Abstract
Restriction factors are natural cellular proteins that defend individual cells from viral infection. These factors include the APOBEC3 family of DNA cytidine deaminases, which restrict the infectivity of HIV-1 by hypermutating viral cDNA and inhibiting reverse transcription and integration. HIV-1 thwarts this restriction activity through its accessory protein virion infectivity factor (Vif), which uses multiple mechanisms to prevent APOBEC3 proteins such as APOBEC3G and APOBEC3F from entering viral particles. Here, we review the basic biology of the interactions between human APOBEC3 proteins and HIV-1 Vif. We also summarize, for the first time, current clinical data on the in vivo effects of APOBEC3 proteins and survey strategies and progress toward developing therapeutics aimed at the APOBEC3-Vif axis.
Keywords: APOBEC3, APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3DE, APOBEC3F, APOBEC3G, APOBEC3H, cytidine, DNA cytidine deaminase, HIV, human immunodeficiency virus, hypermutation, uridine, Vif
Introduction
The past decade has seen an explosion of knowledge on the ability of human innate restriction factors, including the APOBEC3 family of proteins, to counteract infection by human immunodeficiency virus type 1 (HIV-1). All seven APOBEC3 family members have been found to have varying levels of activity against HIV-1 under different experimental conditions (Table 1). This antiretroviral activity is due primarily to the ability of APOBEC3 proteins to deaminate viral cDNA cytidines to uridines and inhibit viral reverse transcription and integration (Ref. 1, 2).
Table 1.
Restriction Activity, susceptibility to Vif and mutational context preferences of APOBEC3 proteins with respect to HIV-1
| Restriction Activity Against HIV-1a | Susceptibility to Vifb | Dinucleotide Preferencec | |
|---|---|---|---|
| A3A |
Active (Ref. 151, 153, 154, 155) Note that two of these references involve fusion of A3A to a helper protein as discussed in the main text. |
Vif-Sensitive (Ref. 154, 155) |
GG; substantial GA (Ref. 151) |
|
Not Active (Ref. 46, 78, 80, 156, 157, 158, 159, 160) |
Not Vif-Sensitive (Ref. 154) |
||
| A3B |
Active (Ref. 46, 76, 78, 155, 157, 160, 161, 162, 163) |
Vif-Sensitive None |
GA (Ref. 78, 162) |
|
Not Active (Ref. 22) Haché et al. 2008 is the lone attempt to detect A3B antiviral activity in a spreading infection system. All references detecting activity represent single-cycle infectivity assays. |
Not Vif-Sensitive (Ref. 76, 78, 155, 162) |
||
| A3C |
Active (Ref. 46, 76, 78, 154, 157, 160, 164) |
Vif-Sensitive (Ref. 122, 131, 132, 154, 164) |
GA; substantial GG Nonspecific compared to others (Ref. 76, 164) |
|
Not Active (Ref. 80, 81, 155, 162) |
Not Vif-Sensitive (Ref. 76, 78) |
||
| A3DE |
Active (Ref. 122, 132, 160, 165, 166) |
Vif-Sensitive (Ref. 122, 132, 165, 166) |
GT; substantial GA (Ref. 166) |
|
Not Active (Ref. 22) Haché et al. 2008 is the lone attempt to detect A3DE antiviral activity in a spreading infection system. All references detecting activity represent single-cycle infectivity assays. |
Not Vif-Sensitive None |
||
| A3F |
Active (Ref. 78, 79, 80, 81) Many others. Restriction by A3F is generally well-accepted, and A3F is often used as a control with A3G. |
Vif-Sensitive (Ref. 78, 79, 80, 81) |
GA (Ref. 78, 79, 80, 81) |
|
Not Active None, but spreading infection systems may show variable strength of inhibition, from strong (Ref. 22) to mixed (Ref. 167). |
Not Vif-Sensitive None, although it should be noted that A3F may be generally less responsive to Vif than A3G, [e.g. (Ref. 79)]. |
||
| A3G |
Active As indicated in main text plus countless reports since. |
Vif-Sensitive As indicated in main text. |
GG (Ref. 26, 27, 28, 77, 79) |
|
Not Active None |
Not Vif-Sensitive None, but see A3G D128K in the main text. |
||
| A3H |
Active (Ref. 73, 74, 75, 165, 168) |
Vif-Sensitive (Ref. 73, 75) [or partially sensitive (Ref. 168)] |
GA (Ref. 74) |
|
Not Active (Ref. 109, 160, 166) Multiple alleles and splice variants exist in the human population, not all of which are active or equally impacted by Vif, as briefly discussed in main text. |
Not Vif-Sensitive (Ref. 74, 165) |
||
Single-cycle activity here is defined as the suppression of infectivity relative to negative restriction controls of at least 50%. Spreading infection activity is defined as the substantial suppression of viral spread for the duration of the experiment shown. Some experiments involved the modification of APOBEC3 proteins by, for example, fusion to a helper protein. Where this occurs, restriction is counted as if the protein were wild-type.
Any data concerning the Vif sensitivity of modified APOBEC3 proteins as described above is disregarded in this table.
References for the dinucleotide preferences of APOBEC3 mutational activity are preferentially chosen from papers with data on mutation of the HIV-1 genome. Studies on other substrates generally agree with those listed here, but variation among papers may occur.
HIV-1 averts restriction by APOBEC3 proteins through the action of its accessory protein virion infectivity factor (Vif), which decreases the encapsidation of certain APOBEC3 proteins [Table 1; (Ref. 3)]. It is not entirely clear, however, to what extent Vif is successful in protecting the integrity of the viral genome. Multiple lines of evidence suggest, in fact, that the neutralization of APOBEC3 proteins by Vif is not absolute. Thus, APOBEC3 proteins may be partially effective at restricting HIV-1 infectivity in vivo despite the presence of Vif. Supporting this view, the effects of hypermutation on the viral genome are readily apparent in patient viral sequences. Recent evidence additionally suggests this mutational activity may have the further undesired effect of providing HIV-1 with a reservoir of genetic diversity that contributes to viral immune evasion and drug resistance.
Here, we briefly review what is known about the basic biology of APOBEC3-Vif interactions. We then summarize current clinical data on correlations between APOBEC3 function and the natural history of HIV-1 infection to ascertain what may occur when HIV-1 and APOBEC3 proteins encounter each other in vivo. Finally, we discuss strategies to harness the restrictive capacity of APOBEC3 proteins to successfully treat HIV-1 infection.
HIV-1 Vif is a viral counterdefense against restriction by APOBEC3 proteins
HIV-1 Vif is required for growth on certain cell types and in vivo
Lentiviruses are a genus within the broader family of retroviruses. In addition to the ubiquitous gag, pol and env genes, lentiviruses possess several accessory genes that have integral roles in viral pathogenesis [recently reviewed by (Ref. 3, 4)]. The prototypical lentivirus, HIV-1, contains two accessory genes involved in the regulation of viral gene expression and four additional accessory genes more directly involved in pathogenesis, one of which is the virion infectivity factor (vif).
The vif gene is located in the center of the HIV-1 genome, overlapping the 3′ end of pol and the 5′ end of the accessory gene vpr. Expressed from a partially spliced, Rev-dependent subgenomic mRNA (Ref. 5, 6), the protein product is 192 residues, highly basic and 23 kDa in size (Ref. 7, 8, 9). Vif is required for productive infection in vivo and for infection of primary CD4+ T cells, monocytes, and macrophages ex vivo (Ref. 10, 11, 12, 13). This requirement, however, is variable in some human T cell lines (Ref. 14), allowing these lines to be grouped as “permissive” for the replication of Vif-deficient HIV-1 (e.g. CEM-SS, SupT1) or “nonpermissive” (e.g. CEM, H9).
When produced in permissive cells, virions from Vif-deficient HIV-1 have no quantitative or qualitative defects relative to wild-type virus except for the absence of the Vif protein itself. (Ref. 11, 14). When produced in nonpermissive cells, however, Vif-deficient HIV-1 virions rarely complete reverse transcription (Ref. 15, 16, 17, 18).
Given the above, two scenarios were considered likely to explain the phenotype of Vif-deficient viruses on nonpermissive cells. 1) Permissive cells contain an endogenous factor that complements the function lacking in a Vif-deficient virus. 2) Nonpermissive cells contain a Vif-suppressible factor with antiviral restriction activity. To determine which hypothesis was correct, two groups tested the infectivity of virions produced from heterokaryons formed by the fusion of permissive and nonpermissive cells (Ref. 19, 20). Under these conditions, the production of infectious virions would support the former hypothesis, while the production of noninfectious virions would support the latter. In fact, such heterokaryons formed from the fusion of permissive and nonpermissive cells produce virions with diminished infectivity, indicating that nonpermissive cells contain a Vif-sensitive dominant restriction factor.
