Abstract
More than 200 phosphorylated 14-3-3-binding sites in the literature were analysed to define 14-3-3 specificities, identify relevant protein kinases, and give insights into how cellular 14-3-3/phosphoprotein networks work. Mode I RXX(pS/pT)XP motifs dominate, although the +2 proline residue occurs in less than half, and LX(R/K)SX(pS/pT)XP is prominent in plant 14-3-3-binding sites. Proline at +1 is rarely reported, and such motifs did not stand up to experimental reanalysis of human Ndel1. Instead, we discovered that 14-3-3 interacts with two residues that are phosphorylated by basophilic kinases and located in the DISC1 (disrupted-in-schizophrenia 1)-interacting region of Ndel1 that is implicated in cognitive disorders. These data conform with the general findings that there are different subtypes of 14-3-3-binding sites that overlap with the specificities of different basophilic AGC (protein kinase A/protein kinase G/protein kinase C family) and CaMK (Ca2+/calmodulin-dependent protein kinase) protein kinases, and a 14-3-3 dimer often engages with two tandem phosphorylated sites, which is a configuration with special signalling, mechanical and evolutionary properties. Thus 14-3-3 dimers can be digital logic gates that integrate more than one input to generate an action, and coincidence detectors when the two binding sites are phosphorylated by different protein kinases. Paired sites are generally located within disordered regions and/or straddle either side of functional domains, indicating how 14-3-3 dimers modulate the conformations and/or interactions of their targets. Finally, 14-3-3 proteins bind to members of several multi-protein families. Two 14-3-3-binding sites are conserved across the class IIa histone deacetylases, whereas other protein families display differential regulation by 14-3-3s. We speculate that 14-3-3 dimers may have contributed to the evolution of such families, tailoring regulatory inputs to different physiological demands.
Keywords: 14-3-3 protein, AGC protein kinase, Ca2+/calmodulin-dependent protein kinase, disrupted-in-schizophrenia 1 (DISC1), evolution
Abbreviations: AANAT, serotonin acetyltransferase; AGC, protein kinase A/protein kinase G/protein kinase C family kinase; AMPK, AMP-activated protein kinase; BAD, Bcl-XL/Bcl-2-associated death promoter; CaMK, Ca2+/calmodulin-dependent protein kinase; CDK5, cyclin-dependent kinase 5; DIG, digoxigenin; DISC1, disrupted-in-schizophrenia 1; DSTT, Division of Signal Transduction Therapy; EST, expressed sequence tag; FOXO, Forkhead box O; GLUT4, glucose transporter 4; GST, glutathione transferase; HA, haemagglutinin; HAP1A, Huntingtin-associated protein 1A; HDAC, histone deacetylase; HEK, human embryonic kidney; KLC, kinesin light chain; MARK, microtubule affinity-regulating kinase; PI4K, phosphoinositide 4-kinase; PKB, protein kinase B; PKC, protein kinase C; PP2A, protein phosphatase 2A; RSK, ribosomal S6 kinase; YAP1, yes-associated protein 1
INTRODUCTION
14-3-3s are dimeric proteins that dock on to phosphorylated serine and threonine residues in hundreds of intracellular target proteins, including enzymes and structural components of metabolism, vesicle and protein trafficking, cytoskeletal regulation, DNA replication, transcription, translation, membrane receptors, reversible ubiquitination and phosphorylation, and effectors of small GTPases [1–11]. Understanding when and how 14-3-3s impact on these targets therefore offers a fantastic opportunity to gain regulatory insights into many areas of eukaryotic biology. We and others are developing differential proteomics methods to characterize the large-scale shifts in the subsets of targets that become phosphorylated and bind to 14-3-3s when specific signalling pathways are activated [12,13]. To fully interpret these global data, however, we must also have a precise understanding of the specificities with which 14-3-3s engage with individual targets.
A 14-3-3 dimer comprises two curved L-shaped monomers, arranged with diagonal symmetry to generate a boat-shaped central groove. The inner corner of each L contains a pocket for binding to phosphorylated residues on target proteins [14,15]. Inspection of the few 14-3-3-binding sites known 13 years ago indicated R(S)X1,2(pS)X(P) as a 14-3-3-binding motif [16,17], whereas screens for optimal binding of synthetic (phospho)peptides to 14-3-3s revealed two canonical motifs, namely mode I RSX(pS)XP and mode II RX(F/Y)X(pS)XP with further subtle preferences and negative determinants for the X residues ([18,19] and http://scansite.mit.edu). There are also mode III sites in which the phosphorylated residue is the penultimate residue in the C-terminal tail. The best-characterized mode III site is in the plant plasma membrane proton pump, where a 14-3-3 dimer binds to the phosphorylated cytoplasmic tails of two adjacent subunits, such that three 14-3-3 dimers stabilize the active hexameric pump [15]. However, this case of a 14-3-3 dimer acting as an adapter that links two phosphorylated proteins appears to be an exception rather than the rule. More commonly, there are two tandem phosphorylated 14-3-3-binding sites in the same protein. The affinity of 14-3-3 binding to dual sites can be tens of fold higher than the single sites, due to co-operative binding [14,19].
In the present study, we collected all of the 14-3-3-binding sites that we could find in the literature to see whether we could define the rules of engagement of 14-3-3s any further. Few publications fulfil every criterion needed to assign a 14-3-3-binding site with the highest confidence. Such criteria could include identification of the in vivo phosphorylated residue, elimination of 14-3-3 binding by dephosphorylation and/or mutagenesis of candidate sites, functional correlation between phosphorylation and 14-3-3 binding in vivo and in vitro, and structural analysis of the target–14-3-3 interaction. It is easy to make mistakes. Loss of 14-3-3 binding when a phosphorylatable residue is mutated could mean that the targeted phosphorylated residue binds directly to 14-3-3, or it could be part of a hierarchical system such that its phosphorylation is essential for phosphorylation of the real 14-3-3-interacting site elsewhere on the protein. For example, a S256A mutation of FOXO3 [Forkhead box O3; also known as FKHR (forkhead in rhabdosarcoma)] inhibits phosphorylation of the Thr24 14-3-3-binding site [20]. We decided not to prejudge the collection of information, except where there were clear uncertainties, but accept that the quality of the dataset is likely to be diluted by wrongly assigned sites. Our aims were to look for patterns in the data, draw inferences about how 14-3-3 proteins work, make predictions that might help in finding novel 14-3-3-binding sites, identify research questions, and provide the foundation of a database that can be updated in future. We checked and corrected one obvious anomaly by identifying two 14-3-3-binding sites on Ndel1 (previously known as NUDEL), instead of the three phosphoSer/Thr-Pro sites that had been previously proposed to interact with 14-3-3 [21]. The deregulation of Ndel1 and associated proteins is strongly implicated in disorders of brain development and cognition, including schizophrenia, and our findings change perspectives on how 14-3-3 impacts this system. More generally, our survey highlights the special mechanical, signalling and evolutionary properties that emerge from the configuration of a 14-3-3 dimer binding to a doubly phosphorylated target.
