Abstract
This study characterized the human apolipoprotein H (APOH, a.k.a. β2-glycoprotein I) promoter and its variants by in vitro functional experiments and investigated their relation with human plasma β2GPI levels. We examined the individual effects of 12 APOH promoter SNPs in the 5' flanking region of APOH (~1.4 kb) on luciferase activity in COS-1 cells and HepG2 cells and their impact on plasma β2GPI levels in 799 U.S. Whites, the DNA-binding properties of APOH promoter using electrophoretic mobility shift assay (EMSA) in HepG2 cells, the effects of serial deletion analysis of APOH 5' flanking region in COS-1 and HepG2 cells, and cross-species conservation of the APOH promoter sequence. The variant alleles of three SNPs (−1219G>A, −643T>C and −32C>A) showed significantly lower luciferase expression (51%, 40% and 37%, respectively) as compared to the wild-type allele. EMSA demonstrated that these three variants specifically bind with protein(s) from HepG2 cell nuclear extracts. Three-site haplotype analysis (−1219G>A, −643T>C, and −32C>A) revealed one haplotype carrying −32A (allele frequency = 0.075) to be significantly associated with decreased plasma β2GPI levels (P < 0.001). Deletion analysis localized the core APOH promoter to ~160 bp upstream of ATG codon with the presence of critical cis-acting elements between −166 and −65. Cross-species conservation analysis of the APOH promoters of 7 species indicated that basic promoter elements are highly conserved across species. In conclusion, we have characterized the functional promoter of APOH and identified functional variants that affect the transcriptional activity of the APOH promoter.
Keywords: APOH, β2-glycoprotein I, promoter, polymorphisms, association
Introduction
Human apolipoprotein H (APOH), also known as β2-glycoprotein I (β2GPI) (in this study, we will use APOH to refer to the gene as used in human genome databases and β2GPI to refer to the protein as commonly used in the rheumatology literature) is a major autoantigen recognized by predominant antiphopholipid antibodies (APA) found in sera of many autoimmune diseases such as primary antiphospholipid syndrome (PAPS) and systemic lupus erythematosus (SLE) [1, 2]. APOH spans 18 kilobases (kb) on chromosome 17q23–24 [3] and encodes for a mature protein of 326 amino acid (aa) residues. β2GPI is a 50-kDa single chain plasma glycoprotein exhibiting internal homology comprised of four contiguous homologous regions of about 60 aa residues, and an additional variable fifth C-terminal domain. The variable configuration of the fifth domain is essential for the binding of β2GPI to anionic phospholipids [4–6]. Primer extensions determined alternate transcription start sites (TSSs) at 31 base pairs (bp) and 21 bp upstream of the APOH translation start codon [3]. TSS 31 bp upstream agreed completely with the consensus for an initiator element (Inr) known to sustain transcription initiation. Previously [7], an atypical TATA box and HNF-1α cis-elements have been identified to be critical for APOH cell type-specific transcriptional regulation leading to differential expression of APOH in humans.
β2GPI is primarily expressed in the liver and sporadically in intestinal cell lines and tissues [8]. The plasma concentration of β2GPI is approximately 20 mg/dL of which a small portion is bound to lipoproteins and the rest exists in lipid free form [9–11]. There is a wide range of interindividual variation in β2GPI plasma levels, ranging from immunologically undetectable to as high as 35 mg/dL with a mean value of 20 mg/dL in Caucasians and 15 mg/dL in African Americans [12], which may have clinical relevance in β2GPI -related pathways. Family and heritability data have provided strong support for the genetic basis of β2GPI plasma variation but the exact molecular basis of this variation remains largely unknown. β2GPI is suggested to regulate thrombin inactivation by heparin cofactor II [13] and thus variation in plasma β2GPI may affect prothrombic tendency in PAPs patients. Thus, it is important to determine the molecular basis of β2GPI plasma variation. Previously we have shown that two SNPs in coding regions (Cys306Gly, Trp316Ser) [12, 14] and one SNP in the promoter (−32 C > A) [15] region of APOH have significant impact on β2GPI plasma variation. Since then we have characterized complete DNA sequence variation in APOH and identified ~ 150 SNPs, including 13 SNPs and 1 deletion (−742delT) in the 5´- region [16].
Variations in the promoter DNA sequence may potentially alter the affinities of existing protein-DNA interactions or recruit new proteins to bind to the DNA, altering the specificity and kinetics of the transcriptional process. Given the importance of promoters in harboring functionally relevant SNPs that regulate gene expression and phenotypic variation, it is important to examine the role of promoter SNPs in relation to disease, gene expression and corresponding plasma levels. Recently we have reported associations of APOH promoter SNPs with SLE risk and carotid plaque formation in SLE patients [17].
The objective of this study was: 1) to characterize a ~ 1.4 kb (1,418 bp) genomic fragment in the 5-region of human APOH to identify the functional promoter; 2) to examine the impact of all 13 reported APOH promoter SNPs in Caucasians (−1284C>G, −1219G>A, −1190G>C, −759 A>G, −700C>A, −643T>C, −38G>A, and −32C>A) and African Americans (−1076G>A, −1055T>G, −627A>C, −581A>C and −363C>T) on APOH gene expression; 3) to determine the association of 8 promoter SNPs in Caucasians on β2GPI levels among U.S. Whites, and (4) to determine the cross-species conservation of the APOH promoter sequence.