APOBEC3G is a Vif-sensitive restrictor of HIV-1 infectivity with DNA cytidine deaminase activity
To identify the cellular factor suggested by cell fusion experiments, one group sought to isolate genes uniquely expressed in a nonpermissive cell line called CEM (Ref. 21). These authors used a technique called suppression subtractive hybridization, which simultaneously suppresses the recovery of transcripts common to CEM and its permissive derivative CEM-SS and enriches the recovery of transcripts that are more abundant in CEM. Among many sequences, a cDNA encoding APOBEC3G (A3G) was isolated, and engineering naturally permissive CEM-SS cells to express A3G rendered these cells nonpermissive for the replication of Vif-deficient but not wild-type virus. A3G therefore accounts for the nonpermissive phenotype of CEM, although a number of studies since then suggest that other APOBEC3 proteins may also contribute to the nonpermissive phenotype (Table 1). Of particular note, passage of Vif-deficient HIV-1 on permissive T cells stably expressing A3G yields viral variants that grow efficiently in the presence of A3G but not in CEM cells (Ref. 22), implying that other factors beyond A3G are also capable of restricting HIV-1.
At approximately the same time, two other groups independently identified A3G, not for its antiviral activity, but rather for its enzymatic activity, which provided a crucial clue about the mechanism by which A3G might restrict HIV-1. One identified A3G as part of a family of polynucleotide cytidine deaminases related to the mRNA editing enzyme APOBEC11, which edits the apolipoprotein B mRNA (Ref. 23). The other hypothesized, based on homology between APOBEC1 and the DNA mutator activation-induced deaminase (AID), which is involved in antibody diversification, that APOBEC1 and its homologs can act on DNA substrates. Indeed, APOBEC1, A3G, A3C and AID were all shown to mutate DNA (Ref. 24, 25).
HIV-1 restriction by A3G is mediated by deamination of viral cDNA cytidine during reverse transcription
Soon after the serendipitous convergence of data showing A3G’s capacity to restrict HIV-1 and deaminate DNA cytidines, several groups showed that the major basis for retroviral restriction is the deamination of minus-strand viral cDNA during reverse transcription (Ref. 26, 27, 28). The proviral DNA of a Vif-deficient virus grown in the presence of A3G becomes heavily mutated with characteristic genomic plus-strand G-to-A transitions. This strand-bias is only explained by deamination of the minus-strand reverse transcript, which after deamination from C-to-U templates A rather than G on the viral plus-strand [Fig. 1 (Ref. 26, 27, 28, 29)]. Furthermore, some catalytically defective A3G mutants are unable to as effectively reproduce the nonpermissive phenotype, which represents important additional evidence for the dependence of restriction on deaminase activity (Ref. 27, 28, 30, 31, 32, 33).
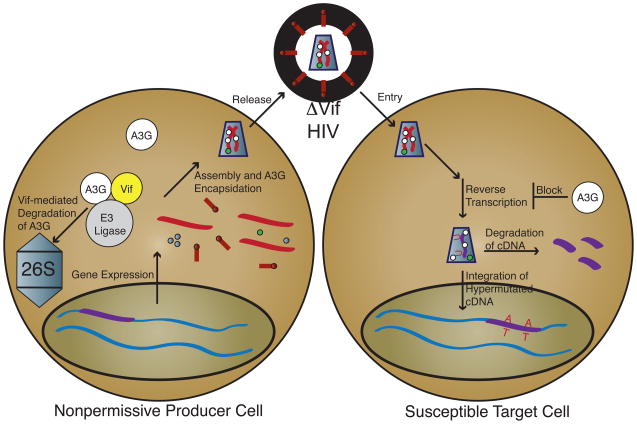
Fig. 1. APOBEC3 and Vif function.
Viral gene expression proceeds normally. During assembly, however, A3G present in a nonpermissive producer cell (white circles) is encapsidated along with normal virion components. To prevent encapsidation, Vif (yellow circle) expressed from a Vif-proficient virus links A3G to an E3 ligase complex (gray circle), which results in the ubiquitination of A3G and its eventual degradation in the proteasome (26S gray capped cylinder). Alternatively, Vif may directly inhibit encapsidation, lower A3G expression and/or inhibit A3G catalytic activity (not shown). Virions with encapsidated A3G bud from a producer cell and enter a target cell normally. Reverse transcription, however, is blocked by the presence of A3G, either by the direct inhibition of cDNA synthesis or the degradation of uridine-containing cDNA. Uridine-containing cDNA templates A on the viral plus-strand, resulting in G-to-A hypermutation. A direct A3G-mediated block to integration has also been proposed (not shown).
Although hypermutation is clearly a crucial part of the retrovirus restriction mechanism, evidence also exists that the catalytic activity of A3G may not wholly explain its ability to restrict HIV-1 (Ref. 31, 34, 35, 36, 37, 38, 39, 40, 41, 42). These studies suggest deaminase-independent effects on various specific steps in reverse transcription or integration (Fig. 1), although at least one report finds some of these same effects to be dependent on catalytic activity (Ref. 43). Although a substantial quantity of evidence suggests deaminase-independent restriction by A3G, the sheer diversity of the steps at which A3G is proposed to exert its deaminase-independent effects makes it highly improbable that all of these phenomena are physiologic. In fact, several groups have suggested that the deaminase-independent effects of A3G may represent overexpression artifacts, as stable expression of physiologic levels of catalytically inactive A3G has little if any effect on HIV-1 (Ref. 30, 32, 44). Thus, it appears that deaminase activity is largely required for the inhibitory activity of A3G under physiologic conditions. Whether this will also be the case for other APOBEC3 proteins, which may have more pronounced deaminase-independent effects (Ref. 31), remains to be seen. Moreover, it is important to note that deaminase-independent mechanisms may be more central to the restriction of many other retroelements including hepatitis B virus and LINE-1 [(Ref. 45, 46, 47, 48) and reviewed recently by (Ref. 49, 50)].
It was proposed early on that the effect of A3G-mediated deamination may be to shunt mutated viral cDNA into a base excision repair pathway that would ultimately result in the degradation of the viral genome (Ref. 26, 51). Under this model, a uracil DNA glycosylase such as UNG2 would recognize and excise cDNA uracils, leaving an abasic site at which an endonuclease such as APE1 would cleave the cDNA backbone. While one report supports such a scenario (Ref. 52), different data sets focusing on uracil DNA glycosylase functions imply either reparative (Ref. 53, 54, 55) or degradative (Ref. 56) effects on the viral genome. Alternatively, uracil DNA glycosylase activity may simply be irrelevant to the viral lifecycle (Ref. 33, 57). Further work will be required to elucidate the potential mechanisms by which viral cDNA is processed following APOBEC3-mediated deamination.
HIV-1 Vif preserves viral infectivity by preventing A3G encapsidation
Many mechanisms are plausible to explain how HIV-1 Vif counteracts the restrictive capacity of A3G. Most authors agree that this occurs by inhibiting A3G packaging, although the details of the mechanism(s) by which this inhibition occurs is debatable. At least part of the mechanism by which Vif overcomes restriction by A3G involves the Vif-mediated degradation of A3G through a proteasome-dependent pathway (Ref. 58, 59, 60, 61, 62, 63). Specifically, Vif binds both to A3G and to an E3 ubiquitin ligase complex consisting of Cullin5, Rbx2 and Elongins B and C. By bringing A3G into proximity with the ubiquitin ligase complex, Vif facilitates the ubiquitination and eventual degradation of A3G through the proteasome [Fig. 1; (Ref. 62, 64, 65)].
Evidence also exists suggesting that Vif can directly inhibit A3G encapsidation (Ref. 60, 66, 67). Such a mechanism need not require degradation as an end point. Other reports indicate a Vif-dependent reduction in A3G expression (Ref. 61, 66), and it is also possible that Vif may function by directly inhibiting the catalytic activity of A3G [Fig. 1; (Ref. 68)].
The mechanisms proposed above are not necessarily mutually exclusive. While details may differ, an overarching message is readily apparent in the current Vif literature: preventing the encapsidation of A3G is crucial for the ability of Vif to preserve the infectivity of HIV-1. In fact, reducing the encapsidation of A3G by simply increasing particle production to titrate out the restriction factor is at least one mechanism by which HIV-1 can resist restriction in the absence of Vif function (Ref. 22).