EXPERIMENTAL
Plasmid construction
The coding region for human Ndel1 (NCBI accession number AY004871.1; GI:12043568) was amplified from IMAGE consortium EST (expressed sequence tag) clone 4820855 using the primers 5′-GAGGATCCATGGATGGTGAAGATATACCAGATTTTTCAAG-3′ and 5′-GTGCGGCCGCTCACACACTGAGAGGCAGCATACC-3′. The PCR product was digested with BamH1 and Not1 and ligated into the pCMV5.HA expression vector [pCMV5 containing an HA (haemagglutinin) tag sequence] for expression of the tagged fusion protein HA–Ndel1. A P322T mutation originating from the EST was mutated back to proline using the mutagenic primers 5′-CAGTAAACGGCTTTGACCCCGCTCCTCCTCCTCC-3′ and 5′-GGAGGAGGAGGAGCGGGGTCAAAGCCGTTTACTG-3′ so that the sequence exactly matches the NCBI accession number AY004871.1; GI:12043568, and the resulting protein matches the Swiss-Prot entry Q9GZM8. S251A and S336A mutants, double-mutant S251A/S336A, triple-mutant S198A/T219A/S231A and the 5×alanine mutant (S198A/T219A/S231A/S251A/S336A) of Ndel1 were generated by PCR mutagenesis using KOD Hot Start DNA polymerase (Novagen). Bacterial expression plasmids expressing GST (glutathione transferase)-tagged wild-type and mutant versions of Ndel1 were generated by subcloning the BamH1/Not1 insert from pCMV5.HA plasmids into pGEX6P-1 (Amersham).
Cell culture, lysis and immunoprecipitations
Human HEK (human embryonic kidney)-293 cells were cultured on 10-cm diameter dishes in medium containing 10% (v/v) FBS (foetal bovine serum), and, at 24 h after transfection with the indicated plasmids, cells were rinsed with ice-cold PBS and lysed in 0.3 ml of ice-cold lysis buffer [50 mM Tris/HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1% (v/v) Triton X-100, 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1 μM microcystin-LR, 0.1% 2-mercaptoethanol, 1 mM benzamidine, 0.2 mM PMSF and one ‘Complete’ protease inhibitor cocktail tablet (Roche) per 50 ml]. For immunoprecipitations, sheep anti-HA antibody (raised against the synthetic peptide YPYDVPDYA) at 3 μg of antibody/mg of lysate was first coupled to Protein G–Sepharose (30 μl of 50% slurry in lysis buffer) by mixing at room temperature (20 °C) for 1 h. Lysate (3 mg) was then added and mixed for a further 2 h at 4 °C. Cells were washed twice with 50 mM Tris/HCl (pH 7.5) and 500 mM NaCl, and twice with 50 mM Tris/HCl (pH 7.5), 1 mM EDTA and 0.1% 2-mercaptoethanol, pelleting Sepharose in between washes at 12000 g. Immunoprecipitates were extracted into SDS-sample buffer, and separated by SDS/PAGE (10% gels).
Antibodies and kinases
Phospho-specific antibodies were raised against the following peptide sequences derived from Ndel1: TPSARIpSALN [residues 245–254 where pS represents phosphorylated Ser251 (pSer)]; and CGSSRPSpSAPGMLP (cysteine plus residues 330–342 where pS represents pSer336). Peptides were coupled separately via the N-terminus and added cysteine residue respectively to BSA and keyhole-limpet haemocyanin, mixed and injected into sheep at Diagnostics Scotland. The antibodies were affinity purified by the DSTT (Division of Signal Transduction Therapy, University of Dundee, Dundee, Scotland, U.K.) on phosphopeptide–Sepharose columns and the flow-throughs collected. In Western blot analysis, the phospho-specific antibodies were used at 1 μg/ml containing 10 μg/ml of the unphosphorylated peptide. Mouse anti-HA and sheep anti-HA antibodies were raised against the peptide YPYDVPDYA. Sheep anti-GST antibody was raised against GST expressed from pGEX-4T and used at 0.1 μg/ml. The kinases CDK5 (cyclin-dependent kinase 5)/p35 and Aurora A were from Upstate, while human Aurora B was produced in the DSTT.
14-3-3 overlays and Western blot analysis
Membranes were incubated in 50 mM Tris/HCl (pH 7.5), 0.15 M NaCl and 0.2% Tween 20 containing 5% (w/v) dried skimmed milk powder (Marvel) and were immunoblotted at 4 °C for 16 h using the antibodies indicated. Detection was performed using HRP (horseradish peroxidase)-conjugated secondary antibodies (Promega) and ECL® (enhanced chemiluminescence reagent; Amersham Biosciences) for all Western blots of recombinant proteins and DIG (digoxigenin)–14-3-3 overlays, which use DIG-labelled 14-3-3 (a mix of the Saccharomyces cerevisiae BMH1 and BMH2 14-3-3 isoforms) in place of primary antibody, as described in [12].
In vitro phosphorylation
GST–Ndel1 proteins were phosphorylated in vitro with the kinases indicated for 30 min at 30 °C. The reaction buffer contained 25 mM Tris/HCl (pH 7.5) and 1 mM EDTA, and reactions were started by adding 0.1 mM ‘cold’ ATP/10 mM magnesium acetate. SDS sample buffer was added to terminate the reactions.
RESULTS AND DISCUSSION
Most reported 14-3-3-binding sites conform to mode I motifs, whereas plants reveal a mode I variant characterized by leucine at −5 and serine at −2
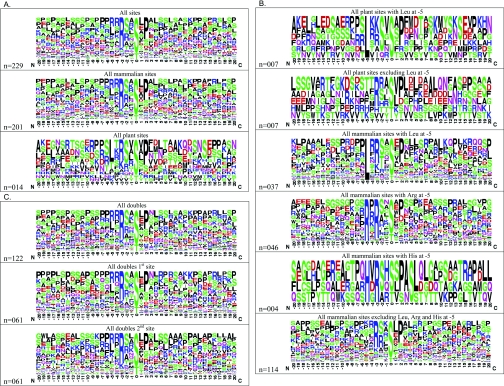
We collated details of sites on target proteins that have been reported to bind directly to 14-3-3 proteins (Supplementary Table 1 at http://www.BiochemJ.org/bj/427/bj4270069add.htm) and searched these data for notable features (Figure 1 and Supplementary Table 2 at http://www.BiochemJ.org/bj/427/bj4270069add.htm). A WebLogo frequency plot of the combined >200 reported sites (Figure 1A) shows that mode I RXX(pS/pT)XP motifs dominate, and leucine and arginine are the most common residues in the −5 position. Serine and proline are the most common residues in the extended sequences flanking the 14-3-3-binding sites. However, the proline residue at +2 is found in only ~50% of target sites. Structural data show that the proline residue at +2 in the canonical motifs twists the peptide back out of the docking site [19,22]. Other residues may also allow such a twist, but it is not clear whether this is always essential for 14-3-3 binding.
