Abstract
The only recognised genetic determinant of the common forms of Alzheimer’s disease (AD) is the ε4 allele of the apolipoprotein E gene (APOE). To identify new candidate genes, we recently performed transcriptomic analysis of 2,741 genes in chromosomal regions of interest using brain tissue of AD cases and controls.
From 82 differentially expressed genes, 1,156 polymorphisms were genotyped in two independent discovery sub-samples (n=945). Seventeen genes exhibited at least one polymorphism associated with AD risk and following correction for multiple testing, we retained the IL-33 gene.
We first confirmed that the IL-33 expression was decreased in the brain of AD cases compared with that of controls. Further genetic analysis led us to select 3 polymorphisms within this gene, which we analysed in three independent case-control studies. These polymorphisms and a resulting protective haplotype were systematically associated with AD risk in non-APOE ε4 carriers. Using a large prospective study, these associations were also detected when analyzing the prevalent and incident AD cases together or the incident AD cases alone. These polymorphisms were also associated with less cerebral amyloid angiopathy (CAA) in the brain of non-APOE ε4 AD cases. Immunohistochemistry experiments finally indicated that the IL-33 expression was consistently restricted to vascular capillaries in the brain. Moreover, IL-33 overexpression in cellular models led to a specific decrease in secretion of the Aβ40 peptides, the main CAA component.
In conclusion, our data suggest that genetic variants in IL-33 gene may be associated with a decrease in AD risk potentially in modulating CAA formation.
Keywords: Alzheimer, IL-33, brain expression, polymorphism, CAA
Introduction
Hereditary early-onset forms of Alzheimer’s disease (AD) have been linked to mutations in three different genes: the amyloid precursor protein (APP) gene on chromosome 21, the presenilin 1 (PS1) gene on chromosome 14 and the presenilin 2 (PS2) gene on chromosome 1 (1). These mutations, however, explain less than 1% of all AD cases (2). The genetics of the vast majority of AD cases (especially late-onset forms, LOAD) appears far more complex and is likely to involve interactions between environmental factors and various susceptibility genes. Only the ε4 allele of the apolipoprotein E (APOE) gene has been consistent shown to be a susceptibility factor for complex inherited sporadic AD (3). Although more than 200 genes have thus far been proposed as genetic determinants of AD, no consensus has yet been reached for even one of them, mainly because of the lack of robustness of the associations observed in independent populations (4).
To remedy this problem and determine the relevance of the different association studies, a data bank (http://www.alzforum.org/res/com/gen/alzgene) was established to make it possible to annotate all of the publications on association studies for AD (5). Beyond such sharing of data in international databases, the study of AD genetics, like that of most multifactorial diseases, has turned towards high- or very high-throughput analyses. The first reports of systematic genome analysis (genome-wide association, or GWA) in case-control studies of AD have been recently published (6–8). High-throughput transcriptomic and proteomic analyses have also led to the development of approaches that make it possible to help to select candidate genes based on their potential function, for example, genes that are differentially expressed in a chromosomal region of interest (9). Even once linkage studies have highlighted the location of genes potentially associated with a disease, it remains difficult to identify candidates within these broad chromosomal regions on the basis of genetic association studies alone. The combination of genome scan studies with more functional approaches, such as differential expression in normal and pathological tissues, may help to pick out relevant candidate genes without any a priori hypotheses about biological functions or involvement in the pathological process.
We applied such a strategy by combining genetic map information with gene expression profiling data. We recently reported results obtained from a customised microarray containing all the genes (n=2,741) located within 9 chromosomal regions of interest (10). Assessment of the levels of gene expression in total RNA from frontal brain tissue of AD patients and controls showed that 106 genes were differentially expressed (11). To speed up the selection of candidate genes, we next developed a high-throughput genetic analysis of 82 of them including 1,156 polymorphisms. Following this systematic approach, we focused on the IL-33 gene.
Materials and methods
Study population
The main characteristics of the populations are described in Table S14 and S15 (supplemental materials and methods). All subjects or, in those with substantial cognitive impairment, a caregiver, legal guardian, or other proxy gave written informed consent for participation in this study. The study protocols for all populations were reviewed and approved by the appropriate Institutional review boards of each country.
French case-control study (12)
All samples were Caucasian from the north of France (AD cases n=734, controls n=636). Clinical diagnosis of probable AD was established according to the DSM-III-R and NINCDS-ADRDA criteria. Caucasian controls were defined as subjects without DMS-III-R dementia criteria and with integrity of their cognitive functions (MMS>25). Presence of family history of dementia was considered as a criterion of exclusion (at least two documented relatives affected). Controls were recruited in retirement homes or from electoral rolls (altruistic volunteers).
UK case-control study (13)
All samples were Caucasian from Greater Birmingham (AD cases n=370, controls n=167). Clinical diagnosis of probable AD was established according to the DSM-III-R and NINCDS-ADRDA criteria. Control subjects were assessed using either DSM-III-R questionnaire or had a MMSE score above 28.
American case-control study (14)
All samples were Caucasian from Pittsburgh area (AD cases n=871, controls n=829). Late-onset AD (LOAD) were from the University of Pittsburgh Alzheimer’s Disease Research Center (ADRC). Clinical diagnoses of the patients were made according to the NINCDS/ADRDA criteria. The ADRC follows a standard evaluation protocol, which includes medical history, general medical and neurological examinations, a psychiatric interview, neuropsychological testing and a MRI scan. Age-and sex-matched 829 controls were recruited from the same Western Pennsylvania region as the cases, and were determined to be cognitively intact following extensive clinical examination (MMSE≥28).
Population-based 3C-study (15)
The 3C Study is a population-based, prospective (4-years follow-up) study of the relationship between vascular factors and dementia. It has been carried out in three French cities: Bordeaux (southwest France), Montpellier (southeast France) and Dijon (central eastern France). A sample of non-institutionalised, over-65 subjects was randomly selected from the electoral rolls of each city. Between January 1999 and March 2001, 9,686 subjects meeting the inclusion criteria agreed to participate. Following recruitment, 392 subjects withdrew from the study. Thus, 9,294 subjects were finally included in the study (2,104 in Bordeaux, 4,931 in Dijon and 2,259 in Montpellier). At the baseline clinical examination, blood samples were obtained from 8,707 individuals.
Brain samples
(for details, see supplemental materials and methods). Brains samples were obtained from 114 patients with early- and late-onset sporadic AD accessioned (16) and 167 Control brains, (17) as previously reported. The extent of cerebral amyloid angiopathy (CAA) in leptomeningeal and intraparenchymal arteries and the proportion of tissue area occupied by Aβ40 and Aβ42 were quantified following immunohistochimistry (16).
Genotyping microarray
Using the HapMap website (see URLs), we defined a minimal set of SNPs capturing a complete genetic information within the genes of interest (using all the referenced SNPs with a frequency >10% and r2<0.8 to define these Tag-SNPs). We finally selected 1,156 SNPs and developed an Affymetrix microarray allowing for the typing of all the SNPs in one experiment for each individual. Experiments were performed by the DNAvision society (Belgium) as described by the supplier using a GeneChip Hybridization Oven 640 plate-form and GeneChip Scanner 3000 7G 4C.