Evidence for APOBEC3 function in vivo and clinical implications
Multiple APOBEC3 family proteins have activity against HIV-1
While discussion to this point has focused on a basic description of the interactions between Vif and A3G, there are, in fact, six additional proteins in the APOBEC3 family – APOBEC3A (A3A), APOBEC3B (A3B), APOBEC3C (A3C), APOBEC3DE (A3DE), APOBEC3F (A3F) and APOBEC3H (A3H) (Ref. 2, 23, 69, 70) – all of which have been implicated in the mutation and restriction of HIV-1 under certain conditions. A summary of references with data concerning the restriction activity and Vif sensitivity of each APOBEC3 protein is provided in Table 1; current in-depth reviews of the differential properties of these APOBEC3 proteins may be found elsewhere [e.g. (Ref. 1, 71)].
While the results apparent in Table 1 do reflect a lack of consensus on the ability of certain APOBEC3 proteins to function against HIV-1, there are areas of agreement. Most notably, it is well-accepted that A3F and, more prominently, A3G have strong activity against HIV-1. Moreover, there is a second tier consisting of A3B and A3DE where antiviral activity is generally reproducible in overexpression single-cycle infectivity assays but may not be apparent in a more physiologic spreading infection system. Further study of these proteins, particularly in spreading infections, will be required to definitively determine their ability to act as dominant restriction factors. It is generally accepted, however, that A3G, A3F, A3DE and perhaps A3H (see below) are Vif-sensitive, while A3B is not, which implies that these Vif-sensitive factors are likely the most relevant for the restriction of HIV-1 in vivo. That said, the fact that A3B expression increases significantly in lymphoid tissue from acutely infected patients along with A3F and A3G (Ref. 72) may also suggest a role for this Vif-insensitive protein.
A3H represents a special case since only one known allele appears to be stable in human cells, and stability is required for its ability to effectively restrict HIV-1 (Ref. 73, 74, 75). It is also presently unclear to what extent this APOBEC3 protein is Vif-sensitive (Table 1), although the fact that some have found it sensitive to Vif suggests that it may be important for the restriction of HIV-1. Perhaps even more muddled is the case of A3C. A3C may be sensitive to Vif, but its restriction activity against HIV-1 is relatively weak, arguing against a role for this APOBEC3 protein in vivo. It should be noted, however, that A3C is quite active against simian immunodeficiency virus (Ref. 76). While some case can thus be made for the ability of six of seven human APOBEC3 proteins to dominantly restrict HIV-1, there is little data to support such a role for A3A, as the references showing the strongest effects require the fusion of A3A to a helper protein that shuttles it into the viral core (See discussion below).
Beyond restriction and Vif sensitivity phenotypes, we now wish to emphasize one key characteristic that helps to distinguish among APOBEC3 proteins – the sequence context in which they typically mutate DNA cytidines to uridines (Table 1). A3G preferentially mutates cytidine residues that are preceded by another cytidine, with an even stronger preference for the context 5′-CCCA-3′ in minus-strand viral cDNA, where the underlined deoxycytidine is the nucleotide preferentially targeted (Ref. 26, 27, 28, 77). In plus-strand reverse transcribed cDNA, this manifests as 5′-TGGG-3′ →5′-TAGG-3′ mutations, or 5′-GG-3′ → 5′-AG-3′ in the less stringent dinucleotide context. Other APOBEC3 proteins, however, generally display a preference for the mutation of cytidines preceded by another deoxyribonucleotide, most often thymidine [e.g. (Ref. 78, 79, 80, 81). This results in 5′-TC-3′ to 5′-TU-3′ transitions, which manifest on plus-strand viral cDNA as 5′-GA-3′ → 5′-AA-3′.
The identification of these different sequence contexts is important for assessing which APOBEC3 proteins act on a given viral substrate in a context where multiple APOBEC3 proteins are expressed such as in vivo. Furthermore, assessing these contexts helps one to appreciate how hypermutation can be detrimental to a virus. Aside from deleterious amino acid or other mutations that may occur in unique sequence contexts, any UGG codon encoding tryptophan is susceptible to the creation of a UAG or perhaps even a UAA stop, particularly in the presence of A3G. Similarly, the AUG start codon may place the wobble base in an APOBEC3-susceptible hotspot depending on the identity of the next base in the sequence, thus making the ablation of initiating methionines another distinct possibility. Extensive mutation may also disrupt HIV-1 genomic RNA secondary structures, which are important for the regulation of HIV-1 gene expression (Ref. 82).
APOBEC3 proteins mutate the HIV-1 genome
Even prior to the discovery of APOBEC3 proteins, the tendency for HIV-1 to acquire G-to-A mutations was evident (Ref. 83, 84, 85). This phenomenon was, in fact, analogous to the original observation of hypermutation in spleen necrosis virus (Ref. 86). These early reports generally noted a preferential sequence context for hypermutation of 5′-GA-3′ and, to a lesser extent, 5′-GG-3′, suggesting that, given what is now known, multiple APOBEC3 proteins account for these early descriptions of hypermutation (Ref. 79).
While more recent studies at times note stronger preferences for one context or another under a given condition (Ref. 87, 88, 89), a theme uniting recent literature on the in vivo hypermutation of HIV-1 is that it frequently occurs in both dinucleotide contexts. One study indicates, for example, that hypermutated sequences separate roughly into thirds – sequences with hypermutation predominantly in a 5′-GA-3′ context, sequences with hypermutation predominantly in a 5′-GG-3′ context and sequences with hypermutation occurring in roughly equal proportions in each context (Ref. 90). Another reports the occurrence of 5′-GG-3′ context mutations in approximately 95% of hypermutated sequences versus the occurrence of 5′-GA-3′ context mutations in approximately 60% of hypermutated sequences (Ref. 91). Furthermore, mutations in both contexts frequently occur in the same cloned sequence. For example, 5′-GG-3′ hypermutations occur within 95% of predominantly 5′-GA-3′ context clones (Ref. 91), while another data set shows a very slight preference for hypermutation in a 5′-GA-3′ context with a strong 5′-GG-3′ component (Ref. 92).
It is reasonable to conclude, therefore, that multiple APOBEC3 proteins are active in vivo and that at least one of these is A3G with its strong 5′-GG-3′ context preference. The identities of the APOBEC3 proteins responsible for the reported 5′-GA-3′ hypermutations, however, remain unknown. While A3F is certainly a likely candidate based on its expression, Vif sensitivity and restriction profiles (see below; Table 1), it is entirely possible that other APOBEC3 proteins account for at least a portion of this hypermutation. A more careful elucidation of the expression and restriction capacities of APOBEC3 proteins in vivo as well as the interactions of these proteins with HIV-1 in vitro will be required to define the true nature of the in vivo APOBEC3 repertoire, which may itself vary from person to person [e.g. (Ref. 93)].
Regardless of which APOBEC3 proteins are actively hypermutating HIV-1, it is clear that their overall effect is significant. Estimates of the frequency of hypermutated sequences in vivo range from approximately 7–20% with wide variation in the proportions seen in any one individual (Ref. 87, 88, 89, 90, 91). In fact, these figures almost certainly underestimate the total proportion of proviral sequences mutated by APOBEC3 proteins. Since a sequence must typically reach a given threshold of G-to-A mutations to be defined as “hypermutated”, methods of quantifying hypermutants fail to account for sequences that have been mutated by APOBEC3 proteins at a lower level. Furthermore, extremely hypermutated sequences may fail to replicate and amplify by PCR while still potentially contributing to the overall mutational load of a viral population through recombination (Ref. 94). As briefly discussed above, the deamination of cDNA cytidines may also create a substrate for the downstream degradation of the viral genome, which would further diminish the apparent proportion of viral genomes affected by APOBEC3 proteins.
APOBEC3 proteins may contribute to viral adaptation
Several authors have previously discussed the theoretical possibility that drug resistance mutations could be attributable to APOBEC3-mediated hypermutation, citing the frequent occurrence of drug resistance mutations in sequence contexts consistent with the action of APOBEC3 proteins (Ref. 95, 96, 97). Combined with the common occurrence of hypermutation in vivo, it would seem that a role for APOBEC3 proteins in aiding the evolution of HIV-1 drug resistance is likely. Nevertheless, the actual data in the literature on this topic are still limited. For example, one study demonstrates that, in the presence of a Vif allele defective for the neutralization of A3G, HIV-1 can readily acquire a particular drug resistance mutation by recombination with a lethally hypermutated genome residing in the same cell (Ref. 94). Computer modeling to assess the likelihood of drug resistance mutation acquisition from A3G action further suggests that HIV-1 does acquire drug resistance mutations from A3G activity at a low rate (Ref. 98). Also of interest, a recent paper on HIV-1 sequence evolution proposes that APOBEC3 proteins may be involved in the acquisition of immune escape mutations early in HIV-1 infection (Ref. 99).