Results
Identification and characterization of the APOH promoter region
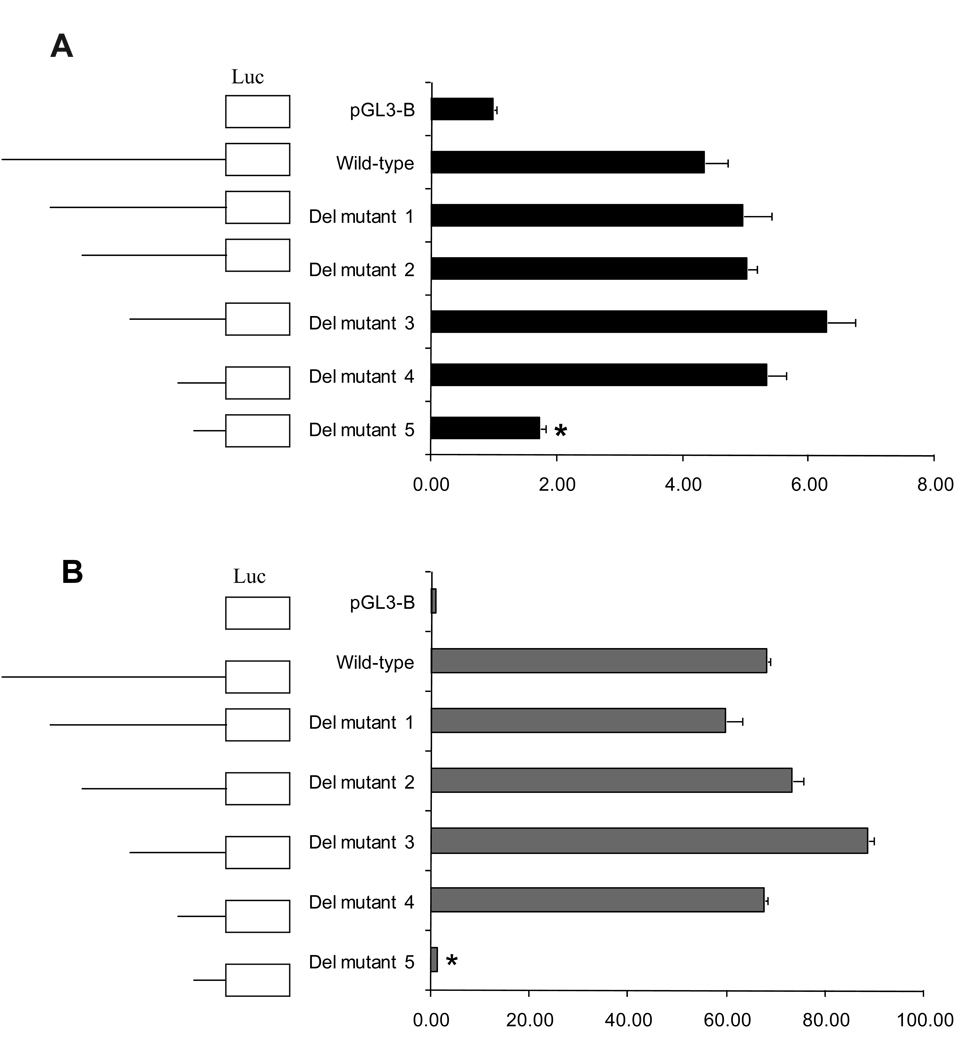
In order to localize the active promoter region and to identify regions that are important for regulation of the human APOH expression, the wild-type 1,418 bp 5'–flanking region of APOH was amplified from genomic DNA and used as template to create a series of five different deletion (del) constructs containing 5'- truncated fragments of APOH promoter fused upstream to a promoterless firefly luciferase (Luc) gene of the pGL3-Basic reporter vector. The sequence of each construct was verified by sequencing (data not shown). Figure 1A shows expression of deletion mutants in COS-1 cells. 5′ deletions of the promoter sequence to −815 (Del mutant 1, −815/+43), and −575 (Del mutant 2, −575/+43) increased promoter activity slightly compared to the wild-type, but the difference was not significant (wild-type vs. Del mutant 1; P = 0.260, wild-type vs. Del mutant 2; P = 0.135). Successive removal of nucleotides from −575 (Del mutant 2, −575/+43), to −325 (Del mutant 3, −325/+43), enhanced promoter activity appreciably (wild-type vs. Del mutant 3; P = 0.019), suggesting the possibility of negative regulatory elements within the −575/−325 regions. The Del mutant 3 construct (−325/+43) conferred maximum luciferase activity in COS-1 cells. A slight decrease in promoter activity was observed after further deletion of sequence from −325 to −166 (Del mutant 4, −166/+43; P = 0.04). However, when the sequence from −166 to −65 was removed (Del mutant 5, −65/+43), promoter activity dropped significantly (P < 0.001) compared to the wild-type. This suggests the presence of a critical element in the region extending between −166 to −65. We replicated the deletion analysis using human HepG2 cell line, since liver is a major site of synthesis of β2GPI and found an overall similar trend as seen in COS-1 cells with Del mutant 3 (−325/+43) showing the highest and Del mutant 5 (−65/+43) showing the lowest (P < 0.001) promoter activity (Figure 1B). A slight difference in trend was observed for the wild-type, mutant 1 (−815/+43), and mutant 2 (−575/+43) constructs, wherein mutant 1 was lower than the wild-type for HepG2, but not in COS-1 cells. Thus, using both COS-1 and HepG2 cell lines, we have identified the region ~166 bp upstream of the translation start site as the basal promoter of human APOH containing key cis-acting elements that regulate APOH expression.
Figure 1.
A. Dual-luciferase reporter gene expression of APOH promoter deletion mutants in COS – 1 cells. Left panel, schematic representation of 5'-deleted fragments of the APOH promoter in conjunction with the luciferase gene in pGL3-basic vector. The nucleotides are numbered from the translation start site (ATG). The effect of wild-type and mutants were measured as the mean of the firefly luciferase levels, which were normalized by the Renilla luciferase activity, which served as the reference for the transfection efficiency. The results presented are from one out of three independent experiments. pGL3-B indicates the promoterless vector. Asterisk (*) indicates that Del mutant 5 has significantly lower luciferase activity than the wild type (P<0.001).
B. Dual-luciferase reporter gene expression of APOH promoter deletion mutants in HepG2 cells. Left panel, schematic representation of 5'-deleted fragments of the APOH promoter in conjunction with the luciferase gene in pGL3-basic vector. The nucleotides are numbered from the translation start site. The effect of wild-type and mutants were measured as the mean of the firefly luciferase levels, which were normalized by the Renilla luciferase activity, which served as the reference for the transfection efficiency. The results presented are from one out of two independent experiments. pGL3-B indicates the promoterless vector. Asterisk (*) indicates that Del mutant 5 has significantly lower luciferase activity than the wild type (P<0.001).
Functional characterization of APOH promoter SNPs
In order to investigate the differential allele-specific effect on promoter activity, pGL3-Basic-APOH promoter constructs harboring individual point mutations for 12 out of 14 APOH promoter sequence variants identified earlier [16] (−1284C>G, −1219G>A, −1190G>C, −1076G>A, −1055T>G, −759A>G, −700C>A, −643T>C, −627A>C, −363C>T −38G>A, and −32C>A) were generated. The relative luciferase activity assessed in three independent experiments performed in triplicate for all the above APOH promoter SNPs is listed in Table 1. The insertion/deletion polymorphism (−742delT) could not be characterized due to repetitive sequences in the surrounding region. Similarly, the −581A>C mutant construct was not successful.
Table 1.