The selected SNPs were analysed in two independent sub-populations obtained by random sampling from the complete French population (n=545) and the American one (n=400) (for the main characteristics, supplemental material, Table S1B). Twenty one SNPs (1.8%) were not successfully genotyped. All other individuals SNPs had a genotype success rate >90%. Seventy SNPs (6%) were not in Hardy-Weinberg equilibrium (P<0.05). However, an important proportion of these SNPs (n=35) were located in vicinity of or within a potential Copy Number variation polymorphism (data not shown), potentially explaining this higher than expected proportion (5%).
SNPs densification in interleukin 33 (IL-33), genotyping and SNPs selection
Besides the 4 Tag-SNPs selected using the HapMap website, thirteen SNPs (frequency>10%) were randomly selected along the gene using NCBI website and denaturing high-performance liquid chromatography (dHPLC) results (for genotyping details and populations, see supplemental materials and methods)
Immunohistochemistry
Brain tissue from the temporal anterior cortex (Brodmann area 38) from nine AD patients and twelve controls was investigated. The AD patients were extensively investigated and followed until death in the Lille Memory Unit, the controls were people devoid of any neurological illness. Positive controls of the staining steps were human tonsils and the colon mucosa from a patient with Crohn’s disease (For immunohistochemistry protocol, see supplemental materials and methods)
IL-33 mRNA quantification
Total RNA was extracted from frozen frontal cortex brain tissue from the 114 AD (29) and 167 control (30) samples using phenol/chloroform protocol (Trizol® reagent, Invitrogen). Quantification of IL-33 mRNA was performed using the quantigene technology (for details, see supplementary materials) (18).
Plasmid constructions
IL-33 cDNA was amplified by PCR using the human IL-33 cDNA as a template and the following oligonucleotides: sense primer contained a Nhe I site (5′-gctagccaagatcacaagaatactga-3′) and an antisense primer contained a BamH I site (5′-ggatccagatgcagttatacagaggg-3′). The PCR fragment was next cloned into a mammalian expression pcDNA3.1(−) vector (Invitrogen, USA). The APP695wt pcDNA3 vector was already described elsewhere (19).
Cell culture and transfection
SKNSH-SY5Y-APP695WT and COS-7 cell lines were maintained in DMEM with 10% serum FCS, 2mM L-Glutamine and 50 U/ml penicillin, and 50 μg/ml streptomycin (and 1% MEM NEAA (Invitrogen, USA) for SY5Y-APPWT) at 37°C in a humidified atmosphere with 5% CO2. Transient transfection of pcDNA3 vectors were performed using Exgen500 (Euromedex, France) in SY5Y-APPWT and Fugen-HD (Roche Diagnostics, Switzerland) in COS-7 according to the manufacturer’s recommendations.
Immunofluorescence (IF)
The SY5Y-APPWT and COS-7 cell line was cultured on poly l-Lys-coated glass coverslips (Chamber Slide System 2 wells (Lab-Tek; Nunc, Rosckilde, Denmark)) for 24 hours. For IL-33 IF, cells were tranfected with IL-33 cDNA. After 48 hours, cells were fixed with in PBS containing 4% paraformaldehyde and labelled with primary antibodies 1/100 (respectively, PAb to IL-33 (human) (Alexis®, Apotech, Switzerland) or anti-ST2 (2A5)) (for details, see supplementary materials).
Analyses of the APP metabolism
Holo-APP, APPs, APP-CFT and Actin were measured by protein blotting as previously reported (19). Aβ40 and Aβ42 peptide concentrations were measured by sandwich ELISA (Human Amyloid β (1–40) Assay Kit (IBL-Hamburg, Germany) and INNOTEST® β-Amyloid (1–42), (Innogenetics, Belgium) respectively). ST2 was measured by protein blotting using 2A5 antibody (1/5000).
Statistical analyses
(for complete statistical methodologies, see supplemental materials and methods). The SAS software release 8.02 was used for statistical analyses (SAS Institute, Cary, NC). The association of the all the SNPs with the risk of AD was estimated by multiple logistic regression models, adjusted for age, gender, APOE status and centre. Haplotypes were estimated using the Haploview and Thesias software. In the French population, in order to take into account of possible inclusion of subjects suffering from mild cognitive impairments, analyses were performed using a MMSE score of 28 as a cut-off. The associations did not differ when the controls exhibiting a MMSE<28 were excluded.
The association of rs1157505, rs11792633 and rs7044343 with age at onset was analysed using a general linear model adjusted for gender and center following APOE stratification. Haplotype associations with age at onset were estimated using the Thesias software (20). For analyses using the 3C-study, logistic regression models were used to analyse the association between the risk of prevalent/incident AD and IL-33 genotypes. Proportional hazards models with delayed entry and age as the time scale were used to analyse the association between the risk of incident AD and the IL-33 genotype (21). Age, gender, centre, educational level, hypertension, body mass index (BMI), diabetes, history of vascular disease and APOE genotype were systematically used as adjusting factors (see supplemental materials and methods). Haplotype associations with the risk of AD were estimated using logistic regression or proportional hazards models using Thesias 3.0 software (20).
Comparison of IL-33 mRNA amounts between AD cases and controls, and the association study of rs1157505, rs11792633 and rs7044343 with CAA score were performed using a non-parametric Wilcoxon test. Variations in Aβ peptides secretion were analysed following normalization to the Mock vector experiments and differences were assessed by two-tailed Student’s t-test.
Results
Initial genetic screening
We used the HapMap database to select an initial set of 1,156 Tag-SNPs from 82 of the 106 differentially expressed genes (the HapMap database had no referenced SNPs for the remaining 24 genes when the customised affymetrix genotyping microarray was designed –HapMap data rel. 20/ phaseII Jan06 on NCBI B35 assembly, dbSNP b125-). These Tag-SNPs were then genotyped in two independent “discovery” sub-samples (French= 545; Americans= 400) randomly selected from two large case-control studies from these populations (12,13).
Seventeen genes had at least one polymorphism associated with the risk of developing AD (nominal p-value<0.05) in the combined “discovery” sub-sample of 945 individuals (Table 1 and supplementary material, Table S1). At this stage, a complementary approach would have been to correlate allele dosage with expression profiles. However, our initial transcriptomic study was performed in a restricted number of brain samples (12 controls and 9 AD patients), not allowing for pertinent statistical analyses. We thus only applied a conservative Bonferroni correction to select the most interesting genes.
Table 1.