Figure 1. WebLogo analyses of 14-3-3-binding sites of the proteins listed in Supplementary Table 1.
(A) The sequences containing each of the phosphorylated 14-3-3-binding sites listed in Supplementary Table 1 were aligned relative to one another, centred on the phosphorylated residue with 20 residues either side (Supplementary Table 2). The sequence logo was created using WebLogo Version 2.8.2 (http://weblogo.berkeley.edu/; [82,83]), with ‘frequency plot’ selected. The height of symbols for each position in the sequence indicates the relative frequency of each amino acid at that position. Also shown are separate sequence logos for 14-3-3-binding sites from mammalian and plant species respectively. (B) WebLogo plots of 14-3-3-binding sites with and without leucine, arginine and histidine residues in the −5 position. (C) WebLogo plots of 14-3-3-binding sites for proteins where pairs of sites have been identified.
Few reported 14-3-3-binding sites have a +1 proline residue, which contrasts with phosphoproteomic studies of cell lysates and subcellular fractions, where phosphoSer-Pro is the most commonly reported phosphorylation motif overall [23]. Proline-directed kinases therefore do not phosphorylate 14-3-3-binding sites, although they can play a regulatory role by phosphorylating the 14-3-3 themselves [24]. Similarly, no reported 14-3-3-binding sites conform to the canonical consensus for phosphorylation by casein kinase II, [namely (pS/pT)(X1)(X2)(D/S/pS) where X1 is not proline], which is probably the second most common type of motif in the mammalian phosphoproteome as a whole [25]. Generally, it has been reported that glutamate and aspartate do not provide good phosphomimetic residues with respect to 14-3-3 binding to target proteins, although there are exceptions such as synaptopodin 2/myopodin [26].
While the optimal mode I and II 14-3-3-binding motifs were defined using phosphopeptides [18,19], target sites in proteins must satisfy the specificity requirements for both 14-3-3s and the protein kinases that create the sites in the first place. Theoretically, kinases and 14-3-3s could each use distinct specificity determinants to home-in on the same site. The data indicate congruence, however, in that basic residues in positions −3 to −5 relative to the phosphorylated site are found in most 14-3-3-binding sites, and basophilic kinases in the AGC [protein kinase A/protein kinase G/PKC (protein kinase C) family kinase] and CaMK (Ca2+/calmodulin-dependent protein kinase) subfamilies of the kinome are most commonly implicated in the phosphorylation of 14-3-3 binding sites [27–29].
Linking specific kinases firmly to target sites is challenging. However, it should be possible to sort 14-3-3-binding sites into motif subsets to aid the task of defining relevant protein kinases. One striking subset comprises the plant 14-3-3-binding sites, for which the distinct motif LX(R/K)SX(pS/pT)XP is most prevalent (Figures 1A and 1B, and Supplementary Table 2). This motif resembles a mode I site, except that that either an arginine or lysine residue can be in the −3 position and there is an additional leucine residue at −5 and a serine residue at −2. Three plant proteins with a leucine at −5 in their 14-3-3-binding proteins, namely nitrate reductase (NIA1 and NIA2), TPS5 (trehalose-phosphate synthase 5) and F2KP (6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase), have been proposed to be co-ordinately regulated in leaves in the dark [30], raising the question of whether the brassinosteroid receptor (BZR1) and vacuolar potassium channel (KCO1) with similar motifs might also be targets of the same kinase(s). A similar motif was also identified in 30 mammalian 14-3-3-binding sites, except that a leucine residue at −5 was accompanied by either a serine or threonine residue at −2 (Figure 1B). Several CaMK enzymes exhibit a preference for a leucine residue at −5, including mammalian PKD (protein kinase D), Chk (checkpoint kinase) 1, Chk2, AMPK (AMP-activated protein kinase), AMPK-related kinases such as C-TAK1/MARK3 (microtubule affinity-regulating kinase 3), and the plant AMPK-related SrRKs [31]. Further experiments are needed to clarify any reason for the apparent coupling between a leucine residue at −5 and a serine residue at −2 in the plant 14-3-3-binding proteins. Because several of the plant proteins have been studied in the context of the same dark-activated signalling pathway [30–32], it seems likely that the −5 leucine and −2 serine guide phosphorylation by the relevant kinase(s), rather than subsequent selection of 14-3-3 isoforms. Defining the points of contact of the −5 leucine with the kinases and 14-3-3s should be informative.
Another subset of 36 14-3-3-binding sites has an arginine residue in the −5 position, which is always accompanied by an arginine residue at −3 (Figure 1B). Arginine residues at −5 and −3 are determinants for phosphorylation by enzymes including PKB (protein kinase B)/Akt, PIM kinases and RSKs (ribosomal S6 kinases), although the presence of these residues does not guarantee phosphorylation by these kinases, and each kinase has further selection criteria [23,32]. None of the plant 14-3-3-binding sites has an arginine at −5. This could reflect the small sample size for the plant proteins, or might be biologically meaningful, given that plants do not have direct counterparts of PKB/Akt, PIM kinases or RSKs [33,34], although they have many other CaMK and AGC protein kinases [35]. We note that PKB/Akt has been assigned responsibility for phosphorylating a number of 14-3-3-binding sites that do not conform to the canonical consensus for this kinase. In the case of the mammalian WW-domain transcriptional regulator YAP1 (yes-associated protein 1), the 14-3-3-binding sites have a histidine residue at −5 and an arginine residue at −3, and phosphorylation of YAP1 was originally attributed to PKB/Akt [36], but later discovered to be catalysed by the Warts/Lats kinases in the NDR branch of the AGC kinases [37,38], as is the related protein TAZ (WWTR1). FAM82A2 and SRPK2 also have reported 14-3-3-binding sites with a histidine residue at −5 (Figure 1B and Supplementary Table 1).
As the dataset of 14-3-3 target sites grows, the sorting into different motif subsets should become more obvious, facilitating the task of matching sites with potential protein kinases.