Dual luciferase results of each APOH promoter construct in COS−1 cells
| SNPs | Wildtype Allele (Mean ± SD) |
Variant Allele (Mean ± SD) |
% Decrease | P −value |
|---|---|---|---|---|
|
−1284C>G |
C 5.06 ± 0.10 5.27 ± 0.06 5.55 ± 0.46 |
G 4.16 ± 0.36 4.56 ± 0.34 4.64 ± 0.46 |
17.79 13.47 16.40 |
0.014 0.023 0.075 |
|
−1219G>A |
G 2.86 ± 0.05 4.10 ± 0.21 3.70 ± 0.12 |
A 1.40 ± 0.01 2.06 ± 0.16 1.81 ± 0.08 |
51.05 49.76 51.08 |
< 0.001 < 0.001 < 0.001 |
|
−1190G>C |
G 3.01 ± 0.19 2.79 ± 0.19 3.93 ± 0.50 |
C 2.16 ± 0.03 1.98 ± 0.23 2.84 ± 0.08 |
28.24 29.03 27.74 |
< 0.01 < 0.01 < 0.01 |
|
−1076G>A |
G 10.01 ± 0.38 10.86 ± 0.53 8.40 ± 0.47 |
A 9.13 ± 0.86 9.98 ± 0.60 7.74 ± 0.07 |
8.79 8.10 7.86 |
0.178 0.129 0.075 |
|
−1055T>G |
T 4.66 ± 0.18 7.66 ± 0.53 3.49 ± 0.09 |
G 3.44 ± 0.17 6.13 ± 0.04 2.53 ± 0.14 |
26.18 19.97 27.51 |
< 0.01 < 0.01 < 0.01 |
|
−759A>G |
A 5.28 ± 0.29 4.82 ± 0.27 4.90 ± 0.12 |
G 4.57 ± 0.11 4.27 ± 0.18 4.38 ± 0.50 |
13.45 11.41 10.61 |
0.017 0.042 0.155 |
|
−700C>A |
C 4.65 ± 0.05 4.90 ± 0.17 4.27 ± 1.32 |
A 4.31 ± 0.10 4.58 ± 0.33 3.99 ± 0.51 |
7.31 6.53 6.56 |
< 0.01 0.214 0.745 |
|
−643T>C |
T 19.91 ± 1.68 5.73 ± 0.07 10.79 ± 0.88 |
C 11.94 ± 0.15 3.20 ± 0.24 6.26 ± 0.39 |
40.03 44.15 41.98 |
0.001 < 0.001 0.002 |
|
−627A>C |
A 3.09 ± 0.15 6.72 ± 0.31 5.75 ± 0.23 |
C 2.85 ± 0.11 6.18 ± 0.12 5.12 ± 0.01 |
7.77 8.04 10.96 |
0.086 0.049 0.009 |
|
−363 C>T |
C 3.82 ± 0.34 2.96 ± 0.49 2.88 ± 0.16 |
T 3.34 ± 0.25 2.42 ± 0.40 2.44 ± 0.26 |
12.57 18.24 15.28 |
0.117 0.212 0.065 |
|
−38G>A |
G 4.56 ± 0.15 3.95 ± 0.20 3.81 ± 0.09 |
A 3.62 ± 0.15 3.21 ± 0.17 3.16 ± 0.03 |
20.61 18.73 17.06 |
0.002 0.009 < 0.001 |
|
−32C>A |
C 18.91 ± 0.38 15.79 ± 1.03 16.71 ± 0.92 |
A 11.92 ± 0.39 10.32 ± 0.17 10.56 ± 0.06 |
36.96 34.64 36.8 |
< 0.001 < 0.001 < 0.001 |
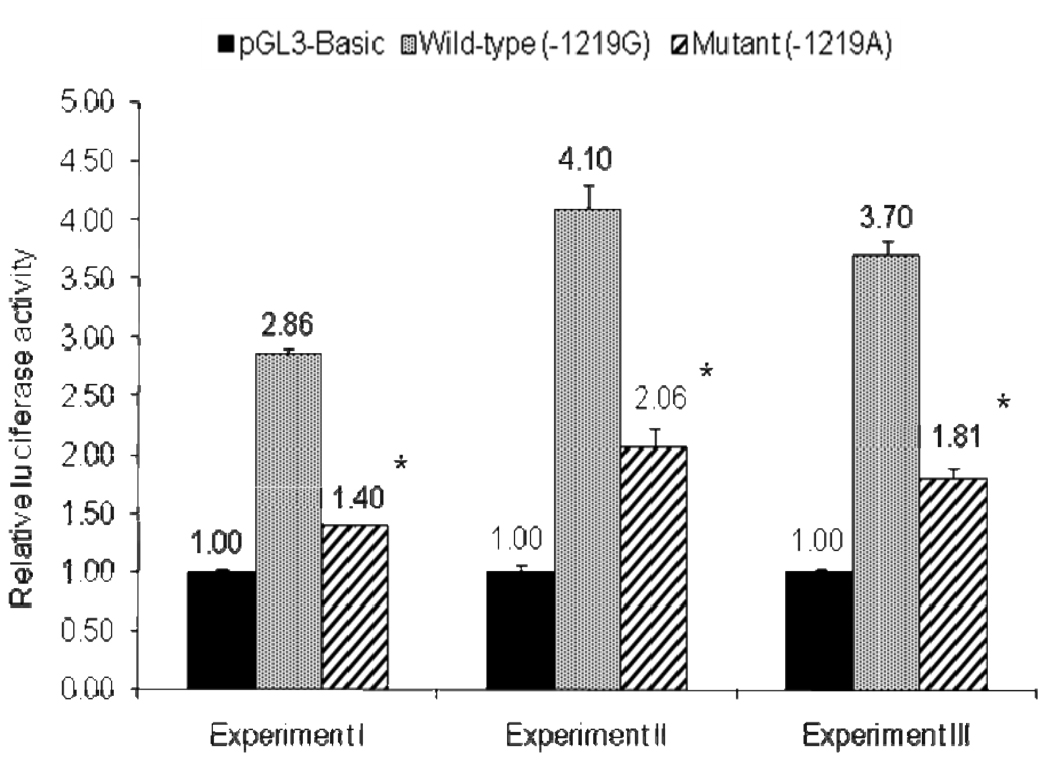
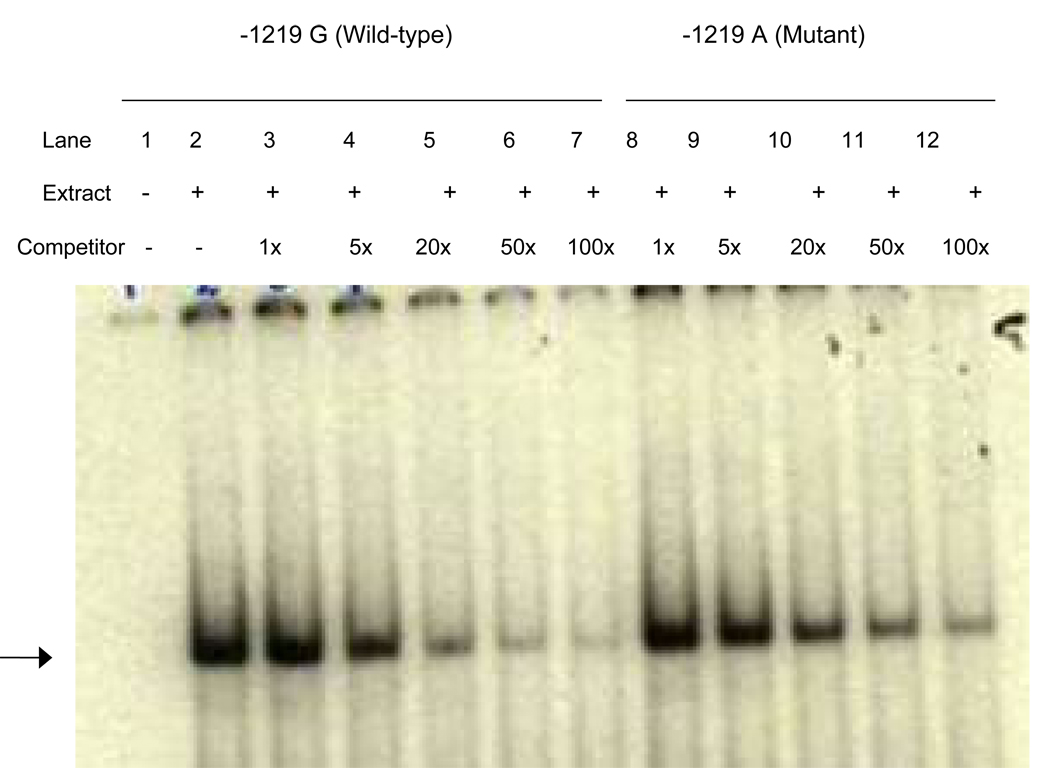
A total of three SNPs were found to be significantly associated with differential gene expression (36% or higher difference at P<0.001), including two previously reported −643T > C [17] and −32C > A [15]. An additional SNP, −1219G>A, showed a significant difference of ~ 51% in luciferase gene expression between wild-type and mutant alleles (Figure 2). EMSA was performed in order to determine whether the APOH promoter −1219G>A SNP affects the binding activity of nuclear factors. Upon incubation of radiolabeled oligonucleotides specific for wild-type (−1219G) and mutant (−1219A) alleles with HepG2 nuclear extracts, DNA-protein complexes were observed, indicating the presence of nuclear factor(s) (Figure 3). Competition assays using increasing amounts of unlabeled wild-type oligonucleotides confirmed the specificity of the binding.