Genes exhibiting Tag-SNPs associated with the risk of developing AD and age at onsert following the systematic genetic analysis of 1,156 SNPs by microarray experiments.
| GENE | RefSeq | Chr. | Chromosomal locationa | Differential expressionb | Number of studied SNPs | Number of associated SNPsc | Association with age at onsetc |
|---|---|---|---|---|---|---|---|
| MYO10 | NM_012334 | 5 | 16,715,016–16,989,385 | + 201% | 103 | 24 | 4 |
| ITGA2 | NM_002203 | 5 | 52,320,913–52,426,365 | + 215% | 26 | 3 | |
| LOC221608 | NM_007047 | 6 | 26,473,377–26,486,525 | − 45% | 13 | 2 | |
| TNXB | NM_019105 | 6 | 32,116,911–32,185,131 | + 100% | 31 | 4 | |
| AGER | NM_001136 | 6 | 32,256,724–32,260,001 | − 53% | 7 | 4 | |
| HLA-DRA | NM_019111 | 6 | 32,515,625–32,520,799 | − 57% | 10 | 5 | |
| PHF1 | NM_002636 | 6 | 33,486,934–33,492,187 | − 47% | 4 | 3 | |
| CCDN3 | NM_001760 | 6 | 42,010,650–42,017,530 | + 157% | 9 | 3 | |
| IL-33 | NM_033439 | 9 | 6,231,678–6,247,982 | − 66% | 4 | 2 | |
| FLJ20375 | AK000382 | 9 | 20,939,639–20,985,947 | − 62% | 55 | 9 | |
| B4GALT1 | NM_001497 | 9 | 33,100,642–33,157,231 | − 43 % | 9 | 5 | |
| CNTFR | NM_001842 | 9 | 34,541,431–34,579,722 | +132 % | 16 | 7 | 2 |
| CA9 | NM_001216 | 9 | 35,663,915–35,671,152 | −50% | 11 | 1 | |
| KIAA1462 | NM_020848 | 10 | 30,343,390–30,376,806 | + 262% | 18 | 3 | 5 |
| CAMK2G | NM_001222 | 10 | 75,242,265–75,304,344 | −50% | 10 | 7 | |
| ANGPT4 | NM_009641 | 20 | 801,604–844,605 | − 65% | 42 | 6 | 5 |
| HSPA12B | NM_175842 | 20 | 4,103,675–4,116,352 | − 42% | 15 | 2 |
Assembly Mar. 2006 (http://genome.ucsc.edu/cgi-bin/hgGateway)
differential expression between brain of AD cases and controls
nominal P-value < 0.05
When such a conservative correction was applied to take into account multiple testing (Threshold, P=4.3×10−5), only one association remained significant. The intronic rs7044343 SNP located within the IL-33 gene on 9p24.1 was associated with AD in the combined sample (OR=0.5, 95% C.I. [0.4–0.7]; P=3×10−5 adjusted for age, gender, centre and APOE status, Table 2). This association was observed in both the French and American sub-samples (OR=0.5, 95% C.I. [0.3–0.7], P=0.0002 and OR=0.6, 95% C.I. [0.4–1.0], P=0.05 adjusted for age, gender, centre and APOE status, Table 2). Another tag-SNP (rs7848215) within the IL-33 gene was also weakly associated with the risk of developing AD in the combined population (nominal P-value of 0.01; Table 2).
Table 2.
Association between the SNPs and the risk of developing AD in the French and American sub-populations
| French |
American |
French + American |
|||||
|---|---|---|---|---|---|---|---|
| SNP | OR [95% C.I.] | Pa | OR [95% C.I.] | Pa | OR [95% C.I.] | Pb | |
| Tag-SNP* | rs7848215 | 1.4 [0.9–2.0] | 0.11 | 1.5 [1.0–2.3] | 0.06 | 1.4 [1.1–1.9] | 0.01 |
| rs16924144 | 0.9 [0.6–1.4] | 0.77 | 0.7 [0.5–1.2] | 0.19 | 0.8 [0.6–1.1] | 0.28 | |
| rs16924159 | 0.9 [0.6–1.3] | 0.58 | 0.6 [0.4–1.0] | 0.05 | 0.8 [0.6–1.0] | 0.08 | |
| rs7044343 | 0.5 [0.3–0.7] | 2.10−4 | 0.6 [0.4–1.0] | 0.05 | 0.5 [0.4–0.7] | 3.10−5 | |
| Supplementary SNPs | rs1157505 | 0.6 [0.4–0.9] | 0.008 | 0.6 [0.4–0.9] | 0.03 | 0.6 [0.4–0.8] | 6.10−4 |
| rs1891385 | 0.9 [0.5–1.4] | 0.62 | 1.5 [0.9–2.5] | 0.16 | 1.1 [0.8–1.6] | 0.58 | |
| rs996029 | 0.7 [0.4–1.4] | 0.36 | 1.4 [0.6–3.1] | 0.38 | 0.9 [0.5–1.6] | 0.79 | |
| rs16924161 | 0.8 [0.5–1.3] | 0.48 | 0.9 [0.5–1.4] | 0.52 | 0.9 [0.6–1.2] | 0.35 | |
| rs10975511 | 0.8 [0.6–1.2] | 0.36 | 0.7 [0.4–1.1] | 0.09 | 0.8 [0.6–1.0] | 0.06 | |
| rs7035413 | 1.3 [0.9–1.9] | 0.20 | 1.7 [1.1–2.6] | 0.02 | 1.4 [1.1–1.9] | 0.01 | |
| rs11792633 | 0.5 [0.3–0.7] | 5.10−4 | 0.8 [0.5–1.2] | 0.23 | 0.6 [0.4–0.8] | 6.10−4 | |
| rs8172 | 0.8 [0.4–1.3] | 0.32 | 0.7 [0.4–1.3] | 0.25 | 0.7 [0.5–1.1] | 0.12 | |
adjusted for age, sex and APOE status
adjusted for age, sex, APOE status and centre
TagSNPs selected in HapMap database
In depth analysis of the IL-33 gene
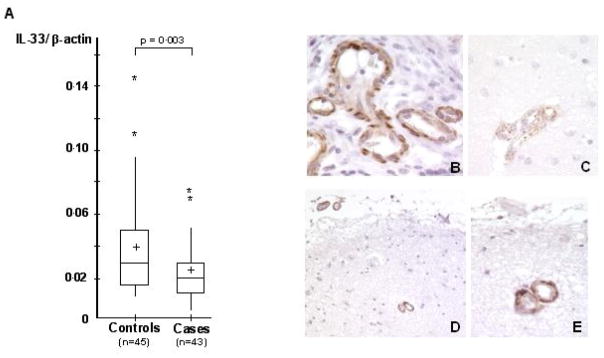
Following these initial observations, we decided to further explore the IL-33 gene. This was supported by our observation of a lower expression in the brain of AD cases compared with one of the controls (11) in our microarray analysis (−66%, P<10−5; Table 1). We first validated this decrease in expression in a larger cohort of brain samples of AD (n=43) and controls (n=45) (−41%, P=0.003, Figure 1A) using direct mRNA quantification (see materials and methods).
Figure 1.
(A) Expression level of IL-33 in the brain of AD cases and controls. All values of IL-33 mRNA were reported as arbitrary units following normalisation by β-actin mRNA quantification. mRNA quantifications (IL-33 and β-actin) were carried out in triplicate in all individuals (n = 45 controls and n=43 cases). Cross: mean of IL-33 expression in cases and controls; Middle line: Median; Upper horizontal line: inclusion of 75% of the individuals; lower horizontal line: inclusion of 25% of the individuals. *: individuals exhibiting extreme values (out of the global distribution). (B) Representative IL-33 immunolabelling in the colon mucosa of a patient with Crohn’s diseas as a positive control; (C) Representative IL-33 immunolabelling in AD brains. In the superficial cortex from an Alzheimer patient, the small meningeal and superficial cortical small vessels show the same pattern of labelling than in the one observed in controls. (D,E) Representative IL-33 immunolabelling in the control brains. This labelling is mostly located in endothelial cells and vascular smooth muscle cells of small vessels, in the nucleus and/or the cytoplasm
Only four Tag-SNPs (rs7848215, rs16924144, rs16924159 and rs7044343) in IL-33 were genotyped in our initial stage, therefore we next searched for supplementary SNPs from the NCBI international database and systematic screening for unknown polymorphisms within promoter, exon, intron/exon boundaries and UTR regions by dHPLC and sequencing. No non-synonymous polymorphisms were found (supplementary materials, Table S2). The construction of a linkage disequilibrium map based on the 4 initial Tag-SNPs and 11 other SNPs genotyped in 220 French controls, led us to finally select 8 supplementary SNPs (supplementary material, Figure S1).