Mode III 14-3-3-binding sites
Unlike most targets, 14-3-3s activate the plant plasma membrane H+-ATPase by binding to a so-called C-terminal mode III binding motif SW(pT)X-COOH in which the phosphorylated residue is the penultimate threonine residue in the C-terminal autoinhibitory domain [15]. It has been suggested that mode III 14-3-3-binding sites are more widespread in nature [39,40], with the mammalian IL (interleukin)-9 receptor α-chain [41], two-pore-domain potassium channels TASK1, 3 and 5 [42], the nicotinic acetylcholine α4β2 receptor [43] and HAP1A (Huntingtin-associated protein 1A) [44] being proposed as mode III interactors. However, to our knowledge, phosphorylation of the penultimate residue has not been demonstrated directly for the channels and receptors, whereas the threonine residue of the mode III site on HAP1A is restricted to the rat version of this protein. Some proteins, such as AANAT (serotonin acetyltransferase) have one mode I and one mode III site [45]. While the mode III sites are at the C-termini, we note that several proteins have 14-3-3-binding sites that are close to their N-termini (Supplementary Tables 1 and 2).
14-3-3 binding to unphosphorylated proteins
Generally, 14-3-3s do not bind appreciably to unphosphorylated proteins, with the exception of the ExoS, which is injected into host mammalian cells by the pathogenic bacterium Pseudomonas aeruginosa and contains an novel binding motif G421LLDALDLAS430, in which the hydrophobic residues (Leu422, Leu423 and Leu428) are essential for binding to cellular 14-3-3s and pathogenesis [46,47]. Similarly, the rolB protein from the hairy root bacterium Agrobacterium rhizogenes interacts with 14-3-3 inside Arabidopsis cells, possibly in a phosphorylation-independent interaction [48]. Phosphorylation-independent interactions between physiological targets and 14-3-3s have also been reported [49,50], although these studies do not exclude the possibility that the reported targets interact with an unknown intermediary protein that is phosphorylated and binds to 14-3-3s.
Two tandem 14-3-3-binding sites within disordered regions and straddling functional domains
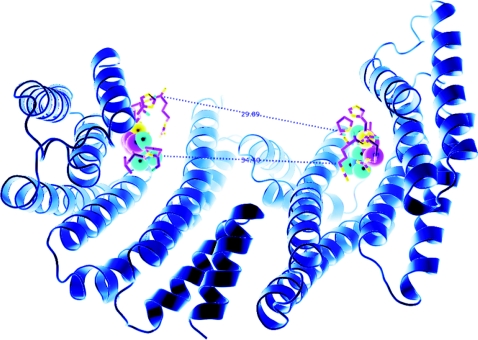
The 14-3-3s are dimeric proteins with diagonal symmetry, such that the two phosphate-binding sites are diagonally opposite each other and ~34.4 Å (1 Å=0.1 nm) apart across the floor of the binding groove for the human 14-3-3ζ dimer (Figure 2), although the spacing may be greater due to some flexibility between the 14-3-3 monomers [51]. In theory, 34 Å could be bridged by a minimum of 15 residues from one phosphorylated residue to the other (inclusive) in fully extended conformation, although more would be needed for a dimer that adopts a more open conformation [51]. For two tandem phosphorylated sites to dock into a single 14-3-3 dimer, the target protein must thread out and in, in anti-parallel orientation, as has been shown in the structure of the 14-3-3 bound to a doubly phosphorylated peptide derived from PKCε [14].
Figure 2. Structure of a 14-3-3 dimer, based on that within the AANAT/14-3-3 structure [22] in which the two phosphate-binding sites are 34.4 Å apart.
If we consider that the two binding sites are arranged in an antiparallel orientation, it is also useful to consider the distance of ~30 Å between the C-α atom of the proline of a six-amino-acid phosphopeptide [RRH(pT)LP derived from AANAT] bound to one side of the binding groove and the C-α atom of the N-terminal residue of the second peptide (arginine). This distance could be covered with a peptide containing approx. 8 amino acids in fully extended conformation, which would make 8+3+4=15 residues from one phosphorylated residue to the other (inclusive of the phosphorylated residues). Allowing (say) 4 amino acid residues for turns, would give a minimum of ~19 amino acid residues. This Figure is provided courteousy of Thomas Obsil (Charles University in Prague, Prague, Czech Republic).
Thus far, approx. 50 proteins have been reported to contain two phosphorylated 14-3-3-binding sites (Figure 1C and Supplementary Tables 1 and 2). For a few targets, there is sufficient structural and kinetic binding data to support the hypothesis that the two phosphorylated sites on the same target bind to either side of a 14-3-3 dimer [14]. It seems reasonable to propose that the configuration of a 14-3-3 dimer engaging with two tandem phosphorylated sites on the same target might be a more general phenomenon. If so, defining the locations of the two sites should give clues about how the 14-3-3 dimer acts mechanically to change the function of the target.
The paired sites can be considered in two broad groups: one set has a short stretch of 40 amino acid residues or less between the phosphorylated sites, which are in regions of predicted disorder in N- and C-terminal tails or between functional domains on the protein, and it may be that the binding of a 14-3-3 dimer imposes order [52,53]. This class includes enzymes whose activities are altered by 14-3-3 binding. For tyrosine hydroxylase, for example, biophysical measurements indicate that the bound 14-3-3 dimer acts as a lever that forces a conformational change which activates the enzyme [54]. In other proteins, the dual 14-3-3-binding sites are further apart and straddle either side of one or more folded domains. Structural analyses show how the two 14-3-3-binding sites at phosphoThr28 and phosphoSer193 of FOXO4 dock its forkhead domain into the central channel of the 14-3-3 dimer, occluding the DNA-binding interface of the transcription factor without causing any dramatic conformational change [55]. There are sometimes much longer distances between two phosphorylated sites on the listed targets, 360 residues for TBC1D1, for example [56], which is too much for the intervening sequences to be enclosed within the central groove of 14-3-3s. In the absence of structural data, one can speculate that the 14-3-3 dimer provides a partial mask, or perhaps the 14-3-3 dimer pins backs the flanking regions to better present the intervening domain for other interactions.
In many papers that define one 14-3-3-interaction motif, there are also data suggestive of a second unidentified motif on the same target. For example, Demmel et al. [57] pinpoint phosphoSer396 as essential for 14-3-3 binding to yeast PI4K (Pik1p; phosphoinositide 4-kinase), and their data indicate that a second unknown binding site exists in the catalytic region of this enzyme. In cases where one phosphate dominates for high-affinity binding, it is possible that a second lower-affinity-binding site on the same target may be needed for the target to slot into place properly so that the 14-3-3 dimer can impose a functional change in the target, as proposed in the ‘gatekeeper’ hypothesis of Yaffe [58]. These secondary low-affinity phosphorylation sites can be difficult to spot by mutagenesis analysis if they do not contribute much to the affinity of binding to 14-3-3s.