Figure 2.
Dual-luciferase reporter gene expression of APOH promoter −1219G>A SNP. (* P < 0.0001). Results are shown for three independent experiments.
Figure 3.
EMSA result for −1219G>A polymorphism. Each sample contains a mixture of 5 µg of nuclear extract derived from human HepG2 cell nuclear extract and 30×mer 32P-labeled wild-type oligonucleotide containing G allele. Arrowhead indicates specific DNA-protein complex associated with the −1219G>A polymorphic site. Lane 1, labeled oligonucleotide without nuclear extract from HepG2 cells; 2, labeled oligonucleotide with nuclear extracts. Lanes 3 to 7 have increasing amounts of G oligo competitor (1×, 5×, 20×, 50×, 100×, respectively); lanes 8 to 12 have increasing amounts of A oligo competitor (1×, 5×, 20×, 50×, 100×, respectively).
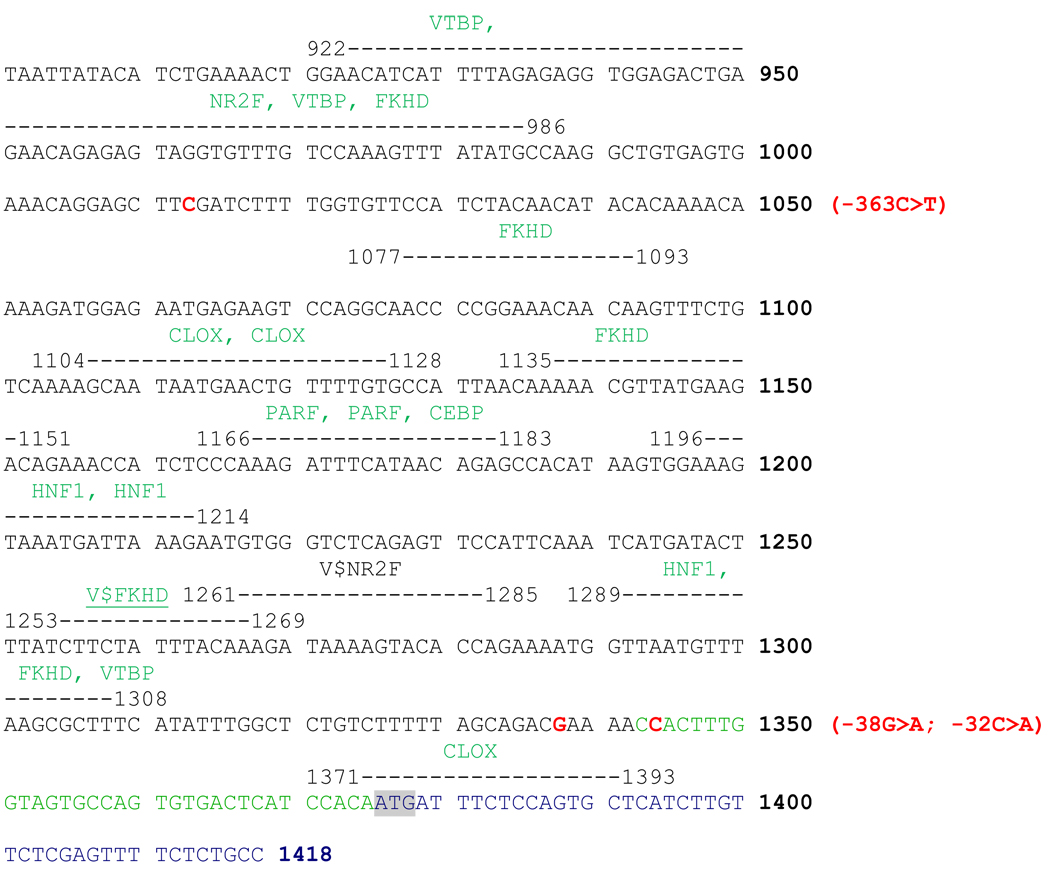
Potential liver-specific transcription factor-binding sites for the three promoter SNPs that showed differential gene expression (−1219 G > A, −643 T > C and −32 C > A) were sought by using MatInspector program from Genomatrix software (http://www.genomatix.de/index.html) [18], which matches by comparing DNA sequences with weighted matrix descriptions of functional binding sites, based on the TRANSFAC database (http://www.biobase.de). Figure 4 shows the locations of these three functional SNPs relative to potential binding sites along with all other SNPs detected in the 5’ flanking region. The list of all the predicted transcription factors, including their consensus sequences and specific binding sites is given in the Supplementary table S1. The program identified binding sites for the −1219G > A and −643T > C SNPs (Figure 4). While the binding site for HNF1 was observed adjacent to the −1219G > A SNP site, the −643T > C SNP region showed binding to CLOX and CLOX homology CCAAT displacement protein (CDP) factors. EMSA results previously reported by us [15] have revealed that the −32C > A SNP disrupts the binding of crude mouse hepatic nuclear extracts and purified TFIID, which is part of the RNA polymerase II preinitiation complex, indicating its functional role in the transcriptional regulation of APOH promoter. However, in silico analysis using MatInspector program for liver-specific factors did not identify any liver-specific transcription factor to bind to the region including the −32C > A SNP.
Figure 4.
MatInspector results for the liver-specific transcription factor binding sites of the APOH promoter. The transcription factors are shown in green along with the exact binding position marked by a dotted line and the APOH promoter SNPs are in red. The ATG start codon is highlighted in grey.
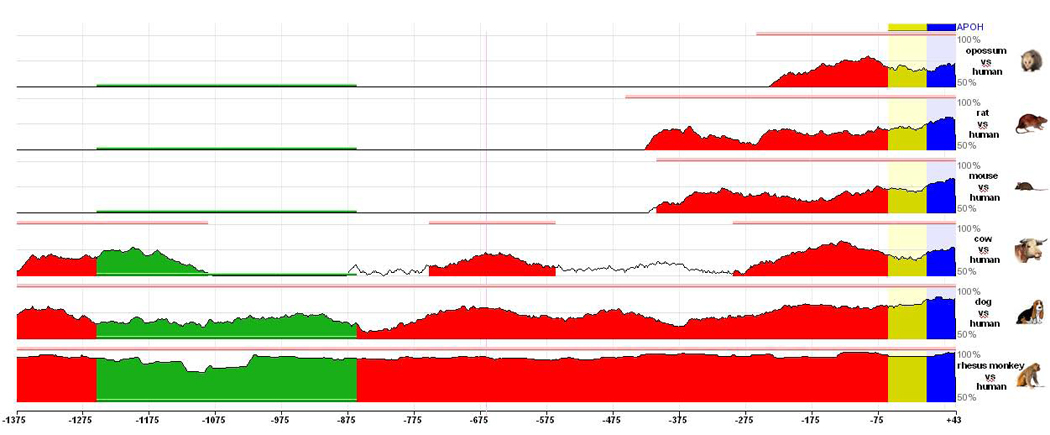
In order to determine the cross-species conservation of the APOH promoter sequence, we used the ECR Browser (http://ecrbrowser.dcode.org/) to visualize the conservation profile of the 5'-region of APOH (1,418 bp; −1375/+43 nucleotides from the translation initiation codon ATG) to identify the Evolutionary Conserved Regions (ECRs). Figure 5 shows the graphical display of the pairwise alignments and comparisons of sequences from 6 other species (monkey, dog, cow, mouse, rat, opossum) to that of human (base genome). Consistent with our deletion analyses, which indicated the presence of critical promoter elements in the region spanning between −166 to −65, the ECR extending from 5’-end of the gene (exon+UTR) to immediately upstream region (~250 bp upstream of the ATG start codon) was highly conserved across all 7 species.