These 8 supplementary SNPs were analysed in the combined “discovery” sub-sample of 945 individuals (supplementary material, Table S3). In addition to the two initial Tag-SNPs we reported associated with AD, associations between three other SNPs and AD risk presented nominal p-values<0.05 (Table 2). The rs7044343 association was still the only one to survive to Bonferroni correction (P=4.3×10−5) but two others intronic SNPs (rs1157505 and rs11792633) exhibited strong associations in the combined sample (Table 2).
We generated 12-site haplotypes and used the most common haplotype as a reference. Only one haplotype including the rare alleles of the intronic rs1157505, rs11792633 and rs704343 SNPs was associated with the risk of developing AD (OR=0.6, 95% C.I. [0.4–0.9], P=0.003, data not shown). Since the rs1157505, rs11792633 and rs704343 SNPs also exhibited restricted linkage desiquilibrium (LD) between them (r2 from 0.19 to 0.64; supplemental material, Figure S1), we thus supposed that these three SNPs were all informative.
We finally detected a significant statistical interaction between the APOE ε4 allele and the rs1157505 or rs11792633 SNPs (respectively, P=0.01 and P=0.04). A similar trend was observed for the rs7044343 SNP (P=0.14). The stratification by APOE status revealed that associations with AD risk were stronger in non-ε4 carriers (allelic association; P=3×10−4 in non-ε4 carriers and P=0.9 in ε4 carriers for rs1157505; P=5×10−4 in non-ε4 carriers and P=0.9 in ε4 carriers for rs11792633; P=5×10−5 in non-ε4 carriers and P=0.9 in ε4 carriers for rs7044343).
IL-33 as a candidate gene for LOAD mainly in non ε4-bearers in case-control studies
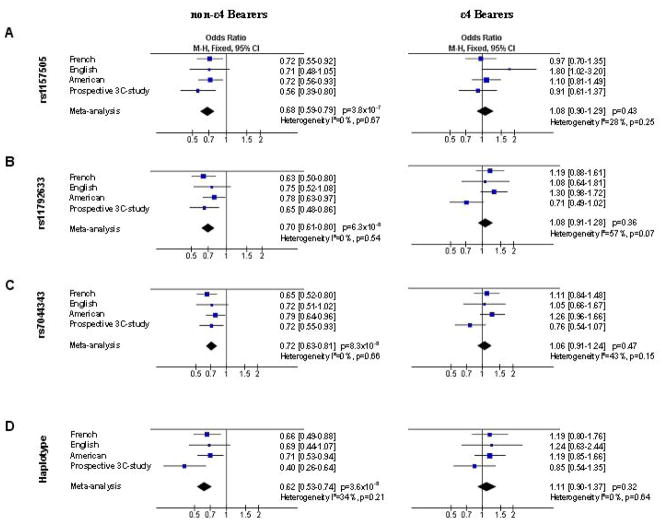
We decided to extend the analyses of the intronic rs1157505, rs11792633 and rs7044343 SNPs to the complete French (n=1,370) and American (n=1,700) case-control studies and to a third independent case-control sample from the UK (n=522). In the combined population, the association of these three SNPs with the risk of developing AD was influenced by the APOE status (supplementary material, Table S4). In accordance with our initial results, we observed a strong interaction between these SNPs and the APOE ε4 allele (P=0.004 for rs1157505, P=0.0001 for rs11792633 and P=0.002 for rs7044343; Table S4). The association of the three SNPs with AD risk was again strictly restricted to non-ε4 carriers, in all the study populations (Figure 2 and supplementary material, Tables S4–7). The minor alleles of rs1157505, rs11792633 and rs7044343 showed consistent protective effects against the risk of developing AD (OR=0.67, 95% C.I. [0.55–0.82], P=6×10−5; OR=0.6395% C.I. [0.52–0.76], P=1×10−6 and OR=0.67 95% C.I. [0.56–0.81], P=3×10−5, respectively (Table S4)). Furthermore, the rs11792633 minor allele was also associated with an older age at onset in non-ε4 carriers (73.3 ± 8.5 years in 12+22 carriers versus 71.9 ± 7.9 in 11 carriers; P=0.02 adjusted for gender and centre).
Figure 2.
Impact of the rs1157505, rs11792633 and rs7044343 SNPs on the risk of developing AD in the American, English, French, prospective-3C study and combined populations according to the APOE status. (A), (B) and (C) Allelic OR for the rs1157505, rs11792633 and rs7044343 SNPs respectively. (D) OR for the haplotype 222 (including the rare alleles of the rs1157505, rs11792633 and rs704343 SNPs).
We next generate 3-site haplotypes from rs1157505, rs11792633 and rs7044343 SNPs. In non-ε4 carriers, the 111 haplotype (resulting from the combination of the three frequent alleles) was over-represented in AD cases compared with controls (63% versus 55%, P=3×10−7) and the converse 222 haplotype (resulting from the combination of the three minor alleles) under-represented (13% versus 18%; P =3×10−4). These findings were consistent in all three case-control populations (supplementary material, Table S8). In the combined sample, the association of these two haplotypes with the risk of developing AD was consistent after permutation tests (respectively, P=2×10−6 and P=0.001 after 1,000 permutations). We estimated that the 222 haplotype was associated with a risk (P=4×10−6; supplementary material, Table S8) of developing AD lower by a factor of 0.65 compared with the 111 haplotype. Haplotypes including at least one minor allele of the rs1157505, rs11792633 or rs7044343 SNPs were also consistently associated with an older age at onset (from 0.9 to 1.7 years), compared with the age at onset attributed to the 111 haplotype (P=0.004 for global effect with Thesias software; data not shown).
IL-33 as a candidate gene for LOAD mainly in non ε4-bearers in the prospective 3C-study
We also analysed the association of the rs1157505, rs11792633 and rs7044343 SNPs in a large prospective population-based study. The rs1157505, rs11792633 and rs7044343 genotype distributions were seen to be in Hardy-Weinberg equilibrium in all AD and control populations (supplementary material Table S9). AD cases and controls differed significantly in terms of allele and genotype distributions, whatever the SNP studied (supplementary material Table S9). A significant decrease in the risk of AD in carriers of at least one rare allele of each SNP was observed, compared with those with no copies of these rare alleles (ORs ranging from 0.61 to 0.64; supplementary material Table S10). These associations were still significant when the analyses using proportional hazards model was limited to incident AD cases (ORs ranging from 0.67 to 0.72, supplementary material Table S10).