14-3-3 dimers as logic gates and coincidence detectors that integrate incoming signals from two different protein kinases
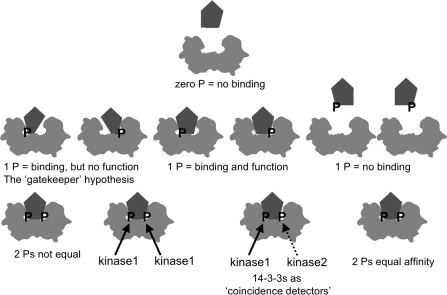
As well as enabling a 14-3-3 dimer to act mechanically as a lever, clamp or mask, docking on to two phosphorylated residues confers a 14-3-3 with the special signalling properties of a digital logic gate that converts two inputs into a single output. If either phosphorylated site is sufficient to generate an output, the system would be an ‘OR’ gate, whereas if two phosphorylated sites must be engaged before the 14-3-3 can generate a mechanical action on the target, the 14-3-3 would be an ‘AND’ gate. An ‘AND’ gate would suppress noise because no action would occur when only one 14-3-3-binding site is phosphorylated, and be a coincidence detector if each site is phosphorylated by a different protein kinase and/or dephosphorylated by a different protein phosphatase. Figure 3 summarizes the variety of configurations in which a 14-3-3 dimer can engage with one or two phosphorylated sites on target proteins.
Figure 3. 14-3-3 dimers read digital codes: a model exploring the signalling implications of 14-3-3 binding to two phosphorylated sites on the same target.
The term ‘digital code’ usually refers to a binary system of zeroes and ones. Because 14-3-3s are dimeric, they can be considered as a coupled binary device that can distinguish patterns of zero, one or two phosphorylated target sites. Generally, zero phosphates means no binding, with the exception of ExoS from P. aeruginosa [47]. One phosphate is sometimes enough to create a sufficiently high-affinity 14-3-3-binding site, and sometimes not. But even when one phosphate dominates for high-affinity binding, sometimes a second lower-affinity-binding site is needed for the target to slot in properly. These second low-affinity phosphorylation sites can be difficult to spot by mutagenesis analysis if they do not contribute much to the affinity of binding to 14-3-3s, but secondary sites may have to be in place for the 14-3-3 to impose a functional change in the target (the ‘gatekeeper’ hypothesis; [58]). The two phosphorylated sites can be inserted by the same kinase, or in one or two cases, different kinases have been found to phosphorylate each site on the same target. The configuration with two kinases turns a 14-3-3 dimer into a coincidence-detecting device where two different signalling inputs have to be received before 14-3-3s can bind to the target and a functional change is induced. In addition, multisite phosphorylation at other sites can enhance or inhibit 14-3-3 interactions, either by influencing the phosphorylation status of the 14-3-3-binding sites, by direct steric modulation of the binding to 14-3-3s, and/or by phosphorylation of the 14-3-3s themselves.
Several authors have highlighted cases where two tandem phosphorylated sites can be phosphorylated by distinct protein kinases, namely TBC1D1, MARK2/Par-1b, BAD (Bcl-XL/Bcl-2-associated death promoter) and myopodin [26,56,59,60]. For example, the two 14-3-3-binding sites on the apoptosis regulator BAD can be phosphorylated variously by ERK1/2 (extracellular-signal-regulated kinase 1/2)-activated kinases, PKB/Akt and PKA. Phosphorylation of either 14-3-3-binding site is sufficient to trigger dissociation of BAD from the anti-apoptotic protein Bcl-XL, inhibiting the pro-apoptotic activity of BAD. For BAD, therefore, the 14-3-3 appears to act as an ‘OR’ gate because phosphorylation of either 14-3-3-binding site induces the interaction and thereby inhibits apoptosis [59].
As a start to breaking down the task of assigning each 14-3-3–target interaction into ‘AND’ gate, ‘OR’ gate and/or ‘coincidence detector’ categories, we examined the 50 targets with paired 14-3-3-binding sites to determine whether they were pairs of similar or different motifs. Several proteins contained RXRXXpS at both sites, including mammalian ADAM2 (a disintegrin and metalloproteinase 2), Mdm2 (murine double minute 2), TBC1D4, FOXO1, FOXO2, FOXO3, BAD and Drosophila FOXO (Supplementary Tables 1 and 2). For others with a single RXRXXpS, the second site is a LXRXXpS motif for TSC2 (tuberous sclerosis complex 2), 2FP36L1 and CDKN1B; IXRXXpS for A-, B- and C-RAF; MXRXXpS for TBC1D1; and QXRXXpS for AKT1S1. There was no obvious pattern for +2 proline residues: some paired sites had proline at +2 in both sites, whereas others had one proline at +2, or none (Figure 1C and Supplementary Tables 1 and 2). While other patterns may emerge as the datasets grow, currently most proteins have different motifs at each 14-3-3-binding site, indicating that the tandem sites on these proteins might be phosphorylated by distinct kinases and operate in ‘coincidence detector’ mode.
More complicated relationships between inputs and outputs could be generated if it matters which of the two sites is phosphorylated first (sequential logic), the 14-3-3-binding sites are under hierarchical control by other post-translational modifications of the target, and if other post-translational modifications inhibit the interaction. For example, the interaction of 14-3-3 proteins with the plant plasma membrane H+-ATPase is inhibited by tyrosine phosphorylation [61], whereas dephosphorylation of Ser376 in response to IR radiation enables 14-3-3s to bind to the nearby phosphoSer378 on p53 [62], and acetylation at Lys9 or Lys10 enhances the phosphoserine-dependent binding of 14-3-3s to histone H3 [63]. In these ways, binding of 14-3-3s will depend on the merging of signals from multiple post-translational modifications that converge on the target.
Multi-protein families that bind to 14-3-3s
14-3-3s, particularly residues within the central groove, are highly conserved in all eukaryotes. Thus far, however, only one pair of protein orthologues share a conserved 14-3-3-binding site across the animal and fungal Kingdoms, namely the Golgi trafficking regulator PI4K (mammalian IIIβ isoform and Saccharomyces cerevisiae Pik1) (Supplementary Table 1). While the Cdc25 (cell division cycle 25) cell-cycle-regulating protein phosphatases in mammalian cells and the fission yeast bind directly to 14-3-3s, the precise site has not been defined in the yeast protein [64] (Supplementary Table 1). Mammalian and plant forms of fructose 2,6-bisphosphate kinase/phosphatase interact with 14-3-3s, although the plant and animal enzymes differ considerably in their overall architectures and biological contexts [65,66].