While the consensus of the data to date is that APOBEC3 proteins are likely to contribute to HIV-1 evolution, many questions remain unanswered. For example, how often does this actually occur in vivo, and what is the relative contribution of APOBEC3 proteins versus other sources of mutation such as reverse transcriptase? Which APOBEC3 proteins are primarily responsible? To what extent does selective pressure contribute to the amplification of mutations caused by APOBEC3 proteins? Much work remains to be done in determining the impact of APOBEC3 proteins on the adaptability of HIV-1. The field would particularly benefit from studies that expand beyond A3G to consider the APOBEC3 proteins likely to cause 5′-GA-3′ context mutations.
Correlation of hypermutation levels, APOBEC3 expression and clinical indicators
While hypermutation clearly occurs in vivo, the data concerning its correlation with clinical parameters of infection are less clear; a summary of pertinent research on this topic is provided in Table 2. For example, one study shows a significant correlation between hypermutation in both 5′-GA-3′ and 5′-GG-3′ contexts and reduced viral load (Ref. 88), consistent with the higher levels of hypermutation found in another group of patients with low viral loads (Ref. 100). Strikingly, the authors of the first study conclude that the drop in viral load attributable to hypermutation is notably greater than that attributable to other known protective alleles such as CCR5Δ32. Corroborating this effect, 17 Kenyan women with significant hypermutation levels have a statistically significant increase in CD4+ cell counts (Ref. 90). When the total adenosine content of patient proviruses is taken as a surrogate of hypermutation, a positive correlation is found between adenosine content and CD4+ cell counts throughout this entire cohort of 240 Kenyan women.
Table 2.
Summary of Linkage Studies between Clinical Indicators and Hypermutation, APOBEC3 Expression or APOBEC3 Polymorphism
| Study | Cohort Description | Relationship Reported |
|---|---|---|
| Association with Hypermutation | ||
| Pace et al. (Ref. 88) |
|
|
| Gandhi et al. (Ref. 91) |
|
|
| Land et al. (Ref. 90) |
|
|
| Ulenga et al. (Ref. 92) |
|
|
| Vázquez-Pérez et al. (Ref. 100) |
|
|
| Piantadosi et al. (Ref. 89) |
|
|
| Association with APOBEC3 Expression | ||
| Jin et al. (Ref. 101) |
|
|
| Cho et al. (Ref. 102) |
|
|
| Biasin et al. (Ref. 104) |
|
|
| Ulenga et al. (Ref. 103) |
|
|
| Vázquez-Pérez et al. (Ref. 100) |
|
|
| Association with APOBEC3 Polymorphisms | ||
| An et al. (Ref. 106) |
|
|
| Do et al. (Ref. 105) |
|
|
| Pace et al. (Ref. 88) |
|
|
| Valcke et al. (Ref. 107) |
|
|
| An et al. (Ref. 108) |
|
|
In contrast, hypermutation does not significantly correlate with viral load in a cohort of Senegalese female sex workers, although the levels of hypermutation do generally correlate with A3G mRNA expression (Ref. 92). Additional evidence suggesting no correlation between hypermutation and improved clinical status has been found by comparing patients on highly active antiretroviral therapy (HAART) with elite suppressors, patients who maintain high CD4+ cell counts and undetectable viral loads (Ref. 91). In contrast with the Senegalese cohort, however, no correlation between A3G expression and the fraction of hypermutated sequences existed in these groups. A third cohort consisting of 28 Kenyan women also shows no correlation between hypermutation and viral load or CD4+ cell counts (Ref. 89).
Others more directly assess the impact of specific APOBEC3 proteins by correlating their mRNA expression levels with clinical indicators; results of these studies are summarized in Table 2. Using this approach, strong negative correlations are found between A3G mRNA levels and viremia, concordant with positive correlations between A3G mRNA levels and CD4+ cell counts (Ref. 100, 101), although one study does not reproduce these associations (Ref. 102). Nevertheless, other data indicate that patients with a low viral set point have significantly higher levels of A3G mRNA both pre- and post-infection than patients with a high viral set point; analysis for A3F yields the same correlation with post-infection A3F mRNA levels but not pre-infection mRNA levels (Ref. 103). Furthermore, the peripheral blood mononuclear cells of HIV-exposed seronegative patients show higher A3G expression and decreased susceptibility to ex vivo infection (Ref. 104), and HIV-exposed seronegative individuals have higher A3G expression levels than healthy controls (Ref. 100).
Thus, the preponderance of evidence indicates a correlation between higher A3G expression and improved clinical status. This correlation may be less apparent or even absent, however, when hypermutation is taken as a surrogate for APOBEC3 action. Failing to find a correlation when substituting hypermutation for APOBEC3 function may not be that surprising, though, given that reducing the probability of successful reverse transcription and integration appears to be an important part of the mechanism by which APOBEC3 proteins restrict HIV-1. Interpretations of data correlating hypermutation with clinical indicators are somewhat confounded because it is not clear what has occurred when no correlation is found. If APOBEC3 proteins are highly effective, the viral genome may simply not persist long enough to integrate. Furthermore, it is possible that some APOBEC3 repertoires and/or individual APOBEC3 alleles may be protective, leading some patients to be selectively excluded from studies due to a lack of apparent HIV-1 infection. Regardless, the positive data in these studies, particularly those focusing on APOBEC3 expression, do indicate promise for therapies aimed at the APOBEC3-Vif axis. The field would benefit from further research correlating clinical indicators with both hypermutation and the expression of more APOBEC3 family members in larger patient cohorts. Individuals known to be relatively resistant to HIV-1 should also be closely examined for APOBEC3 sequence and copy number variations.
APOBEC3 polymorphisms may be important for restriction activity against HIV-1
Relatively little is known about the functional relevance of human APOBEC3 polymorphisms, but several attempts have been made to correlate clinical status with APOBEC3 variations, as summarized in Table 2. One survey of the A3G alleles in a French cohort divided into rapid and slow progressors reports 29 polymorphisms, two of which affect the amino acid sequence, but none of these correlates with the rate at which these groups progress to disease (Ref. 105). Others find that the H186R allele identified in this cohort does, however, correlate with CD4+ cell depletion and rapid progression to AIDS-defining conditions in African-American patients (Ref. 106), although there is no apparent association with risk of transmission. Another allelic variant, C40693T in intron 4 of A3G, is associated with an increased risk of infection in a cohort of Caucasian patients (Ref. 107).
While these associations are suggestive, their functional basis is far from clear. For example, there is no apparent difference between the wild-type and H186R alleles of A3G in an in vitro single-cycle infectivity assay (Ref. 105), and no experimental data are available for the C40693T variant. As the authors of the latter study rightly point out, however, the close association of A3G with other APOBEC3 proteins on chromosome 22 raises the distinct possibility that a given association may actually reflect linkage with an undetected variation elsewhere in the locus rather than a direct effect of the identified polymorphism (Ref. 107).
In the case of A3H, activity against HIV-1 is strongly influenced by polymorphisms. Four major haplotypes of human A3H exist, but only one of these appears to be expressed stably and able to inhibit HIV-1 (Ref. 73, 74, 75). This active variant, A3H haplotype II, is present in roughly 50% of individuals of African descent, 10–18% of individuals of European descent and 3–4% of individuals of Asian descent (Ref. 73). Another common APOBEC3 polymorphism is an A3B deletion, which occurs in approximately 20% of the world’s population (Ref. 93). In contrast with A3H Haplotype II, however, deletion of A3B is relatively rare in African and European populations but common in East Asian (36.9%), Amerindian (57.7%) and Oceanic (92.9%) populations.