Figure 5.
ECR Browser conservation profile of the 5'-region of APOH (1,418 bp; −1375/+43 nucleotides from the translation initiation codon ATG). Sequence elements of significant length (≥ 100 nucleotides) that are conserved above a certain level of sequence identity (≥ 65%) between the two compared genomes are highlighted as ECRs (pink rectangles at the top of the graphs). The horizontal axis represents positions in the base genome (human) and the vertical axis represents % identity between the base and aligned genomes (monkey, dog, cow, mouse, rat and opossum). The color-coding used by ECR Browser is: blue for coding exons, yellow for UTRs, red for intergenic regions, and green for transposable elements and simple repeats.
APOH promoter SNPs and plasma β2GPI levels
The distribution of plasma β2GPI levels showed only a modest difference (17.90 ± 4.15 mg/dl vs. 18.72 ± 4.68; P = 0.054) in mean plasma β2GPI levels between cases (n = 241) and controls (n = 206), therefore the association analyses were done using the combined case + control cohort data. Stepwise regression analysis revealed that age, BMI and ever smoking were the significant determinants of the plasma β2GPI levels. Only −32C>A SNP showed significant associations with the adjusted mean plasma β2GPI levels in both single-site (P < 0.001) and multiple regression (P < 0.001) analyses. Mean plasma β2GPI levels were higher in homozygotes of the wild-type allele, CC (mean = 18.62 mg/dL) compared to both the heterozygotes, CA (mean = 16.24 mg/dL) and homozygotes of less common allele, AA (mean = 13.90 mg/dL). Eight-site haplotype analysis including 6 APOH promoter SNPs (present in Whites) and 2 coding SNPs identified a total of 11 haplotypes with a frequency of > 1% (Table 2). Since data for plasma β2GPI levels was available only for the White population we excluded the five SNPs present in Blacks. Out of the eight SNPs present in Whites, −1284C>G SNP was excluded due to its rare presence (MAF< 0.01) and −700C>A SNP which is in high linkage disequilibrium to −759A>G as shown previously [17]. Three haplotypes (H5, H6, H10) showed significant association with plasma β2GPI levels (P < 0.001). The haplotype (H5) harbored minor alleles for the −1190G>C, −32C>A, and Trp316Ser SNPs. The other two significant haplotypes were predominantly defined by the minor alleles of the two coding polymorphisms, (H6: Cys306Gly; H10:Trp316Ser; respectively) that are already known to be major determinants of plasma β2GPI levels. Although the −32C>A SNP was significant in single-site analysis, the other haplotype (H7) defined by minor alleles only at −1190G>C and −32C>A SNPs and not for Trp316Ser did not show significant association, suggesting that the effect of the −1190G>C and −32C>A SNPs is dependent upon Trp316Ser polymorphism. None of the individual haplotypes harboring less common alleles for the −643T>C (H2 and H9) and −1219G>A SNPs (H4) that significantly decrease gene expression in vitro showed significant impact on plasma β2GPI levels. Three-site haplotype analysis (data not shown) with the functionally relevant (based on dual-luciferase and EMSA data) −1219G>A, −643T>C, and −32C>A SNPs were consistent with the individual SNP results. That is, only the haplotype carrying −32A was significantly associated with decreased plasma β2GPI levels (P < 0.001).
Table 2.
Haplotype analysis* of APOH SNPs for plasma β2GPI levels
| HAPLOTYPE^ | rs8178819 (−1219G>A) |
rs3760290 (−1190G>C) |
rs817820 (−759A>G) |
rs3760292 (−643T>C) |
(−38G>A) | rs8178822 (−32C>A) |
rs1801689 (Cys306Gly) |
rs1801690 (Trp316Ser) |
CASES + CONTROLS | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| freq | coef | se | P | |||||||||
| base haplotype | G | G | A | T | G | C | T | G | 0.384 | - | - | - |
| H1 | G | C | G | T | G | C | T | G | 0.156 | −0.218 | 0.408 | 0.539 |
| H2 | G | G | A | C** | G | C | T | G | 0.098 | 0.791 | 0.534 | 0.139 |
| H3 | G | C | A | T | G | C | T | G | 0.081 | 0.301 | 0.538 | 0.576 |
| H4 | A** | C | G | T | G | C | T | G | 0.062 | −0.046 | 0.675 | 0.946 |
| H5 | G | C | A | T | G | A**** | T | C*** | 0.042 | −4.632 | 0.737 | < 0.001 |
| H6 | G | G | A | T | G | C | G*** | G | 0.038 | −5.439 | 0.739 | < 0.001 |
| H7 | G | C | A | T | G | A**** | T | G | 0.023 | −1.271 | 1.031 | 0.218 |
| H8 | G | G | A | T | A | C | T | G | 0.017 | 0.280 | 1.128 | 0.804 |
| H9 | G | G | G | C** | G | C | T | G | 0.013 | −0.097 | 1.255 | 0.938 |
| H10 | G | C | A | T | G | C | T | C*** | 0.013 | −4.748 | 1.247 | < 0.001 |
| H11 | G | G | G | T | G | C | T | G | 0.011 | −0.727 | 1.453 | 0.617 |
| rare haplotype | - | - | - | - | - | - | - | - | 0.062 | −1.604 | - | 0.024 |
R software (haplo.stats package) for β2GPI plasma levels; p-values were calculated from coefficients (coef) and standard errors (se) Regression model included disease, age, BMI, ever smoking
Only the haplotypes with more than 0.01 total frequencies are shown
Alleles found to decrease gene expression in vitro
Alleles found to be significantly associated with low plasmaβ2GPI levels in univariate analysis
Alleles found to decrease gene expression in vitro and also associated with low plasmaβ2GPI levels in univariate analysis
Discussion
The goals of this study were (i) to clone and characterize a 1,418 bp fragment of the 5´- region of APOH, (ii) to functionally characterize the APOH promoter SNPs present in the 1,418 bp fragment, (iii) to examine the effect of the APOH promoter SNPs on plasma β2GPI levels, and (iv) to determine the cross-species conservation of the APOH promoter sequence.
To identify regions of the APOH promoter that affect its basal transcription, several 5' - promoter deletion mutants were linked to the luciferase reporter gene and assayed. Promoter constructs containing either −1375/+43 (wild-type) or −166/+43 (Del mutant 4) of upstream sequence had similar high levels of basal transcriptional activity when transfected into either COS-1 or HepG2 cell lines. These results indicates that all of the necessary machinery for driving basal APOH expression is localized in this −166/+43 sequence. Further deletion from −166 to −65 revealed regions within the APOH promoter that are important for its function. This deletion resulted in ~ 60% decrease in transcriptional activity in COS-1 cells and an even more pronounced (~98%) decrease in HepG2 cells, indicating the presence of an activator motif(s) within this sequence. These results are consistent with the previous deletion analysis [7] that identified the proximal promoter region necessary for hepatic-specific APOH expression. The smallest APOH 5´deletion mutant (−65/+43) used in this study differed from the prior study [7] as it lacked both the critical cis-elements (TATTA and HNF-1α) identified within this region, whereas the smallest deletion mutant used in the previous study [7] lacked only the TATTA element. Despite this difference, our study replicates the key findings in which the smallest 5´deletion mutant almost completely abolished luciferase activity by ~98% (present study) and ~91% (Wang and Chiang) [7] in HepG2 cells, emphasizing the vital role of the TATTA cis-element in APOH transcription. Our cross-species conservation analysis of APOH promoters from different species indicates that basic promoter elements are highly conserved across the 7 species examined.