As summarized in Supplementary Table 1, the class IIa HDACs (histone deacetylases; comprising HDACs 4, 5 6 and 9) share identical 14-3-3-binding sites that regulate their nuclear–cytoplasmic shuttling and are phosphorylated by various protein kinases, including AMPK [67]. Similarly, the 14-3-3-binding sites are conserved within FOXO transcription factors (1, 3 and 4) [55], and these families presumably evolved with differences in other structural and functional aspects of the proteins.
In contrast, 14-3-3s display distinct modes of binding to two related Rab GTPase-activating proteins, namely TBC1D4 (AS160) and TBC1D1, involved in trafficking of the GLUT4 (glucose transporter 4) transporter to the cell surface for uptake of glucose into cells. Both AS160 and TBC1D1 contain two 14-3-3-binding sites straddling either side of the PTB2 (phosphotyrosine-binding domain 2) domain on these proteins. The phosphoThr642 14-3-3-binding site on AS160 is similar to phosphoThr596 on TBC1D1, and these residues appear to be phosphorylated by PKB/Akt in response to insulin and IGF1 (insulin-like growth factor 1). In contrast, the phosphoSer341 site on AS160 is distinct from the phosphoSer237 14-3-3-binding site on TBC1D1, with the latter being phosphorylated by AMPK and/or enzyme with similar specificity [56]. We hypothesize, therefore, that differential expression and regulation of AS160 and TBC1D1 may help explain why GLUT4 trafficking is more or less sensitive to insulin and AMPK activators in different tissue types.
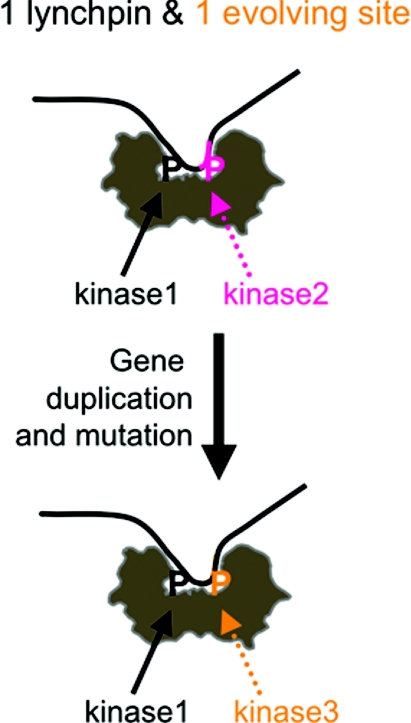
TBC1D1 is the more ancient protein [56], raising the question of how the phosphoSer237 region of TBC1D1 transmuted into phosphoSer341 of AS160. In one possible scenario, we propose that the dimeric 14-3-3 may have facilitated this evolutionary event (Figure 4). Suppose that a copy of TBC1D1 arose by gene duplication. In the copy, if one 14-3-3-binding site (Thr642/Thr596) remained unchanged, this might provide ‘good enough’ regulation by 14-3-3 to avoid serious loss of control, thereby allowing the second site to vary. In time, a new and useful consensus for a different protein kinase arose at the second site, and the resulting AS160 became a paralogue with distinct regulatory properties from the original TBC1D1.
Figure 4. Hypothesis for the role of 14-3-3 dimers in driving evolutionary change of 14-3-3-binding sites in paralogous proteins.
If a copy of a 14-3-3-binding protein were to arise by gene duplication, we hypothesize that because the 14-3-3s are dimers, the function of the copy could be kept under sufficient control if one 14-3-3-binding site remained unchanged ‘the lynchpin site’, while allowing sequence divergence to occur at the second 14-3-3-binding site. If a useful consensus for phosphorylation by a different kinase were to arise at the evolving site, the result would be the generation of a paralogue with distinct regulatory inputs from the original. Whether or not 14-3-3s have actually driven evolution in this way, this theory should stimulate experimentation to compare the regulatory details of 14-3-3 binding to paralogous members of multi-protein families.
Physiologically, the availability of differentially regulated target paralogues could allow the signalling inputs into a given process to be tailored to suit the particular demands of different organs and tissues. We note that Supplementary Table 1 lists representatives of several multi-protein families. In the large 14-3-3-phosphoproteomics studies, further protein families are also being identified to display affinity for binding to 14-3-3 [1–13]. Comparing the regulatory details for 14-3-3 binding to each representative of these protein families will be an interesting focus for future research.
Variants of a protein can also be generated by alternative splicing of mRNA transcripts. We note that in the 14-3-3-binding phosphoproteome there are at least two cases where a 14-3-3-binding site exists in one splice variant of a protein, but not another. Thus the KCN2 gene generates many forms of KLC1 (kinesin light chain 1) with different C-termini due to differing exon selection that may specify loading of different cargoes on to the kinesin motor complex. A residue equivalent to the 14-3-3-binding phosphoSer582 in KLC2 is found only in KLC1 variants expressing exon 17 [68]. Similarly, only variants of tumour protein D52 that express exon 6 contain the 14-3-3-binding site identified on this protein [69].
14-3-3 binds to phosphoSer251 and phosphoSer335 in the DISC1 (disrupted-in-schizophrenia 1)-interacting site of human Ndel1
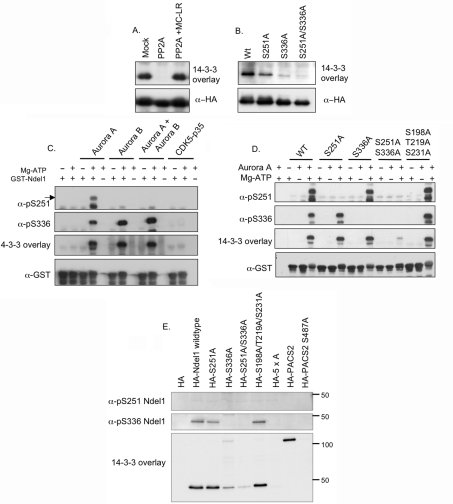
One obvious anomaly in our survey was Ndel1 (formerly named NUDEL), for which mutation of three residues (Ser198, Thr219 and Ser231), each with a proline residue at +1 and phosphorylated by the proline-directed kinase CDK5/p35, had been reported to prevent Ndel1 binding to 14-3-3s (Supplementary Table 1) [70]. We therefore re-examined the binding of 14-3-3 to recombinant Ndel1 expressed in HEK-293 cells, and first confirmed that HA–Ndel1 isolated from transfected HEK-293 cells binds directly to 14-3-3, and this interaction is abolished by dephosphorylation with PP2A (protein phosphatase 2A) (Figure 5A). Ndel1 has a potential mode I 14-3-3-binding site at Ser336 [SSRPS(S336*)AP, where * indicates the residue that would have to be phosphorylated to become a potential mode I site]. In addition, Ndel1 is phosphorylated by Aurora A at Ser251 [SARI(pS251)ALN], which is consistent with Ndel1 and Aurora A co-localizing at the centrosome during prophase [71] and lies within a fragment of Ndel1 (residues 189–256) that can interact with 14-3-3ε in a yeast two-hybrid assay [70]. Mutation of Ser336 to alanine markedly decreased the binding of 14-3-3, while the S251A mutation also reduced the interaction, and the double S251A/S336A mutant displayed only trace binding to 14-3-3 (Figure 5B).