Whether these or other unidentified allelic variants are of significance for susceptibility to HIV-1 infection or disease progression is unknown, although at least one recent report has found a statistically significant increased risk of HIV-1 infection and progression in individuals with homozygous A3B deletions (Ref. 108). It is important to point out, however, that most APOBEC3 proteins have evolved under positive selection, implying that the challenges posed by ancient and presumably divergent pathogens have resulted in the rapid evolution of these restriction factors (Ref. 109, 110). While this selective pressure may not have been applied by HIV-1 given its relatively recent arrival in the human population, selection for APOBEC3 proteins better able to restrict other pathogens may have inadvertently selected cross-reactivity for the restriction of HIV-1. Thus, it is reasonable to predict that, despite the relative dearth of data presently available, variation at the genetic level will account for some effect on HIV-1 in vivo. Furthermore, it is possible that ongoing selection by the HIV-1 epidemic itself may result in the expansion of protective APOBEC3 alleles not yet identified.
Studies of APOBEC3 effects in specific primary cell types
CD4+ T cells are the primary reservoir of HIV-1 infection in vivo, but relatively little is known about the APOBEC3 repertoire found in these cells. One report suggests that A3G forms the basis of a largely uncharacterized, deaminase-independent target cell restriction of incoming HIV-1 virions in resting CD4+ T cells (Ref. 111), an effect enhanced by treatment with interferon α, which upregulates A3G expression (Ref. 112). The reproducibility of this phenomenon, however, has been questioned (Ref. 113, 114). Regardless, evidence indicates that among activated CD4+ T cells, the Th1 subset of helper T cells expresses greater levels of A3G and A3F, which are further enhanced by the autocrine action of interferon γ (Ref. 115). Greater expression, in turn, results in greater encapsidation of APOBEC3 proteins in HIV-1 produced from these cells and consequently lower viral infectivity than in the Th2 subset of helper T cells, even for Vif-proficient viruses. In general, most APOBEC3 family proteins with the exception of A3A and, to a lesser extent, A3B appear to be well-expressed in primary CD4+ T cells; among T cell subsets, however, A3G is somewhat better expressed than A3F (Ref. 116).
Studies with primary myeloid lineage cells have also noted a prominent effect of interferon treatment on the ability of APOBEC3 proteins to restrict HIV-1, linking higher expression of A3G and A3A to the relative resistance of monocytes versus differentiated macrophages to infection (Ref. 117). Another report indicates that A3G and A3F are responsible for preventing HIV-1 from infecting immature dendritic cells, an effect which augments as dendritic cells mature and increase their A3G expression (Ref. 118). Expression of interferon α and, to a lesser extent, interferon γ have also been linked to increased APOBEC3 levels and increased resistance to HIV-1 infection; this effect is so dramatic, in fact, that the upregulation of APOBEC3 proteins may effectively overwhelm normal Vif function and enable restriction of wild-type HIV-1 in macrophages (Ref. 117, 119). Of note, interferon α clearly upregulates all APOBEC3 proteins in macrophages and dendritic cells with the possible exceptions of A3B and A3C, indicating that these are part of the normal interferon response (Ref. 116). This upregulation is particularly astounding for A3A (Ref. 116, 117). Given the common expression of many APOBEC3 proteins in primary cell types and their interferon inducibility, it is tempting to speculate that the combination of different APOBEC3 proteins may yield emergent properties not readily apparent in the majority of in vitro studies to date, which almost exclusively focus on the effects of single APOBEC3 proteins.
Strategies for the clinical application of basic APOBEC3-Vif biology
Therapeutic strategies center on increasing the encapsidation of APOBEC3 proteins
Strategies aimed at the therapeutic use of APOBEC3 proteins to restrict HIV-1, despite their diversity, are united by one goal: to increase the amount of APOBEC3 proteins encapsidated in HIV-1 virions (Fig. 2). If substantial quantities of APOBEC3 proteins are packaged in a particular virion, even in a Vif-proficient virus (see below), the genomic RNA of that virion is not likely to survive to become a replication-competent provirus. Moreover, while the mutagenic capacity of APOBEC3 proteins carries with it at least some risk for contributing to viral adaptation (Ref. 94, 98), particularly when amplified, the risk associated with A3G mutagenic activity specifically may be tolerable relative to the potential benefits of unleashing its antiviral activity (Ref. 98). To increase the packaging of APOBEC3 proteins, two general strategies are tenable – indirectly preventing the degradation of APOBEC3 proteins or directly enhancing the packaging of APOBEC3 proteins.
Fig. 2. Methods of enhancing APOBEC3-mediated restriction.
Methods of enhancing restriction by APOBEC3 proteins are united in the common goal of delivering more APOBEC3 proteins to the viral core. Strategies, as discussed in the main text, include i) Indirectly increasing APOBEC3 encapsidation by protecting APOBEC3 proteins from degradation. This may occur by chemically shielding APOBEC3 proteins. Alternatively, it may be possible to target inhibitors to a Vif surface that interacts with APOBEC3 proteins or with cellular proteins involved Vif-mediated degradation. Finally, a degradation-resistant variant of an APOBEC3 protein such as A3G D128K may be employed. ii) Directly increasing the encapsidation of APOBEC3 proteins. This may occur by increasing the expression of APOBEC3 proteins, particularly through interferon (orange octagon) upregulation. Alternatively, fusion to a privileged molecule with core access (white circle labeled “Help”) may shuttle APOBEC3 proteins to the viral core with great enough efficiency to overcome Vif-mediated degradation. All methods will theoretically result in greater APOBEC3 encapsidation (white circles) and restriction of HIV-1, even in the presence of Vif.
Indirectly increasing encapsidation by preventing the degradation of APOBEC3 proteins: A pharmaceutical approach
Prevention of the degradation of APOBEC3 proteins as depicted in Fig. 2 would be most simply accomplished by blocking the interaction of APOBEC3 proteins with Vif. To that end, a great deal of effort has been expended attempting to elucidate the important interacting regions on Vif and on APOBEC3 proteins [(Ref. 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133) and others cited elsewhere]. While a thorough discussion of the many details involved in these investigations is beyond the scope of this review, a schematic summary of important regions on Vif and on A3G with particular attention to their putative interaction domains is provided in Fig. 3, and biologically relevant regions of A3G are highlighted on a recently-published full-length structural model of this restriction factor (Ref. 134) in Fig. 4.
Fig. 3. Important domains in Vif and A3G.
While many papers have identified residues critical to Vif function, only putative continuous interaction domains in Vif (A) and A3G (B) are depicted here for the sake of clarity. Exceptions are made for two important lysines in Vif, K22 and K26, as well as the C-terminal zinc-binding domain of Vif. Vif and A3G are internally to scale, but Vif is depicted at twice its actual size relative to A3G. HIV-1 Vif residues shown are those found in HIV IIIB (EU541617.1). A3G corresponds to reference sequence NP_068594. Note that K26 and 23SLV25 of Vif are required for neutralization of A3G and A3F, but these residues may not mediate direct interaction with A3F or A3G. Similarly, one group finds that while A3G D128 is important in permitting the degradation of A3G, it does not mediate direct binding to Vif. See main text for references and discussion.
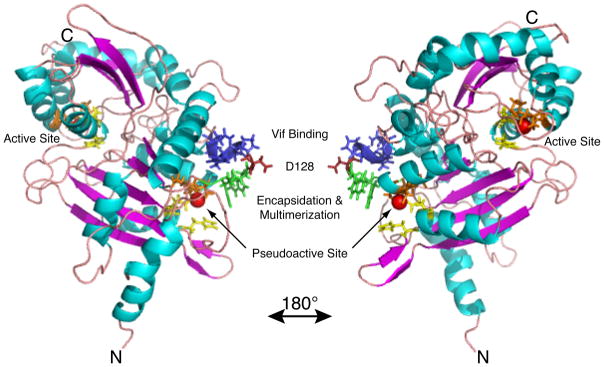
Fig. 4. Important domains of A3G on a full-length model structure.
Important domains of A3G identified by genetic analysis are highlighted on a full-length model structure of A3G (Ref. 134) with coloration carried out in Pymol. See main text for references and discussion. Sidechains are shown for key regions, and the right panel represents a rotation of 180 degrees about the y-axis. Red Spheres = Active site zinc. Green Sidechains = 124YYFW127, multimerization/encapsidation determinant (overlaps 126–127 start of Vif binding region 126FWDPDYQ132); Red Sidechain = D128, a key residue for Vif sensitivity; Blue Sidechains = 129PDYQ132, the remainder of the putative Vif binding site. Yellow Sidechains = Residues H65 & E67 and H257 & E259, the HxE of the pseudocatalytic and catalytic N- and C-terminal domains, respectively. Orange Sidechains = 96PC97 & C100 and 287PC288 & C291, the PCxxC of the active and pseudoactive N- and C-terminal domains, respectively.