We stratified our population by the APOE ε4 allele. As previously described, the association of the rs1157505 with AD risk was more pronounced in non-ε4 bearers (supplementary material Table S10 and P=0.09 for interaction). In contrast, no difference was observed for the rs11792633 and rs7044343 polymorphisms (supplementary material Table S10 and respectively, P=0.70 and P=0.75 for interaction). We next generated 3-site haplotypes and used the most common haplotype as a reference. Only the haplotype including the rare alleles of the rs1157505, rs11792633 and rs704343 SNPs was associated with the risk of developing AD. Stratification by APOE status revealed that this association with AD risk was restricted to non-ε4 carriers (Figure 2 and supplementary material Table S11). Again, this association was significant when the analyses using proportional hazards model was limited to incident AD cases (supplementary material Table S11).
Lastly, the association of the IL-33 SNPs with the risk of other types of dementia was also assessed but no association was found, whatever the SNP studied (supplemental materials, Table S12).
IL-33 as a candidate gene for LOAD mainly in non ε4-bearers: evidence from combined analyses
We finally performed combined analyses of all the studies using the review manager software release 5.0 (http://www.cc-ims.net/RevMan/) to test for heterogeneity between the different case-control studies and to estimate the overall effect (Mantel-Haentzel fixed odds ratio). The different associations were highly homogeneous among the different studies allowing for combined analyses (see tests for heterogeneity, Figure 2).
The rare allele distributions of the rs1157505, rs11792633 and rs704343 SNPs were decreased in AD cases as compared to controls in all the studies, with ORs ranging from 0.68 to 0.72 (OR =0.68, 95% C.I. [0.59–0.79], P =3.8×10−7; OR =0.70, 95% C.I. [0.61–0.80], P =6.3×10−8; OR =0.72, 95% C.I. [0.63–0.81], P =8.3×10−8, respectively for the rs1157505, rs11792633 and rs704343 in the combined population; Figure 2A, 2B, 2C). Similarly, the haplotype including the rare alleles of the rs1157505, rs11792633 and rs704343 SNPs was associated with the risk of developing AD, with ORs ranging from 0.40 to 0.71 (OR =0.62, 95% C.I. [0.53–0.74], P =3.6×10−8 in the combined population; Figure 2D).
Comparison between our analysis and published GWAs
Recently, two GWA analyses in AD case-control studies were published (7,8). Since data for both GWAs is in part publicly available, we compared our results with those reported in these GWAs. Nine SNPs in IL-33 were analysed in the 500K affymetrix microarray which was used in both GWAs but none of them overlapped with the SNPs we analysed. However, we noticed that the rs10975519 SNP used on the microarray was in LD with the rs7044343 SNP (r2=0.74). In the Remain’s study (7), in accordance with our data, the rare allele of the rs10975519 SNPs was associated with a decrease in the risk of developing AD in non-ε4 bearers (OR=0.70, 95% C.I. [0.52–0.93], P=0.01; supplemental material, Table S13). Whereas the study of Reiman et al. supports that the IL-33 gene may be a genetic determinant of AD in non-ε4 bearers (7), the Li’s study do not allow us for assessing this point (8). The individual APOE genotype data was unfortunately not publicly available and so we were not able to assess the association of the IL33 SNPs with AD risk according to the APOE status.
The IL-33 gene, a determinant of CAA in non ε4-bearers, is strictly expressed in the vascular cells of the cerebral capillaries
Finally, to help define the involvement of IL-33 in AD, we analysed the impact of the rs1157505, rs11792633 or rs7044343 SNPs on pathological hallmarks of AD in a collection of 114 brain samples of confirmed AD cases. We investigated cerebral amyloid angiopathy (CAA) and parenchymal amyloid load (both Aβ40 and Aβ42). No SNP was individually associated with parenchymal amyloid load (both Aβ40 and Aβ42) whatever the APOE status (data not shown). On the other hand, after stratification by APOE status, the three SNPs were associated with cerebral amyloid angiopathy (CAA) in non-ε4 carriers. Individuals with at least one copy of the minor rs1157505, rs11792633 and rs7044343 alleles had lower whole-brain CAA scores (Table 3), with the decrease most pronounced in the temporal and frontal areas (Table 3). However, the limited number of non-ε4 carriers did not allow us to assess the association of the 111 and 222 haplotypes with CAA scores. It should nonetheless be noted that, in this cohort of confirmed AD cases, the haplotype frequencies in non-ε4 carriers were similar of those observed in the probable AD cases (111 haplotype: 60.0% and 222 haplotype: 10.0%).
Table 3.
Association of rs1157505, rs11672633 and rs7044343 with CAA in the brain of non-ε4 AD cases.
| CAAa |
|||||||
|---|---|---|---|---|---|---|---|
| n | Temporal | Frontal | Parietal | Occipital | Total | ||
| rs1157505b | 11 | 18 | 1.8 ± 0.7 | 1.7 ± 0.7 | 1.9 ± 2.4 | 2.3 ± 0.7 | 7.8 ± 2.5 |
| 12+22 | 8 | 1.5 ± 0.5 | 1.4 ± 0.9 | 1.7 ± 1.0 | 2.0 ± 1.1 | 6.6 ± 2.9 | |
| Variation | −17% | −18% | −19% | −13% | −15% | ||
| P | ns | ns | ns | ns | ns | ||
| rs11792633b | 11 | 10 | 2.2 ± 0.6 | 2.1 ± 0.6 | 2.2 ± 1.0 | 2.5 ± 0.7 | 9.0 ± 2.2 |
| 12+22 | 14 | 1.4 ± 0.5 | 1.4 ± 0.6 | 1.8 ± 0.8 | 2.0 ± 0.7 | 6.5 ± 2.1 | |
| Variation | −36% | −33% | −18% | −20% | −28% | ||
| P | 0.007 | 0.006 | ns | ns | 0.01 | ||
| rs7044343b | 11 | 10 | 2.2 ± 0.6 | 2.1 ± 0.6 | 2.2 ± 1.0 | 2.4 ± 0.8 | 8.9 ± 2.4 |
| 12+22 | 16 | 1.5 ± 0.5 | 1.4 ± 0.7 | 1.8 ± 0.9 | 2.1 ± 0.8 | 6.7 ± 2.4 | |
| Variation | −32% | −33% | −18% | −13% | −25% | ||
| P | 0.01 | 0.01 | ns | ns | 0.04 | ||
Cerebral Amyloid Angiopathy (CAA), non-parametric Wilcoxon test.
Frequent allele is coded 1 and minor allele is coded 2
In agreement with the genotype/phenotype associations observed with CAA, immunohistochemistry experiments showed that IL-33 immunoreactivity was restricted to the endothelium and vascular smooth muscle cells of small arteries located in the arachnoid, pia mater and superficial cortex in the brains of AD cases and controls (Figure 1B).