Figure 5. Identification of Ser251 and Ser336 as 14-3-3-binding sites in human Ndel1.
(A) HEK-293 cells were transfected with a plasmid to express HA–Ndel1 and lysed after 24 h. HA–Ndel1 was immunoprecipitated from lysates (3 mg) and incubated in vitro for 30 min at 30 °C in the absence or presence of 50 m-units/ml PP2A with or without its inhibitor microcystin-LR (MC-LR) at 5 μM, as indicated. The samples were separated by SDS/PAGE, transferred on to nitrocellulose and probed for direct binding to DIG–14-3-3s in a far-Western overlay assay, with anti-HA immunoblot as the loading control. (B) HA–Ndel1 (Wt, wild-type), HA–S251A-Ndel1, HA–S336A-Ndel1 and HA–S251A/S336A-Ndel1 double mutant proteins were expressed in HEK-293 cells. Proteins were immunoprecipitated from lysates (3 mg) and analysed for direct binding to DIG–14-3-3s in a far-Western overlay assay with an anti-HA immunoblot as a loading control. (C) GST–Ndel1 (4 μg) expressed and purified from Escherichia coli was phosphorylated in vitro in the absence or presence of Mg-ATP with 5 units/ml Aurora A, 1 unit/ml Aurora B and 2.5 units/ml CDK5/p35 for 30 min at 30 °C. Samples (25%) of the reactions were separated by SDS/PAGE, transferred on to nitrocellulose and immunoblotted with the indicated antibodies. The arrow indicates the signal representing phosphoSer251 Ndel1. (D) Wild-type and mutant GST–Ndel1 proteins expressed and purified from E coli were phosphorylated in vitro with 5 units/ml Aurora A kinase in the absence or presence of Mg-ATP for 30 min at 30 °C. After separation by SDS/PAGE and transfer on to nitrocellulose, membranes were immunoblotted with the antibodies indicated. (E) HEK-293 cells were transfected with plasmids encoding the wild-type and indicated mutant HA-tagged Ndel1 proteins. After 24 h, cells were lysed and HA proteins were immunoprecipitated from lysates (3 mg). The immunoprecipitates were separated by SDS/PAGE, transferred on to nitrocellulose and analysed by immunoblotting with the specified antibodies. HA–PACS2 and HA–PACS2-S487A mutant proteins extracted from transfected HEK-293 cells were used as positive and negative controls respectively, for 14-3-3 binding in the overlay assay (Ser487 is equivalent to the phosphoSer437 14-3-3-binding site in [84]; Supplementary Table 1). Numbers on the right-hand side show the migration position of molecular mass standards (in kDa). 5xA represents the HA–Ndel1-S198A/T219A/S231A/S251A/S336A mutant protein.
HEK-293 cells do not have active CDK5 because they lack its activators p35 and p39, and we therefore tested the effect of in vitro phosphorylation of Ndel1 with the candidate kinases Aurora A, Aurora B and CDK5/p35. Aurora B was included because Ndel1 at the kinetochore has been shown to play a role in M-phase progression [72], and Aurora B is one of the major kinases known to regulate M-phase progression. While incubation with Mg-ATP and each of the three kinases induced an upwards band shift of the GST–Ndel1 substrate, indicative of phosphorylation, a 14-3-3-binding signal was only seen after phosphorylation by the Aurora kinases, either singly or in combination (Figure 5C). In contrast, while CDK5/p35 phosphorylates Ndel1, it did not induce 14-3-3 binding appreciably above background levels. Use of phospho-specific antibodies, and mutated GST–Ndel1, confirmed that 14-3-3 binding correlated with phosphorylation at Ser251 and Ser336, and was almost eliminated by mutation of Ser251 and Ser336 (Figure 5D). In contrast, the 14-3-3-binding signal of the Ser198/Thr219/Ser231 triple alanine mutant was comparable with the wild-type Ndel1 (Figure 5D).
Having confirmed the specificity of the anti-phosphoSer251 and anti-phosphoSer336 antibodies (Figures 5C and 5D), we checked whether phosphorylation of Ser198, Thr219 and/or Ser231 was responsible for the residual 14-3-3-binding signal seen with the S251A/S336A double mutant isolated from HEK-293 cells (Figure 5B). HA–Ndel1 was not phosphorylated on Ser251 under these cellular conditions, but binding to 14-3-3 mainly reflected the phosphorylation of Ser336. Note that Ser336 is only in isoform 1 of Ndel1 (Q9GZM8) and whether other isoforms of Ndel1 also interact with 14-3-3 is unclear.
When Ser251 and Ser336 were mutated, there was still a residual 14-3-3-binding signal that could be eliminated by also mutating Ser198, Thr219 and Ser231 into alanine residues (Figure 5E). This background signal probably explains why Toya-oka et al. [70] pinpointed phosphoSer198, phosphoThr219 and phosphoSer231 as the 14-3-3-binding sites following their exciting discoveries linking Ndel1 and 14-3-3s to the aetiology of lissenscephaly (smooth brain) [70,73]. Briefly, the N-terminal coiled-coil domain of Ndel1 binds to Lis1, a protein whose deletion or mutation causes lissencephaly due to defective migration of cortical neurons [21]. Children with deletions in Lis1 and the adjacent 14-3-3ε gene have a more severe disorder called MDS (Miller–Dieker syndrome), which is phenocopied in mice with haplodeficiency in Lis1 and 14-3-3ε [70]. Our findings place the 14-3-3 interaction at the C-terminal part of Ndel1, which has endo-oligopeptidase activity [74] and interacts with multiple protein partners including DISC1. Disruption of the DISC1 gene segregates with schizophrenia, other psychiatric disorders and cognitive variation [75–80]. 14-3-3 genes, in particular 14-3-3η, have also been implicated in schizophrenia and bipolar disorder [81]. Solving the regulation of 14-3-3 binding to Ndel1 should therefore give insights into the dynamics of processes that underlie lissencephaly and schizophrenia.