In surveying this literature, is worthwhile to note that several discontinuous surfaces on Vif have been implicated in the binding of specific APOBEC3 proteins (Fig. 3). This indicates that the APOBEC3 interaction surfaces on HIV-1 Vif are structurally complex and that three-dimensional structures of Vif bound to APOBEC3 proteins will likely be required both to understand and to effectively target this interaction. In contrast, what is known of the Vif interaction surfaces on APOBEC3 proteins indicates that, while such surfaces may be distinct on different APOBEC3 proteins (Ref. 122, 129), they are relatively continuous. It may be possible, therefore, to protect APOBEC3 proteins by simply designing a molecule that binds the host protein and shields it from interaction with Vif, a strategy that would likely hamper the ability of HIV-1 to develop resistance to this putative therapy (Ref. 2). Conversely, it may be possible to inhibit Vif interaction with either APOBEC3 proteins or with the proteasomal machinery using a molecule that binds Vif itself.
Although no one has yet made public a small molecule inhibitor of the APOBEC3-Vif interaction, a significant recent development is the identification of RN-18, a small molecule antagonist of HIV-1 Vif (Ref. 135). Interestingly, RN-18 decreases Vif levels and thereby increases A3G (and A3F and A3C but not A3B) levels when these two are present together. This small molecule is also able to inhibit the growth of Vif-proficient HIV-1 in cultured nonpermissive cells. Thus, while further research is necessary to understand the mechanism by which this and other putative inhibitors function, RN-18 has provided proof of principle that the APOBEC3-Vif axis can be targeted by small molecules.
Another promising development in the APOBEC3 field is the growing body of structural information on the deaminase domain of A3G (Ref. 134, 136, 137, 138) and related proteins such as APOBEC2 (Ref. 139). As progress continues toward the expansion of our structural knowledge of A3G and other APOBEC3 proteins [reviewed recently by (Ref. 140)], it may one day be possible to visualize the APOBEC3-Vif interaction structurally and thereby rationally design interaction inhibitors. Half of this putative interaction surface is depicted in the A3G full-length model presented in Fig. 4. In the meantime, the field would benefit from further genetic study of the APOBEC3-Vif interaction, both for the sake of reproducing current results and identifying mutants that may further our mechanistic understanding of the APOBEC3-Vif interaction in anticipation of complementary structural data. For more detail on such efforts, see Further reading and resources below.
Indirectly increasing packaging by preventing the degradation of APOBEC3 proteins: A gene therapy approach
While small molecules remain undoubtedly the most technologically feasible means by which to manipulate the APOBEC3-Vif axis, progress in the use of gene therapy approaches for the treatment of HIV-1 [e.g. (Ref. 141, 142, 143)] suggests that similar methods may one day incorporate aspects of APOBEC3-mediated restriction. While the application of APOBEC3 biology to HIV-1 therapy in this manner must be preceded by further advances in the safety and efficacy of both gene delivery and cell therapy, the strategies discussed in this and the following section merit discussion if for no other reason than to note how they reinforce the basic concepts of APOBEC3-Vif biology.
Because Vif is required for the spread of HIV-1 in nonpermissive cells and in vivo, one obvious way to go about killing the virus would be to find an APOBEC3 protein that is not neutralized by Vif. In fact, nature has already provided just such a variant, as a number of studies on the ability of the Vifs carried by the lentiviruses of different species to neutralize their cognate APOBEC3 proteins have revealed that quite small changes in APOBEC3 proteins can dramatically affect their sensitivity to Vif. Specifically, substitution of the human A3G aspartate 128 (e.g. Fig. 4) with lysine renders A3G resistant to Vif-mediated degradation (Ref. 67). The D128K substitution may protect A3G by preventing Vif binding (Ref. 128, 144, 145, 146), although this view is not universally supported (Ref. 120, 147). Whatever the mechanism, D128K is not destabilized by HIV-1 Vif, and this leads to increased packaging of A3G and the restriction of Vif-proficient HIV-1. Despite the resistance of A3G D128K to Vif-mediated degradation, there are no known instances of this allele occurring naturally in humans.
An alternative method of protecting A3G from degradation that has been reported is to fuse it to the ubiquitin-associated domain 2 (UBA2), a stabilization signal that protects proteins from proteasomal degradation (Ref. 148). Although this fusion is only partially resistant to Vif-mediated degradation, it does appear to have a modestly improved effect on the restriction of wild-type HIV-1 relative to that of wild-type A3G. This study combined with those above on A3G D128K provide proof of principle that relatively simple modifications to APOBEC3 proteins may allow them to resist Vif-mediated degradation. As our technical facility with therapeutic gene delivery develops, these may one day prove viable strategies to engineer HIV-1 resistant T cells to counter infection.
Directly increasing encapsidation by increasing APOBEC3 expression: A pharmaceutical approach
In much the same way that protecting A3G from degradation might leave enough A3G available to encapsidate and effectively restrict HIV-1, increasing the expression of A3G might overwhelm Vif and result in the encapsidation of nonpermissive levels of A3G as suggested in Fig. 2. Proof of principle for this concept has been shown in macrophages, where the induction of APOBEC3 proteins by interferon α treatment is so effective that it greatly restricts the infectivity of Vif-proficient HIV-1 (Ref. 119), although it should be noted that the many effects of interferon induction make it difficult to rule out effects nonspecific to APOBEC3 action. In theory, then, methods aimed at selectively increasing the expression of APOBEC3 proteins in the natural reservoirs of HIV-1 infection should be able to potently inhibit viral replication, perhaps additionally allowing the broader immune system to do a more effective job of controlling infection and supporting the suppression of HIV-1 by current antiretroviral therapies. An important advantage of such methods is that they would likely be facilitated by small molecules and would work by enhancing innate immune processes that already occur naturally. That said, it is unclear what the consequences of increasing APOBEC3 expression in vivo may be, since there is at least a theoretical possibility that increasing APOBEC3 levels may overwhelm cellular regulatory mechanisms and result in hypermutation of the human genome, possibly impacting the development of cancer [e.g. (Ref. 25, 149)].
Directly increasing packaging by directly facilitating APOBEC3 access to the viral core: A gene therapy approach
Alternatively, selectively directing APOBEC3 proteins to the HIV-1 viral core may permit the restriction of even Vif-proficient HIV-1 (Fig. 2). Evidence from several groups shows that fusing A3G to a fragment of Vpr sufficient for encapsidation (Ref. 150), fusing A3A to a full-length Vpr protein (Ref. 151) or fusing A3G to the Nef7 mutant derivative of HIV-1 Nef (Ref. 152) results in enhanced APOBEC3-mediated restriction. All of these methods lead to the incorporation of APOBEC3 proteins into the viral core via the viral peptides to which they are fused, which themselves have natural core access. This process is furthermore so efficient that Vif cannot overcome it.
An important lesson is also apparent in the ability of A3A to restrict under conditions in which it is fused to a molecule that grants it core access. While this APOBEC3 protein does not normally restrict HIV-1 or reach the viral core, forcing it past that barrier by fusion to Vpr or A3G results in A3A-mediated restriction (Ref. 151, 153). Thus, it is not sufficient for an A3A to simply find its way into HIV-1 virions. Rather, it must be specifically targeted to the viral core to have sufficient access to the viral genome and carry out its antiviral effects. Whether the localization of other APOBEC3 proteins within virions is an important determinant of their relative abilities to function remains a subject for future research.
Conclusions and outstanding research questions
Controversies aside, research to date has yielded significant insight into the mechanisms by which A3G and HIV-1 interact in the balance between restriction and infection. Given what is known about APOBEC3 protein expression and hypermutation profiles in vivo and in primary ex vivo cell types, it is likely that multiple APOBEC3 proteins are active against HIV-1 in vivo, and that these may display distinct properties. More work is required to define the APOBEC3 proteins that impact HIV-1 in vivo and the exact mechanisms by which this occurs.
Clinical studies may also prove useful in the continuing characterization of the APOBEC3-Vif axis. As discussed above, incorporating the quantification of the expression levels of more APOBEC3 proteins and correlating those to hypermutation levels while increasing cohort sizes would improve our understanding of the in vivo functions of APOBEC3 proteins. Further characterization of APOBEC3 polymorphisms may also be significant, since it is possible that protective APOBEC3 alleles await discovery. To this end, further studies of the full APOBEC3 genomic locus in uninfected individuals, infected individuals and individuals with an atypical response to HIV-1 such as long-term nonprogressors and those uninfected but repeatedly exposed may hold a key to novel treatments aimed at the APOBEC3-Vif axis.
