007 Jul 20;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008 Jun;40(6):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009 Jan;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 14.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009 Jan;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunstad J, Schofield P, Paul RH, et al. BDNF Val66Met polymorphism is associated with body mass index in healthy adults. Neuropsychobiology. 2006;53(3):153–156. doi: 10.1159/000093341. [DOI] [PubMed] [Google Scholar]
- 16.Grant SF, Bradfield JP, Zhang H, et al. Investigation of the Locus Near MC4R With Childhood Obesity in Americans of European and African Ancestry. Obesity (Silver Spring) 2009 Mar 5; doi: 10.1038/oby.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakonarson H, Grant SFA, Bradfield JP, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448(7153):591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.