About one third of promoter variants exert a functional effect on gene expression [19]. The functional importance of the APOH promoter SNPs was predicted by allelic differences in expression of the luciferase reporter gene. In this study we “functionally” validated SNPs in the APOH promoter based on two experimental approaches (reporter assays and EMSA). For this purpose, we tested 12 of the 14 sequence variants located within the 1,418 bp of the 5'-flanking region of APOH for allele-specific regulatory effects on expression of the dual-luciferase reporter gene and by EMSA for SNPs within transcription factor binding sites. Of the 12 SNPs examined, three SNPs at positions −1219G>A, −643T>C and −32C>A showed a significant decrease in luciferase expression (~50%, ~40% and ~36%, respectively) in COS-1 cells. The −32C>A SNP is a part of the core APOH promoter region (−166 bp upstream from ATG) identified in this study and has been previously shown to play a key role in the transcription initiation process by serving as a site for the binding of transcription factor II D (TFIID) [15]. Although 5´- serial deletion of APOH promoter identified the basal transcriptional activity restricted to the region ~160 bp upstream of ATG codon, it does not eliminate the possibility of the functional roles of the −643T>C and −1219G>A SNPs as part of the extended APOH promoter transcriptional machinery. To further substantiate the functional relevance of the three APOH promoter SNPs (−1219G>A, −643T>C, and −32C>A), EMSAs revealed strong in vitro protein binding for both wild-type and mutant type oligos for each SNP using nuclear extracts of HepG2 cells. However, no significant differential binding was observed for the two alleles for all SNPs. In silico analysis using MatInspector program for the prediction of liver-specific transcription factor binding sites revealed potential binding sites for the −1219G>A and −643T>C SNPs (Fig. 4). Binding of an important liver-enriched transcription factor, HNF1, was observed adjacent to the −1219 G > A polymorphic site, which could explain for the functional relevance of this SNP. HNF1 plays a prominent role in regulating genes that are expressed in hepatocytes [20]. The −643T>C SNP region binds to CLOX and CLOX homology CCAAT displacement protein (CDP) factors, that have been previously reported as transcriptional repressors [21]. This could probably explain the decrease in reporter gene expression observed by the mutant allele.
In addition to characterizing the basal APOH promoter and its functional variants, the effect of the APOH promoter SNPs on plasma β2GPI levels were examined for a subgroup of the Pittsburgh white population (SLE cases, n = 241; and controls, n = 206). In univariate analysis, only the previously reported −32C>A SNP showed a significant effect after adjustment for covariates. None of the other APOH promoter SNPs used in this study had a significant effect on plasma β2GPI levels. Our previous report [17] suggested a role for the −643T>C polymorphism protecting against carotid plaque formation in autoimmune-mediated atherosclerosis in SLE patients and the −1219G>A SNP showed a moderate effect on lupus nephritis. Functional role for the two SNPs was established using promoter gene assays and EMSA. Despite the functional effects of the −1219 G > A and −643 T > C SNPs on gene expression, their lack of association with plasma β2GPI levels is interesting. Although in vitro luciferase assays measuring promoter activity suggest that the two polymorphisms show an effect on gene expression, this may not entirely be a true reflection of the complexity of regulation that occurs in vivo. The regulation of human gene expression is a critical, highly coordinated, and complex process. The core promoter is generally within 50 bp of the transcription start site, where the preinitiation complex forms and the general transcription machinery assembles [22]. The extended promoter can contain specific regulatory sequences that control spatial and temporal expression of the downstream gene. The transcription machinery, which consists of interconnected co-regulatory protein complexes in a regulatory network, is responsible for mRNA synthesis from a given promoter. Control of gene regulation could occur at various stages, including level of transcription, post-transcriptional regulation, alternative splicing, translation, post-translational modifications and secretion of β2GPI, all of which may have an effect on the quantitative measure of plasma β2GPI levels. Alternatively, it is also possible that a change in promoter activity does not necessarily result in a quantitative change at the protein level. Whether the APOH promoter SNPs (−643T>C and −1219G>A) could influence the promoter activity by either the former or latter methods is beyond the scope of in vitro experiments. Further studies will be needed to explore the mechanism for these associations.
APOH promoter SNPs explain a small proportion of the variance in APOH expression, thus the ability of these SNPs to influence plasma β2GPI levels may be obscured by the strong effects of other factors (undefined promoter elements which are in strong LD with the promoter SNPs and other regulatory factors that affect in vivo gene expression) in aggregate. However, given the reporter gene expression data on promoter activity and EMSA results indicating possible binding to transcription factors, there is clearly a functional effect of the two polymorphisms on APOH regulation that are worthy of further investigation. However, haplotype analysis including APOH promoter SNPs alone or in conjunction with previously known coding SNPs affecting plasma β2GPI levels (Cys306Gly and Trp316Ser, Table 1) gave us no new insights into determining the genetic basis of plasma β2GPI levels. The significant haplotypes were defined predominantly by the minor alleles at the coding SNPs, which are already known to have a major effect on β2GPI levels. Consistent with the univariate data, none of the haplotypes defined by the minor alleles at APOH promoter SNPs reached significance. Although the −32C>A SNP was significant in the univariate analysis, the individual haplotype (H7) harboring the minor allele −32A was not significant, indicating that the effect of the −32C>A SNP is dependent upon the presence of the Trp316Ser coding SNP, which is in strong LD with the −32C>A SNP as shown in haplotype (H5). Three-site haplotype analysis with only the APOH promoter functionally relevant SNPs (−643T>C, −1219G>A and −32C>A) showed a highly significant effect for haplotype defined by the −32A allele and also a moderate effect for the −1219A allele. Another questionable mechanism for the lack of association of APOH promoter SNPs on plasma β2GPI levels in this study is the modified capture-ELISA method that was used to determine the plasma β2GPI levels, wherein, the analyzed antibodies could have been targeted against only a small number of the antigenic sites in β2GPI. Therefore, given both the method and also the small sample size, further studies are warranted in larger cohorts using improvised methods (antibody titers measured against other/additional β2GPI sites) that will help better to delineate the molecular basis of plasma β2GPI levels.
Materials and Methods
Construction of APOH promoter luciferase reporter gene vector (wild-type and individual mutant constructs)
A 1,418 bp fragment of the human APOH 5´- region (−1375/+43 nucleotides from the translation initiation codon ATG) containing the promoter and the first untranslated exon was polymerase chain reaction (PCR) amplified using forward (5'-TGGCAGCACACTCTTCTTAT-3') and reverse (5´- GTTCTCGAGTTTTCTCTGCC-3´) primers. This APOH promoter fragment was amplified from an individual who had wild-type alleles for all 13 SNPs (−1284C>G, −1219G>A, −1076G>A, −1055T>G, −1190G>C, −759 A>G, −700C>A, −643T>C, −627A>C, −581A>C, −363C>T, −38G>A, −32C>A) and no deletion at −742 site. The PCR condition consisted of denaturation at 95°C for two minutes, followed by 35 cycles of denaturing at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for one minute, before a final extension at 72°C for 10 minutes. The PCR-generated fragment was cloned into the pCR-2.1-TOPO vector (Invitrogen Corporation, Carlsbad, CA) using the supplier’s standard protocol. The size and orientation of the DNA insert was confirmed by restriction analysis (HindIII and SacI). The promoter fragment was then excised out of the TOPO vector using enzymes KpnI and EcoRV and ligated into the KpnI-SmaI restricted pGL3-Basic firefly luciferase (Luc) reporter plasmid and transformed into top 10 chemically competent cells (Invitrogen Corporation, Carlsbad, CA). Following transformation, the positive clones were confirmed by sequencing.