IL-33 overexpression is associated with decreased Aβ40 secretion
The observation that the less common alleles of certain IL-33 SNPs might be associated with lower CAA levels — composed mainly of Aβn-40 peptides— (22, 23) suggests that IL-33 may modulate mechanisms involved in Aβ production and more specifically Aβn-40. To explore this question, we undertook cell biology experiments to determine the specific role of IL-33 in Aβn-40 production. We used two well-characterised cellular models to address this point. (19, 24)
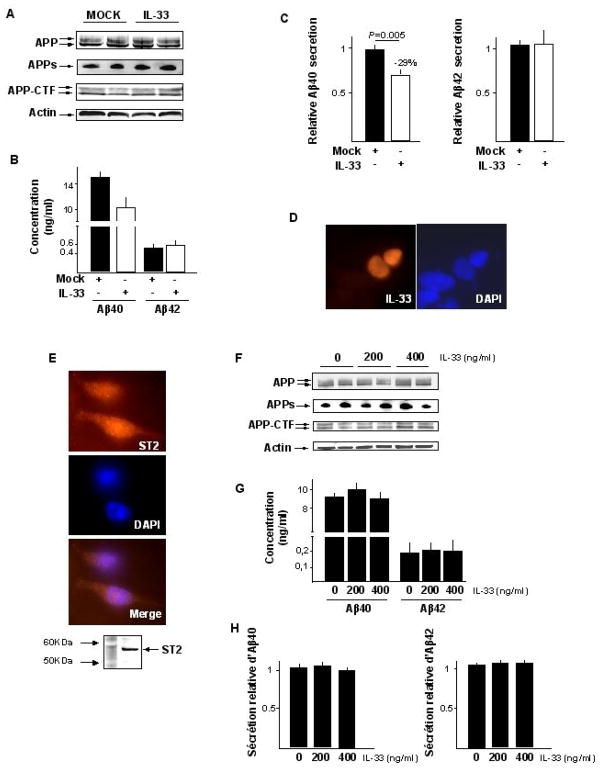
Overexpression of IL-33 in a SKNSH-SY5Y neuroblastoma cell line stably expressing APP695wt led to a significant 30% decrease in secretion of the Aβ1–40 peptides (P=0,005) without modifying the production of Aβ1–42 or other APP fragments (figure 3A and 3B). A similar significant specific decrease in Aβ1–40 secretion was observed when we used COS-7 cells co-transfected with eukaryotic IL-33 and APP695wt expression vectors (P=0,01; supplementary figure S2A and S2B). IL-33 is a dual function protein that may act as both a pro-inflammatory cytokine and an intracellular nuclear factor with transcriptional regulatory properties.(25–28). To determine its function in our in vitro models, we first conducted immunofluorescence experiments that showed that the protein was located exclusively in the nucleus in both SKNSH-SY5Y and COS-7 cells transfected with an IL-33 expression vector (figure 3D and supplementary figure S2D). Secondly, we exposed the SY5Y-APP695wt cell line to a recombinant human IL-33 protein in cell culture medium. Although the SY5Y-APP695wt cell line expressed the cell surface receptor ST2, which mediates IL-33 pro-inflammatory capacities (26, 27) (figure 3E), this treatment did not modify the level of Aβ secretion (figure 3G and 3H). Together, these results indicated that the reduction in Aβ1–40 secretion caused by IL-33 is likely mediated by pathways independent of its cell surface receptor but depending instead on its transcription-regulating properties in the nucleus (26, 29).
Figure 3.
Overexpression of IL-33 in the neuroblastoma SKNSH-SY5Y cell line stably transfected with APP695wt. (A) Representative experiment of APP metabolism variations following transfection of the IL-33 expression vector (APPs, APP soluble; APP-CTF, APP carboxy terminal fragments); (B) Representative experiment of Aβ40 and Aβ42 secretions following transfection of the IL-33 expression vector; (C) Mean variations of the Aβ40 and Aβ42 secretions from three independent experimentations in duplicates; (D) IL-33 accumulates in nucleus (colocalisation with dense regions of DAPI staining, indicating association with heterochromatin). No signal was detected using an empty vector (Mock) (Data not shown); (E) detection of the ST2 receptor expression in the SY5Y-APP695wt cell line by immunofluorescence experiments and western blot; (F) Representative experiment of APP metabolism variations in response to human recombinant IL-33 protein; (G) Representative experiment of Aβ40 and Aβ42 secretions in response to human recombinant IL-33 protein; (H) Mean variations of the Aβ40 and Aβ42 secretions from three independent experimentations in duplicates.
Discussion
The integrated strategy we developed to look for new genetic determinants of AD (supplemental material, Figure S3) led us to identify the IL-33 gene as a potential genetic determinant of AD. This gene is located on chromosome 9p24, a chromosomal region of interest in AD defined by genome scan studies using family-based populations (10). We postulated that a “true” genetic factor should systematically be associated with the risk of developing the disease whatever the kind of populations analysed: family-based studies, population-based studies or case-control studies.
A series of consistent observations in three independent case-control studies and a prospective population-based study established that: (i) the minor alleles of the intronic rs1157505, rs11792633 and rs7044343 SNPs within IL-33 were all associated with a reduced risk of AD risk in non-ε4 carriers; (ii) these minor alleles defined a 3-site protective haplotype among non-ε4 carriers. We thus observed significant and consistent effects in pooling together a large number of AD cases and controls. Remarkably, the associations of the rs1157505, rs11792633 and rs7044343 SNPs with the risk of developing AD, were highly homogeneous between all the case-control studies (Figure 3). The lack of significance observed in the English population was very likely a consequence of a lack of statistical power (4% power to detect an OR of 0.7 assuming an α level of 0.05). Additional genetic studies involving prospective cohorts as well as both family-based and large case-control samples, will be of course required to fully characterise the association of the IL-33 gene with AD. However, it is encouraging that coherent observations from a recent GWA support our findings.
Beyond these association studies, we also established a potential link between IL-33 and the AD physiopathological process. We determined that the minor alleles of rs1157505, rs11792633 and rs7044343 were associated with a lesser degree of CAA in the brains of non-ε4 AD patients and expression of IL-33 was consistently restricted to the cerebral vascular network. We also found a decrease in expression of IL-33 in the brains of AD cases compared to controls which replicates our initial finding in a small number of brains that we used with the microarray approach (11).
Although our findings suggest IL-33 modifies risk for AD, it is not clear how genetic variants in IL-33 might affect its function to alter risk. We were not able to establish an association between the IL-33 mRNA level in the AD or control brains and the SNPs or haplotypes studied (data not shown). The lack of power due to the number of brains available for study as well as inter-individual variations in the measure of the IL-33 mRNA levels may partly explain this absence of genotype/phenotype association. Furthermore, it is not possible to exclude that the difference in expression found in autopsied brain, is not representative of differences at early stages of the disorder or that SNPs may modulate unknown splicing events modifying IL-33 biological properties.
Because we observed that the protective genetic variants in IL-33 were associated with a lesser degree of CAA, which contains mainly Aβ40, (22, 23), we examined how IL-33 might specifically modulate Aβ40 production in two well-characterised cellular models previously used to study APP metabolism.(19, 24). Again, our observations consistently linked IL-33 expression with AD pathology: (i) overexpression of IL-33 in cellular models led to a systematic decrease in Aβ40 secretion but without modifying production of either Aβ42 or other APP fragments; (ii) this decrease in Aβ40 secretion is likely to be a consequence of transcriptional activity by IL-33, independently of its inflammatory properties. The latter finding, moreover, appears to be related to two other observations: that IL-33 expression appears to be restricted to the vascular capillaries and that smooth muscle cells are involved in the production of vascular Aβ peptides. Cultures of smooth muscle cells isolated from CAA of human, canine or murine origins have confirmed the involvement of these cells in vascular Aβ peptide production, by demonstrating the abundant secretion of Aβ40, Aβ42 and Aβ intracellular or extracellular deposits.(24, 25). The data, considered together, indicate that local secretion of Aβ40 peptides in the vascular brain network — perhaps independently of neuronal secretion — may be a key determinant of AD pathology.