Complementary studies focusing on the structures of APOBEC3 proteins and of Vif are also likely to be of great benefit. It may be possible to identify, characterize, and ultimately commercialize Vif inhibitors such as RN-18 in the absence of structural information. Nevertheless, the ability to rationally design pharmaceuticals aimed at the APOBEC3-Vif axis will be significantly facilitated by a deep understanding of the physical interactions of APOBEC3 proteins and Vif. Furthermore, such understanding will undoubtedly be required to effectively design molecules that minimize the risk of selecting HIV-1 strains resistant to this hypothetical new class of antiretroviral drugs. To that end, however, the APOBEC3 family is particularly attractive, as it comprises multiple potential endogenous inhibitors of HIV-1 infectivity, at least two of which may be able to function in the long-term restriction of HIV-1 (Ref. 22). With further study, the APOBEC3 proteins may one day form the basis of the next generation of antiretroviral therapies.
Further reading and resources
Internet resources
-
Los Alamos National Laboratory HIV Sequence Database: This website provides not only an excellent collection of HIV sequence information, but also a number of analysis tools (e.g. Hypermut, a program designed to detect and analyze hypermutated sequences) and HIV reference materials.
-
Stanford University Drug Resistance Database: This database is especially useful for analyzing drug resistance mutations in patient sequences, a topic relevant to the emerging consensus that APOBEC3 proteins affect HIV sequence evolution and adaptation.
-
Retroviruses: This is a free, online edition of an older but highly influential textbook on the properties of retroviruses, including HIV.
Reviews
-
This is a comprehensive review of APOBEC3 biology.
○ Chiu, Y. L. and Greene, W. C. (2008) The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol 26, 317–353
-
This is an excellent review of the roles of accessory proteins, including Vif, in HIV biology. Its exploration of the common theme of accessory proteins mediating the degradation of host proteins through the proteasomal machinery is particularly insightful.
○ Malim, M. H. and Emerman, M. (2008) HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398
-
This is a review dedicated to the interaction surfaces of Vif and APOBEC3 proteins.
○ Smith, J.L., et al. (2009) Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends in Pharmacological Sciences In press
-
These are several recent reviews on Vif and APOBEC3 proteins that provide more mechanistic detail than is given here.
○ Goila-Gaur, R. and Strebel, K. (2008) HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5, 51
○ Henriet, S., et al. (2009) Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC3G and APOBEC3F restriction factors. Microbiol Mol Biol Rev 73, 211–232
○ Malim, M. H. (2009) APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci 364, 675–687
Acknowledgments
We thank the reviewers and A.M. Land for constructive comments and assistance in improving this manuscript. We also thank E. Harjes and H. Matsuo for providing the full-length model of A3G. This work was funded by the National Institute of Allergy and Infectious Diseases (R01 AI064046 to RSH). JSA was supported by a Kirschstein National Research Service Award for Individual Predoctoral MD-PhD Fellows and by the University of Minnesota Medical Scientist Training Program (National Institute on Drug Abuse, F30 DA026310; National Institute of General Medical Sciences, T32 GM008244).
Footnotes
The manuscript posted here is a revised version incorporating comments from peer review but prior to editorial input from ERMM. The definitive version has been published by Expert Reviews in Molecular Medicine: 12: e4 (22 January 2010), pp. 1–26 (© Cambridge University Press 2010) and is available at the following address: http://journals.cambridge.org/action/displayAbstract?fromPage=online&aid=7103604
The acronym APOBEC derives from APOBEC1 and stands for “apolipoprotein B mRNA editing, catalytic polypeptide”. Human APOBEC3 proteins are designated “apolipoprotein B mRNA editing, catalytic polypeptide-like 3” followed by the letter, A-H, identifying the specific protein.
References
- 1.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 2.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 3.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Gramberg T, Sunseri N, Landau NR. Accessories to the crime: recent advances in HIV accessory protein biology. Curr HIV/AIDS Rep. 2009;6:36–42. doi: 10.1007/s11904-009-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrett ED, Tiley LS, Cullen BR. Rev activates expression of the human immunodeficiency virus type 1 vif and vpr gene products. J Virol. 1991;65:1653–1657. doi: 10.1128/jvi.65.3.1653-1657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz S, Felber BK, Pavlakis GN. Expression of human immunodeficiency virus type 1 vif and vpr mRNAs is Rev-dependent and regulated by splicing. Virology. 1991;183:677–686. doi: 10.1016/0042-6822(91)90996-o. [DOI] [PubMed] [Google Scholar]
- 7.Kan NC, et al. Identification of HTLV-III/LAV sor gene product and detection of antibodies in human sera. Science. 1986;231:1553–1555. doi: 10.1126/science.3006245. [DOI] [PubMed] [Google Scholar]
- 8.Lee TH, et al. A new HTLV-III/LAV protein encoded by a gene found in cytopathic retroviruses. Science. 1986;231:1546–1549. doi: 10.1126/science.3006243. [DOI] [PubMed] [Google Scholar]
- 9.Sodroski J, et al. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science. 1986;231:1549–1553. doi: 10.1126/science.3006244. [DOI] [PubMed] [Google Scholar]
- 10.Desrosiers RC, et al. Identification of highly attenuated mutants of simian immunodeficiency virus. J Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strebel K, et al. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 12.Gabuzda DH, et al. Role of Vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabuzda DH, et al. Essential role of vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J Acquir Immune Defic Syndr. 1994;7:908–915. [PubMed] [Google Scholar]
- 14.Fisher AG, et al. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 15.Simon JH, Malim MH. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courcoul M, et al. Peripheral blood mononuclear cells produce normal amounts of defective Vif- human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J Virol. 1995;69:2068–2074. doi: 10.1128/jvi.69.4.2068-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goncalves J, et al. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sova P, Volsky DJ. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J Virol. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon JH, et al. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 21.Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 22.Haché G, et al. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr Biol. 2008;18:819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 24.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 25.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 26.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 27.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lecossier D, et al. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 30.Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes RK, et al. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 32.Miyagi E, et al. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schumacher AJ, et al. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shindo K, et al. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J Biol Chem. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- 35.Newman EN, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 36.Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bishop KN, et al. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwatani Y, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo F, et al. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J Virol. 2006;80:11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, et al. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology. 2007;365:92–100. doi: 10.1016/j.virol.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 41.Li XY, et al. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J Biol Chem. 2007;282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- 42.Luo K, et al. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mbisa JL, et al. HIV-1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schumacher AJ, Nissley DV, Harris RS. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc Natl Acad Sci U S A. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turelli P, et al. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 46.Bogerd HP, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muckenfuss H, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 48.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 49.Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci. 2007;32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Bonvin M, Greeve J. Hepatitis B: modern concepts in pathogenesis--APOBEC3 cytidine deaminases as effectors in innate immunity against the hepatitis B virus. Curr Opin Infect Dis. 2008;21:298–303. doi: 10.1097/QCO.0b013e3282fe1bb2. [DOI] [PubMed] [Google Scholar]
- 51.Harris RS, et al. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat Immunol. 2003;4:641–643. doi: 10.1038/ni0703-641. [DOI] [PubMed] [Google Scholar]
- 52.Yang B, et al. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J Biol Chem. 2007;282:11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- 53.Chen R, et al. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J Biol Chem. 2004;279:28419–28425. doi: 10.1074/jbc.M403875200. [DOI] [PubMed] [Google Scholar]
- 54.Mansky LM, et al. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 In vivo mutation rate. J Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Priet S, et al. HIV-1-associated uracil DNA glycosylase activity controls dUTP misincorporation in viral DNA and is essential to the HIV-1 life cycle. Mol Cell. 2005;17:479–490. doi: 10.1016/j.molcel.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 56.Schröfelbauer B, et al. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol. 2005;79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase APOBEC3G. J Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 59.Marin M, et al. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 60.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 61.Stopak K, et al. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 62.Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 63.Mehle A, et al. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 64.Mehle A, et al. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu Y, et al. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kao S, et al. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mariani R, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 68.Santa-Marta M, et al. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J Biol Chem. 2005;280:8765–8775. doi: 10.1074/jbc.M409309200. [DOI] [PubMed] [Google Scholar]