Constructs bearing mutant/minor alleles for each APOH promoter SNP were generated by PCR using the wild-type APOH promoter/luciferase report construct (~1.4 kb 5' region of APOH promoter inserted into the pGL3-Basic luciferase reporter vector) as template using the QuickChange II Site-directed Mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol.
Construction of APOH promoter deletion mutants
A series of 5´- deletion mutants of the ~ 1.4 bp APOH promoter fragment were subcloned into a new Luc reporter vector (pGL3-Basic). For this purpose, the original wild-type construct carrying the 1,418 bp APOH promoter fragment served as a parental template for designing PCR primers to amplify several truncated APOH promoter fragments. We designed five APOH deletion mutant constructs differing in ~ 200 bp between each fragment as follows:
APOH Deletion fragment 1 (APOH del FR #1): It is the largest (858 bp) of all 5 fragments. The position of this region with respect to the translational start site is +43 to −815.
APOH Deletion fragment 2 (APOH del FR #2): This fragment contains 618 bp. The location of this deletion mutant from the translational start site is +43 to −575.
APOH Deletion fragment 3 (APOH del FR #3): The third fragment (368 bp) position with respect to the translational start site is +43 to −325.
APOH Deletion fragment 4 (APOH del FR #4): The fourth fragment is further truncated to position −166 and is sized 209 bp.
APOH Deletion fragment 5 (APOH del FR #5): It is the smallest of all 5 fragments (109 bp). The position of this region with respect to the translational start site is +43 to −65.
Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi) was used to design PCR primers containing linker sites for the restriction enzymes - KpnI and BamHI at the 5´ and 3´ ends of each deleted fragment respectively. The PCR products were gel purified (Qiagen, Valencia, CA) and then digested with KpnI and BamHI restriction enzymes. The digested fragments were again gel purified. The promoter-less pGl3-Basic vector (Promega Corporation, Madison, WI) was digested with KpnI and BglII, gel purified and Calf Intestinal Alkaline Phosphatase (CIP) treated in order to prevent self ligation of the empty vector. The APOH-PCR DNA was then ligated to the gel purified and CIP-treated pGL3-Basic vector by T4 DNA ligase to generate the fusion vector construct carrying APOH-upstream truncated sequence fused to the in-frame luciferase reporter gene. The ligated product was then transformed into competent E. coli followed by screening of recombinant plasmids by colony PCR technique. The positive clones were further confirmed by restriction digestion and DNA sequencing.
Cell culture, transient transfection and dual-luciferase reporter gene assay
Dual-Luciferase reporter gene assays (Promega, Madison, WI) were performed to measure the in vitro promoter activity between wild-type and mutant constructs carrying minor allele at individual SNP sites for each of the APOH promoter SNPs. The wild-type and mutant APOH promoter constructs along with along the Renilla luciferase control vector (pRL-TK) (Promega, Madison, WI) were used to transiently co-transfect COS-1 (African green monkey kidney) from the American Type Culture Collection (ATCC CRL-1650, Rockville, MD) and HepG2 cells (Human hepatocellular liver carcinoma; ATCC HB-8065). COS-1 cells were cultured at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Gibco, Invitrogen) supplemented with 10% fetal bovine serum, 2mM glutamine, 100 IU/ml penicillin and 100 µg/ml streptomycin. HepG2 cells were grown in Eagle's minimal essential medium (EMEM, ATCC) supplemented with 10% fetal bovine serum, and penicillin/streptomycin. A day prior to transfection, 1.6 × 105 cells were seeded in each well of a 12-well plate with 1mL of antibiotic-free DMEM/EMEM media. Transfection was performed by Lipofectamine 2000 reagent (Invitrogen Corporation, Carlsbad, CA) as per manufacturer's instructions. After 48 hrs, the cells were lysed and assayed for light outputs using the dual-luciferase reporter system (Promega Corporation, Madison, WI). Firefly and Renilla luciferase were measured with either TD-20/20 Luminometer (Turner Design, Sunnyvale, CA) or the Tecan Infinite 200 plate reader (Tecan Trading, Switzerland) according to the manufacturer's instructions. The luciferase data (firefly/renilla) was normalized to the average activity of the promoter-less empty vector to yield data reflecting fold-activity increase over baseline levels for each APOH promoter construct. Triplicate wells for each transfection condition were assayed (intra-experiment variation), and three independent transfections were carried out (inter-experiment variation).
Electrophoretic mobility shift assay (EMSA)
EMSA was performed for the APOH promoter SNPs to analyze the binding of nuclear proteins from HepG2 nuclear extracts. To make double-stranded probes and competitors, equal amounts of complementary oligos (Sigma Genosys, TX and Operon Biotechnologies, AL) corresponding to the wild-type or mutant alleles for each APOH SNP were heated at 95°C for 5 min and then annealed for an hour at room temperature. The wild-type oligonucleotide was 5'-end-labeled with [γ-32P] ATP using T4 polynucleotide kinase (New England Biolabs, MA) and purified by the QIAquick Purification kit (Qiagen, Valencia, CA). To allow DNA-protein binding, the mixture of unlabeled and labeled oligos were incubated with 1 µL (5.68µg) of human HepG2 cell nuclear extracts for 20 minutes at room temperature in gel shift binding buffer (1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 10 mM TRIS-HCl pH 7.5, 20% glycerol). For the competition experiments, unlabeled competitor DNA was added in 1×, 5×, 20×, 50×, and 100× excess volumes of the labeled probe and was incubated with the HepG2 nuclear extract (Active Motif, CA) for 10 min before the addition of the labeled probe The DNA-protein complexes were then separated on 5% polyacrylamide gel at 120 volts for two hours, the gels were dried and exposed overnight for autoradiography on X-ray films. For setting up of EMSA experimental procedures, an earlier published positive shift assay for the APOH promoter SNP −32C>A was reproduced and used as a positive control.
Subjects
For genetic association of APOH promoter SNPs with plasma β2GPI levels, we genotyped 345 Caucasian women with SLE from the Pittsburgh Lupus Registry and 454 Caucasian healthy control women from the Central Blood Bank of Pittsburgh by Pyrosequencing. Details regarding the phenotypic characteristics of this lupus case-control cohort along with genetic screening have been published elsewhere [17]. Plasma β2GPI levels were determined by the modified capture-ELISA method as described previously [12]. Data for plasma β2GPI levels were available only for a subgroup of Caucasian SLE cases (n = 241) and controls (n = 206). This study was approved by the University of Pittsburgh Institutional Review Board and all participants provided written informed consent.
Statistical analysis
All computations were performed using the R statistical software package (version 2.3.1, http://www.r-project.org). The haplotype analysis was performed using Haploview programs to check for individual haplotype associations with plasma β2GPI levels. Age, BMI, ever smoking and case-control status were used as covariates. A P- value of less than 0.05 was considered as suggestive evidence of association. Student’s t-test was used to determine the significance of reporter gene expression difference between the wild-type and mutant constructs.