The presence of CAA is often found in AD brains but it is hard to establish whether its appearance is related to the initial manifestations of the disorder or is a process that occurs later during the development of the disease. Interestingly, ApoE is also thought to play a prominent role in the deposition of Aβ peptides in cerebral vessels, as two items of supporting evidence suggest: (i) the APOE ε4 allele is a major risk factor for CAA (30–33) and (ii) transgenic mice models show that ApoE4 strongly favours the formation of CAA rather than parenchymal plaques (34). As a consequence, our data in addition to others seem to point out the potential importance of this lesion in the AD process (35). Furthermore, our evidence that allelic variants of both IL-33 and APOE genes modulate CAA may explain in part the systematic interaction we observed between these two genes on the risk of AD or CAA. The limited association between the IL-33 SNPs with risk of AD or CAA in ε4 carriers may thus be due to the major impact of the ε4 allele on AD risk and Aβ deposition, which might mask the effect of IL-33. Moreover, in the population-based 3C-study, we observed that the IL-33 gene may be a genetic determinant of AD but not of other types of dementia (such as mixed/vascular dementia in particular, data not shown). Although we studied only 124 individuals suffering from other types of dementia, which limits the relevance of this observation, it is interesting to note that vascular dementia has occasionally been associated with CAA (36, 37) (present in >80% of AD case series) (38, 39). Altogether, these observations may support a limited association between the IL-33 SNPs and AD risk by assuming that IL-33 is involved in CAA formation.
The cellular pathways controlled by the IL-33 protein that may specifically modulate Aβ40 secretion have not yet been identified. We only know that this protein, identified in 2003, is a protein that may have a dual function possibly acting as both a cytokine stimulating the Th2-associated cytokines production and an intracellular nuclear factor with transcription regulating properties (26–29). Although our data suggest that IL-33 is involved in AD as a nuclear factor, we cannot rule out the possibility that it also influences the AD process as a cytokine and participates in lymphocyte T recruitment (40, 41). Such an observation may be relevant since T-cell deficiency has been associated with deficits in spatial learning and memory (42) and that immune cells that infiltrate the brain appear to modulate the AD process in animal models (43, 44).
In conclusion, there is clear evidence of an association between the risk of developing AD and the IL-33 gene. The National Center for Biotechnology Information (NCBI) database currently lists at least 178 SNPs (validated or not) in this region. As a consequence, only important, systematic and ambitious efforts in sequencing and genotyping (even rare variants) in combination with replication in large independent populations and with functional analyses of intermediate phenotypes will help determine the exact implication of IL-33 in AD. However, the characterisation of IL-33 as a genetic determinant of AD indicates a potential relevant link between CAA formation, neurovascular dysfunction, alteration of immune cell functions and inflammatory process, all of which contribute to AD.
Supplementary Material
Acknowledgments
We thank Jo Ann Cahn for her helpful contribution in the writing of the manuscript. The authors thank Dr. Morisada Hayakawa for providing us the monoclonal ST2 antibody and Ryan Minster for technical support. Julien Chapuis was supported by the Ministère de l’enseignement supérieur et de la Recherche (MESR). Franck Hansmannel was supported by the Alzheimer’s association (Grant IIRG-06-25487). Faiza Bensemain was supported by the France Alzheimer Association. Geoffroy Laumet was supported by the Pasteur Institute of Lille and the region Nord-Pas de Calais. This work was funded by Genoscreen, INSERM (ATC-vieillissement), the Pasteur Institute of Lille, the genopole of Lille, the CPER-neuroscience and the US National Institute on Aging grants AG13672 and AG05133 (I.K. and S.T.D.).
The Three-City Study was performed as part of a collaboration between the Institut National de la Santé et de la Recherche Médicale (INSERM), the Victor Segalen–Bordeaux II University and Sanofi-Synthélabo. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study was also funded by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, MGEN, Institut de la Longévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Aquitaine and Bourgogne Regional Councils, Fondation de France and the jont French Ministry of Research/INSERM “Cohortes et collections de données biologiques” program. Lille Génopôle received an unconditional grant from Eisai.
Footnotes
Conflicts of interest
Dr. Lambert reports being a consultant to Genoscreen
References
- 1.Cruts M, Van Broeckhoven C. Molecular genetics of Alzheimer’s disease. Ann Med. 1998;30:560–565. doi: 10.3109/07853899809002605. [DOI] [PubMed] [Google Scholar]
- 2.Campion D, Dumanchin C, Hannequin D, Dubois B, Belliard S, Puel M, Thomas-Anterion C, Michon A, Martin C, Charbonnier F, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet. 1999;65:664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 4.Lambert JC, Amouyel P. Genetic heterogeneity of Alzheimer’s disease: complexity and advances. Psychoneuroendocrinology. 2007;32:S62–S70. doi: 10.1016/j.psyneuen.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 6.Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, et al. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–73. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 7.Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, et al. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, et al. Candidate Single-Nucleotide Polymorphisms From a Genomewide Association Study of Alzheimer Disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 9.Li YJ, Oliveira SA, Xu P, Martin ER, Stenger JE, Scherzer CR, Hauser MA, Scott WK, Small GW, Nance MA, et al. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003;12:3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- 10.Lambert JC, Testa E, Cognat V, Soula J, Hot D, Lemoine Y, Gaypay G, Amouyel P. Relevance and limitations of public databases for microarray design: a critical approach to gene predictions. Pharmacogenomics J. 2003;3:235–241. doi: 10.1038/sj.tpj.6500184. [DOI] [PubMed] [Google Scholar]
- 11.Bensemain F, Hot D, Ferreira S, Dumont J, Bombois S, Maurage CA, Huot L, Hermant X, Levillain E, Hubans C, et al. Evidence for induction of the ornithine transcarbamylase expression in Alzheimer’s disease. Mol Psychiatry. 2007 doi: 10.1038/sj.mp.4002089. in press. [DOI] [PubMed] [Google Scholar]
- 12.Chapuis J, et al. Association study of the vascular endothelial growth factor gene with the risk of developing Alzheimer’s disease. Neurobiol Aging. 2006;27:1212–1215. doi: 10.1016/j.neurobiolaging.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Chapuis J, et al. Association study of the GAB2 gene with Alzheimer’s disease. Neurobiol Dis. 2008;30:103–6. doi: 10.1016/j.nbd.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Kamboh MI, Minster RL, Feingold E, DeKosky ST. Genetic association of ubiquilin with Alzheimer’s disease and related quantitative measures. Mol Psychiatry. 2006;11:273–279. doi: 10.1038/sj.mp.4001775. [DOI] [PubMed] [Google Scholar]