- 69.Conticello SG, et al. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 70.Wedekind JE, et al. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 71.Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci. 2009;364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Q, et al. Microarray analysis of lymphatic tissue reveals stage-specific, gene expression signatures in HIV-1 infection. J Immunol. 2009;183:1975–1982. doi: 10.4049/jimmunol.0803222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.OhAinle M, et al. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harari A, et al. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan L, et al. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. Faseb J. 2009;23:279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu Q, et al. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 77.Yu Q, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 78.Bishop KN, et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 79.Liddament MT, et al. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 80.Wiegand HL, et al. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng YH, et al. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janini M, et al. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J Virol. 2001;75:7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vartanian JP, et al. Selection, recombination, and G----A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991;65:1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vartanian JP, et al. G-->A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci U S A. 1994;91:3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kieffer TL, et al. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pace C, et al. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J Virol. 2006;80:9259–9269. doi: 10.1128/JVI.00888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piantadosi A, et al. Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif. J Virol. 2009;83:7805–7814. doi: 10.1128/JVI.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Land AM, et al. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. J Virol. 2008;82:8172–8182. doi: 10.1128/JVI.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gandhi SK, et al. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82:3125–3130. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ulenga NK, et al. The level of APOBEC3G (hA3G)-related G-to-A mutations does not correlate with viral load in HIV type 1-infected individuals. AIDS Res Hum Retroviruses. 2008;24:1285–1290. doi: 10.1089/aid.2008.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kidd JM, et al. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Berkhout B, de Ronde A. APOBEC3G versus reverse transcriptase in the generation of HIV-1 drug-resistance mutations. Aids. 2004;18:1861–1863. doi: 10.1097/00002030-200409030-00022. [DOI] [PubMed] [Google Scholar]
- 96.Haché G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006;8:148–157. [PubMed] [Google Scholar]
- 97.Pillai SK, Wong JK, Barbour JD. Turning up the volume on mutational pressure: is more of a good thing always better? (A case study of HIV-1 Vif and APOBEC3) Retrovirology. 2008;5:26. doi: 10.1186/1742-4690-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jern P, et al. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wood N, et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vazquez-Perez JA, et al. APOBEC3G mRNA expression in exposed seronegative and early stage HIV infected individuals decreases with removal of exposure and with disease progression. Retrovirology. 2009;6:23. doi: 10.1186/1742-4690-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jin X, et al. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol. 2005;79:11513–11516. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cho SJ, et al. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol. 2006;80:2069–2072. doi: 10.1128/JVI.80.4.2069-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ulenga NK, et al. Relationship between human immunodeficiency type 1 infection and expression of human APOBEC3G and APOBEC3F. J Infect Dis. 2008;198:486–492. doi: 10.1086/590212. [DOI] [PubMed] [Google Scholar]
- 104.Biasin M, et al. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: a possible role in the resistance to HIV of HIV-exposed seronegative individuals. J Infect Dis. 2007;195:960–964. doi: 10.1086/511988. [DOI] [PubMed] [Google Scholar]
- 105.Do H, et al. Exhaustive genotyping of the CEM15 (APOBEC3G) gene and absence of association with AIDS progression in a French cohort. J Infect Dis. 2005;191:159–163. doi: 10.1086/426826. [DOI] [PubMed] [Google Scholar]
- 106.An P, et al. APOBEC3G genetic variants and their influence on the progression to AIDS. J Virol. 2004;78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Valcke HS, et al. APOBEC3G genetic variants and their association with risk of HIV infection in highly exposed Caucasians. Aids. 2006;20:1984–1986. doi: 10.1097/01.aids.0000247124.35129.e1. [DOI] [PubMed] [Google Scholar]
- 108.An P, et al. APOBEC3B deletion and risk of HIV-1 acquisition. J Infect Dis. 2009;200:1054–1058. doi: 10.1086/605644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.OhAinle M, et al. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chiu YL, et al. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 112.Chen K, et al. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol. 2006;80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Santoni de Sio FR, Trono D. APOBEC3G-depleted resting CD4+ T cells remain refractory to HIV1 infection. PLoS One. 2009;4:e6571. doi: 10.1371/journal.pone.0006571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kamata M, Nagaoka Y, Chen IS. Reassessing the role of APOBEC3G in human immunodeficiency virus type 1 infection of quiescent CD4+ T-cells. PLoS Pathog. 2009;5:e1000342. doi: 10.1371/journal.ppat.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vetter ML, et al. Differences in APOBEC3G expression in CD4+ T helper lymphocyte subtypes modulate HIV-1 infectivity. PLoS Pathog. 2009;5:e1000292. doi: 10.1371/journal.ppat.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koning FA, et al. Defining APOBEC3 Expression Patterns in Human Tissues and Hematopoietic Cell Subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peng G, et al. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110:393–400. doi: 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pion M, et al. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J Exp Med. 2006;203:2887–2893. doi: 10.1084/jem.20061519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peng G, et al. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Simon V, et al. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang W, et al. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PLoS ONE. 2008;3:e3963. doi: 10.1371/journal.pone.0003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.He Z, et al. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 124.Tian C, et al. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J Virol. 2006;80:3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yamashita T, et al. Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes Infect. 2008;10:1142–1149. doi: 10.1016/j.micinf.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 126.Yang S, Sun Y, Zhang H. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J Biol Chem. 2001;276:4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huthoff H, et al. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Russell RA, et al. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol. 2009;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang L, et al. Function analysis of sequences in human APOBEC3G involved in Vif-mediated degradation. Virology. 2008;370:113–121. doi: 10.1016/j.virol.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 131.Pery E, et al. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol. 2009;83:2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen G, et al. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J Virol. 2009;83:8674–8682. doi: 10.1128/JVI.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dang Y, et al. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol. 2009;83:8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Harjes E, et al. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nathans R, et al. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen KM, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 137.Holden LG, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Furukawa A, et al. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. Embo J. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Prochnow C, et al. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 140.Bransteitter R, Prochnow C, Chen XS. The current structural and functional understanding of APOBEC deaminases. Cell Mol Life Sci. 2009;66:3137–3147. doi: 10.1007/s00018-009-0070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mitsuyasu RT, et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 143.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bogerd HP, et al. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mangeat B, et al. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 146.Schröfelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci U S A. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu H, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li L, et al. APOBEC3G-UBA2 fusion as a potential strategy for stable expression of APOBEC3G and inhibition of HIV-1 replication. Retrovirology. 2008;5:72. doi: 10.1186/1742-4690-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yamanaka S, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ao Z, et al. Vpr14–88-Apobec3G fusion protein is efficiently incorporated into Vif-positive HIV-1 particles and inhibits viral infection. PLoS ONE. 2008;3:e1995. doi: 10.1371/journal.pone.0001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aguiar RS, et al. Vpr.A3A chimera inhibits HIV replication. J Biol Chem. 2008;283:2518–2525. doi: 10.1074/jbc.M706436200. [DOI] [PubMed] [Google Scholar]
- 152.Green LA, Liu Y, He JJ. Inhibition of HIV-1 infection and replication by enhancing viral incorporation of innate anti-HIV-1 protein A3G: a non-pathogenic Nef mutant-based anti-HIV strategy. J Biol Chem. 2009;284:13363–13372. doi: 10.1074/jbc.M806631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goila-Gaur R, et al. Targeting APOBEC3A to the viral nucleoprotein complex confers antiviral activity. Retrovirology. 2007;4:61. doi: 10.1186/1742-4690-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marin M, et al. Human immunodeficiency virus type 1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J Virol. 2008;82:987–998. doi: 10.1128/JVI.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rose KM, et al. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res Hum Retroviruses. 2005;21:611–619. doi: 10.1089/aid.2005.21.611. [DOI] [PubMed] [Google Scholar]
- 156.Bogerd HP, et al. Equine infectious anemia virus resists the antiretroviral activity of equine APOBEC3 proteins through a packaging-independent mechanism. J Virol. 2008;82:11889–11901. doi: 10.1128/JVI.01537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bogerd HP, et al. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen H, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 159.Gooch BD, Cullen BR. Functional domain organization of human APOBEC3G. Virology. 2008;379:118–124. doi: 10.1016/j.virol.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kinomoto M, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bogerd HP, et al. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364:486–493. doi: 10.1016/j.virol.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Doehle BP, Schafer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 163.Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem. 2006;281:36624–36631. doi: 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- 164.Langlois MA, et al. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dang Y, et al. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J Biol Chem. 2008;283:11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dang Y, et al. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Han Y, et al. APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication. PLoS Pathog. 2008;4:e1000095. doi: 10.1371/journal.ppat.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li MM, Wu LI, Emerman M. The Range of Human APOBEC3H Sensitivity to Lentiviral Vif Proteins. J Virol. 2009 doi: 10.1128/JVI.01344-09. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]