Supplementary Material
Acknowledgement
This study was supported by the National Heart, Lung, and Blood Institute Grant HL 54900.
Footnotes
Supporting information
Table S1 List of liver-specific transcription factors for APOH promoter (Matinspector)
References
- 1.Galli M, Comfurius P, Maassen C, Hemker HC, de Baets MH, van Breda-Vriesman PJ, Barbui T, Zwaal RF, Bevers EM. Anticardiolipin antibodies (ACA) directed not to cardiolipin but to a plasma protein cofactor. Lancet. 1990;335:1544–1547. doi: 10.1016/0140-6736(90)91374-j. [DOI] [PubMed] [Google Scholar]
- 2.McNeil HP, Simpson RJ, Chesterman CN, Krilis SA. Anti-phospholipid antibodies are directed against a complex antigen that includes a lipid-binding inhibitor of coagulation: beta 2-glycoprotein I (apolipoprotein H) Proc Natl Acad Sci U S A. 1990;87:4120–4124. doi: 10.1073/pnas.87.11.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okkels H, Rasmussen TE, Sanghera DK, Kamboh MI, Kristensen T. Structure of the human beta2-glycoprotein I (apolipoprotein H) gene. Eur J Biochem. 1999;259:435–440. doi: 10.1046/j.1432-1327.1999.00063.x. [DOI] [PubMed] [Google Scholar]
- 4.Sheng Y, Sali A, Herzog H, Lahnstein J, Krilis SA. Site-directed mutagenesis of recombinant human beta 2-glycoprotein I identifies a cluster of lysine residues that are critical for phospholipid binding and anti-cardiolipin antibody activity. J Immunol. 1996;157:3744–3751. [PubMed] [Google Scholar]
- 5.Sanghera DK, Wagenknecht DR, McIntyre JA, Kamboh MI. Identification of structural mutations in the fifth domain of apolipoprotein H (beta 2-glycoprotein I) which affect phospholipid binding. Hum Mol Genet. 1997;6:311–316. doi: 10.1093/hmg/6.2.311. [DOI] [PubMed] [Google Scholar]
- 6.Mehdi H, Naqvi A, Kamboh MI. A hydrophobic sequence at position 313–316 (Leu-Ala-Phe-Trp) in the fifth domain of apolipoprotein H (beta2-glycoprotein I) is crucial for cardiolipin binding. Eur J Biochem. 2000;267:1770–1776. doi: 10.1046/j.1432-1327.2000.01174.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang HH, Chiang AN. Cloning and characterization of the human beta2-glycoprotein I (beta2-GPI) gene promoter: roles of the atypical TATA box and hepatic nuclear factor-1alpha in regulating beta2-GPI promoter activity. Biochem J. 2004;380:455–463. doi: 10.1042/BJ20031610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Averna M, Paravizzini G, Marino G, Lanteri E, Cavera G, Barbagallo CM, Petralia S, Cavallaro S, Magro G, Grasso S, Notarbartolo A, Travali S. Liver is not the unique site of synthesis of beta 2-glycoprotein I (apolipoprotein H): evidence for an intestinal localization. Int J Clin Lab Res. 1997;27:207–212. doi: 10.1007/BF02912460. [DOI] [PubMed] [Google Scholar]
- 9.Polz E, Kostner GM. The binding of beta 2-glycoprotein-I to human serum lipoproteins: distribution among density fractions. FEBS Lett. 1979;102:183–186. doi: 10.1016/0014-5793(79)80955-2. [DOI] [PubMed] [Google Scholar]
- 10.Gambino R, Ruiu G, Pagano G, Cassader M. The binding of apolipoprotein H (beta2-glycoprotein I) to lipoproteins. Prostaglandins Other Lipid Mediat. 1999;57:351–359. doi: 10.1016/s0090-6980(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 11.Ağar C, de Groot PG, Levels JH, Marquart JA, Meijers JC. Beta2-glycoprotein I is incorrectly named apolipoprotein H. J Thromb Haemost. 2009;7:235–236. doi: 10.1111/j.1538-7836.2008.03223.x. [DOI] [PubMed] [Google Scholar]
- 12.Kamboh MI, Manzi S, Mehdi H, Fitzgerald S, Sanghera DK, Kuller LH, Atson CE. Genetic variation in apolipoprotein H (beta2-glycoprotein I) affects the occurrence of antiphospholipid antibodies and apolipoprotein H concentrations in systemic lupus erythematosus. Lupus. 1999;8:742–750. doi: 10.1191/096120399678840909. [DOI] [PubMed] [Google Scholar]
- 13.Rahgozar S, Giannakopoulos B, Yan X, Wei J, Cheng Qi J, Gemmell R, Krilis SA. Beta2-glycoprotein I protects thrombin from inhibition by heparin cofactor II:potentiation of this effect in the presence of anti-beta2-glycoprotein I autoantibodies. Arthritis Rheum. 2008;58:1146–1155. doi: 10.1002/art.23387. [DOI] [PubMed] [Google Scholar]
- 14.Mehdi H, Aston CE, Sanghera DK, Hamman RF, Kamboh MI. Genetic variation in the apolipoprotein H (beta2-glycoprotein I) gene affects plasma apolipoprotein H concentrations. Hum Genet. 1999;105:63–71. doi: 10.1007/s004399900089. [DOI] [PubMed] [Google Scholar]
- 15.Mehdi H, Manzi S, Desai P, Chen Q, Nestlerode C, Bontempo F, Strom SC, Zarnegar R, Kamboh MI. A functional polymorphism at the transcriptional initiation site in beta2-glycoprotein I (apolipoprotein H) associated with reduced gene expression and lower plasma levels of beta2-glycoprotein I. Eur J Biochem. 2003;270:230–238. doi: 10.1046/j.1432-1033.2003.03379.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q, Kamboh MI. Complete DNA sequence variation in the apolipoprotein H (beta-glycoprotein I) gene and identification of informative SNPs. Ann Hum Genet. 2006;70:1–11. doi: 10.1111/j.1529-8817.2005.00211.x. [DOI] [PubMed] [Google Scholar]
- 17.Suresh S, Demirci FY, Jacobs E, Kao AH, Rhew EY, Sanghera DK, Selzer F, Sutton-Tyrrell K, McPherson D, Bontempo FA, Kammerer CM, Ramsey-Goldman R, Manzi S, Kamboh MI. Apolipoprotein H promoter polymorphisms in relation to lupus and lupus-related phenotypes. J Rheumatol. 2009;36:315–322. doi: 10.3899/jrheum.080482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoogendoorn B, Coleman SL, Guy CA, Smith K, Bowen T, Buckland PR, O'Donovan MC. Functional analysis of human promoter polymorphisms. Hum Mol Genet. 2003;12:2249–2254. doi: 10.1093/hmg/ddg246. [DOI] [PubMed] [Google Scholar]
- 20.Noda C, Ichihara A. Regulation of liver-specific gene expression. Cell Struct Funct. 1993;18:189–194. doi: 10.1247/csf.18.189. [DOI] [PubMed] [Google Scholar]
- 21.Pattison S, Skalnik DG, Roman A. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5' end of the human papillomavirus type 6 long control region. J Virol. 1997;71:2013–2022. doi: 10.1128/jvi.71.3.2013-2022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.