- 15.3C Study Group. Vascular factors and risk of dementia: design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology. 2003;22:316–25. doi: 10.1159/000072920. [DOI] [PubMed] [Google Scholar]
- 16.Tian J, Shi J, Smallman R, Iwatsubo T, Mann DM. Relationships in Alzheimer’s disease between the extent of Abeta deposition in cerebral blood vessel walls, as cerebral amyloid angiopathy, and the amount of cerebrovascular smooth muscle cells and collagen. Neuropathol Appl Neurobiol. 2006;32:332–340. doi: 10.1111/j.1365-2990.2006.00732.x. [DOI] [PubMed] [Google Scholar]
- 17.Berr C, et al. Neuropathological epidemiology of cerebral aging: a study of two genetic polymorphisms. Neurobiol Aging. 2001;22:227–235. doi: 10.1016/s0197-4580(00)00227-x. [DOI] [PubMed] [Google Scholar]
- 18.Canales RD, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 19.Vingtdeux V, et al. Intracellular pH regulates amyloid precursor protein intracellular domain accumulation. Neurobiol Dis. 2007;25:686–696. doi: 10.1016/j.nbd.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Tregouet DA, Tiret L. Cox proportional Hazards survival regression in haplotype-based association analysis using the stochastic-EM algorithm. Eur J Hum Genet. 2004;12:971–974. doi: 10.1038/sj.ejhg.5201238. [DOI] [PubMed] [Google Scholar]
- 21.Barberger-Gateau P, Raffaitin C, Letenneur L, Berr C, Tzourio C, Dartigues JF, et al. Dietary patterns and risk of dementia: the Three-City cohort study. Neurology. 2007;6920:1921–30. doi: 10.1212/01.wnl.0000278116.37320.52. [DOI] [PubMed] [Google Scholar]
- 22.Joachim CL, Duffy LK, Morris JH, Selkoe DJ. Protein chemical and immunocytochemical studies of meningovascular beta-amyloid protein in Alzheimer’s disease and normal aging. Brain Res. 1988;474:100–11. doi: 10.1016/0006-8993(88)90673-7. [DOI] [PubMed] [Google Scholar]
- 23.Fryer JD, et al. Apolipoprotein E markedly facilitates age-dependent cerebral amyloid angiopathy and spontaneous hemorrhage in amyloid precursor protein transgenic mice. J Neurosci. 2003;23:7889–1896. doi: 10.1523/JNEUROSCI.23-21-07889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoe HS, Tran TS, Matsuoka Y, Howell BW, Rebeck GW. DAB1 and Reelin effects on amyloid precursor protein and ApoE receptor 2 trafficking and processing. J Biol Chem. 2006;281:35176–85. doi: 10.1074/jbc.M602162200. [DOI] [PubMed] [Google Scholar]
- 25.Frackowiak J, et al. Extracellular deposits of A beta produced in cultures of Alzheimer disease brain vascular smooth muscle cells. J Neuropathol Exp Neurol. 2005;64:82–90. doi: 10.1093/jnen/64.1.82. [DOI] [PubMed] [Google Scholar]
- 26.Carriere V, et al. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz J, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 28.Baekkevold ES, et al. Molecular characterization of NF-HEV, a nuclear factor preferentially expressed in human high endothelial venules. Am J Pathol. 2003;163:69–79. doi: 10.1016/S0002-9440(10)63631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moulin D, et al. Interleukin (IL)-33 induces the release of pro-inflammatory mediators by mast cells. Cytokine. 2007;40:216–225. doi: 10.1016/j.cyto.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Herzig MC, Winkler DT, Burgermeister P, Pfeifer M, Kohler E, Schmidt SD, Danner S, Abramowski D, Stürchler-Pierrat C, Bürki K, et al. Abeta is targeted to the vasculature in a mouse model of hereditary cerebral hemorrhage with amyloidosis. Nat Neurosci. 2004;7:954–960. doi: 10.1038/nn1302. [DOI] [PubMed] [Google Scholar]
- 31.Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci. 1993;90:9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidal R, Calero M, Piccardo P, Farlow MR, Unverzagt FW, Méndez E, Jiménez-Huete A, Beavis R, Gallo G, Gomez-Tortosa E, et al. Senile dementia associated with amyloid beta protein angiopathy and tau perivascular pathology but not neuritic plaques in patients homozygous for the APOE-epsilon4 allele. Acta Neuropathol. 2000;100:1–12. doi: 10.1007/s004010051186. [DOI] [PubMed] [Google Scholar]
- 33.Chalmers K, Wilcock GK, Love S. APOE epsilon 4 influences the pathological phenotype of Alzheimer’s disease by favouring cerebrovascular over parenchymal accumulation of A beta protein. Neuropathol Appl Neurobiol. 2003;29:231–238.21. doi: 10.1046/j.1365-2990.2003.00457.x. [DOI] [PubMed] [Google Scholar]
- 34.Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, Holtzman DM. Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci. 2005;25:2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicoll JA, Yamada M, Frackowiak J, Mazur-Kolecka B, Weller RO. Cerebral amyloid angiopathy plays a direct role in the pathogenesis of Alzheimer’s disease. Pro-CAA position statement. Neurobiol Aging. 2004;25:589–597. doi: 10.1016/j.neurobiolaging.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Esiri MM, Wilcock GK, Morris JH. Neuropathological assessment of the lesions of significance in vascular dementia. J Neurol Neurosurg Psychiatry. 1997;63:749–53. doi: 10.1136/jnnp.63.6.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalaria RN, Kenny RA, Ballard CG, Perry R, Ince P, Polvikoski T. Towards defining the neuropathological substrates of vascular dementia. J Neurol Sci. 2004;226:75–80. doi: 10.1016/j.jns.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Jellinger KA, Attems J. Prevalence and pathogenic role of cerebrovascular lesions in Alzheimer disease. J Neurol Sci. 2005;37:229–230. doi: 10.1016/j.jns.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 39.Ellis RJ, Olichney JM, Thal LJ, Mirra SS, Morris JC, Beekly D, et al. Cerebral amyloid angiopathy in the brains of patients with Alzheimer’s disease: the CERAD experience, Part XV. Neurology. 1996;46:1592–1596. doi: 10.1212/wnl.46.6.1592. [DOI] [PubMed] [Google Scholar]
- 40.Komai-Koma M, Xu D, Li Y, McKenzie AN, McInnes IB, Liew FY. IL-33 is a chemoattractant for human Th2 cells. Eur J Immunol. 2007;37:2779–2786. doi: 10.1002/eji.200737547. [DOI] [PubMed] [Google Scholar]
- 41.Ali S, Huber M, Kollewe C, Bischoff SC, Falk W, Martin MU. IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells. Proc Natl Acad Sci. 2007;104:18660–18665. doi: 10.1073/pnas.0705939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziv Y, Ron N, Butovsky O, Landa G, Sudai E, Greenberg N, Cohen H, Kipnis J, Schwartz M. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci. 2006;9:268–275. doi: 10.1038/nn1629. [DOI] [PubMed] [Google Scholar]
- 43.Buckwalter MS, Coleman BS, Buttini M, Barbour R, Schenk D, Games D, Seubert P, Wyss-Coray T. Increased T cell recruitment to the CNS after amyloid beta 1–42 immunization in Alzheimer’s mice overproducing transforming growth factor-beta 1. J Neurosci. 2006;26:11437–41. doi: 10.1523/JNEUROSCI.2436-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Britschgi M, Wyss-Coray T. Immune cells may fend off Alzheimer disease. Nat Med. 2007;13:408–9. doi: 10.1038/nm0407-408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.