Concluding remarks
Collating all reported 14-3-3-binding sites was a valuable exercise. Even though the datasets are incomplete and flawed, tangible rules about how 14-3-3 dimers engage with their phosphoprotein targets are emerging. The running head title ‘two is the key to 14-3-3’ highlights that the collective data suggest how a 14-3-3 dimer engaging with dual phosphorylation sites has special mechanical and signalling properties, and may even have facilitated evolutionary change in target protein families. These features should have useful predictive power for dissecting the cellular regulation of the many phosphorylated proteins that have been isolated from cell extracts by various 14-3-3-affinity capture and release procedures [1–11]. Further sorting of 14-3-3-binding motifs into different categories should facilitate the daunting task of defining relevant protein kinases, and understanding how networks of hundreds of phosphoprotein–14-3-3 interactions control the behaviour of every eukaryotic cell.
A future plan is to create and maintain a database that shows the location of 14-3-3-binding sites in relation to functional domains on the target proteins, residue conservation across species and links to relevant protein kinases. Towards this goal, authors are invited to submit additions and corrections for Supplementary Table 1 to c.mackintosh@dundee.ac.uk.
Online data
AUTHOR CONTRIBUTION
Carol MacKintosh and Catherine Johnson conceived the study and wrote the paper. Catherine Johnson, Rachel Toth and David Campbell performed the biochemical experiments. Carol Mackintosh, Catherine Johnson, Sandra Crowther and Margaret Stafford collated and analysed sequences, and proposed hypotheses.
ACKNOWLEDGEMENTS
We thank Thomas Obsil (Charles University in Prague, Prague, Czech Republic) for providing Figure 2, Alastair Aitken and Sam Clokie (University of Edinburgh, Edinburgh, Scotland, U.K.), and Bob MacKintosh who helped compile sequence data, Kirsten McLeod for tissue culture support, the DSTT antibody and protein production team co-ordinated by James Hastie for purification of antibodies and bacterially expressed GST-fusion proteins, and Rachel Naismith for secretarial assistance.
FUNDING
This work was supported by the U.K. Medical Research Council via a Developmental Pathway Funding scheme award and core funding, and the companies who support the DSTT at the University of Dundee, namely AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck Serono and Pfizer.
References
- 1.Aitken A. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Bridges D., Moorhead G. B. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 2005;2005:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- 3.Darling D. L., Yingling J., Wynshaw-Boris A. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 2005;68:281–315. doi: 10.1016/S0070-2153(05)68010-6. [DOI] [PubMed] [Google Scholar]
- 4.Dougherty M. K., Morrison D. K. Unlocking the code of 14-3-3. J. Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- 5.Hermeking H., Benzinger A. 14-3-3 proteins in cell cycle regulation. Semin. Cancer Biol. 2006;16:183–192. doi: 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Mhawech P. 14-3-3 proteins: an update. Cell Res. 2005;15:228–236. doi: 10.1038/sj.cr.7290291. [DOI] [PubMed] [Google Scholar]
- 7.Michelsen K., Yuan H., Schwappach B. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 2005;6:717–722. doi: 10.1038/sj.embor.7400480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pozuelo Rubio M., Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., MacKintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzivion G., Gupta V. S., Kaplun L., Balan V. 14-3-3 proteins as potential oncogenes. Semin. Cancer Biol. 2006;16:203–213. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 10.van Heusden G. P., Steensma H. Y. Yeast 14-3-3 proteins. Yeast. 2006;23:159–171. doi: 10.1002/yea.1338. [DOI] [PubMed] [Google Scholar]
- 11.Wilker E., Yaffe M. B. 14-3-3 proteins: a focus on cancer and human disease. J. Mol. Cell. Cardiol. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Dubois F., Vandermoere F., Gernez A., Murphy J., Toth R., Chen S., Geraghty K. M., Morrice N. A., MacKintosh C. Differential 14-3-3-affinity capture reveals new downstream targets of PI 3-kinase signaling. Mol. Cell. Proteomics. 2009;8:2487–2499. doi: 10.1074/mcp.M800544-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yip M. F., Ramm G., Larance M., Hoehn K. L., Wagner M. C., Guilhaus M., James D. E. CaMKII-mediated phosphorylation of the myosin motor Myo1c is required for insulin-stimulated GLUT4 translocation in adipocytes. Cell Metab. 2008;8:384–398. doi: 10.1016/j.cmet.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Kostelecky B., Saurin A. T., Purkiss A., Parker P. J., McDonald N. Q. Recognition of an intra-chain tandem 14-3-3 binding site within PKCε. EMBO Rep. 2009;10:983–989. doi: 10.1038/embor.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ottmann C., Marco S., Jaspert N., Marcon C., Schauer N., Weyand M., Vandermeeren C., Duby G., Boutry M., Wittinghofer A., et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell. 2007;25:427–440. doi: 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 16.Aitken A. 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- 17.Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 18.Obenauer J. C., Cantley L. C., Yaffe M. B. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 20.Rena G., Prescott A. R., Guo S., Cohen P., Unterman T. G. Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem. J. 2001;354:605–612. doi: 10.1042/0264-6021:3540605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wynshaw-Boris A., Gambello M. J. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 2001;15:639–651. doi: 10.1101/gad.886801. [DOI] [PubMed] [Google Scholar]
- 22.Obsil T., Ghirlando R., Klein D. C., Ganguly S., Dyda F. Crystal structure of the 14-3-3ζ:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell. 2001;105:257–267. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- 23.Ubersax J. A., Ferrell J. E., Jr Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 24.Han J. Y., Jeong E. Y., Kim Y. S., Roh G. S., Kim H. J., Kang S. S., Cho G. J., Choi W. S. c-Jun N-terminal kinase regulates the interaction between 14-3-3 and Bad in ethanol-induced cell death. J. Neurosci. Res. 2008;86:3221–3229. doi: 10.1002/jnr.21759. [DOI] [PubMed] [Google Scholar]
- 25.Salvi M., Sarno S., Cesaro L., Nakamura H., Pinna L. A. Extraordinary pleiotropy of protein kinase CK2 revealed by weblogo phosphoproteome analysis. Biochim. Biophys. Acta. 2009;1793:847–859. doi: 10.1016/j.bbamcr.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Faul C., Dhume A., Schecter A. D., Mundel P. Protein kinase A, Ca2+/calmodulin-dependent kinase II, and calcineurin regulate the intracellular trafficking of myopodin between the Z-disc and the nucleus of cardiac myocytes. Mol. Cell. Biol. 2007;27:8215–8227. doi: 10.1128/MCB.00950-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linding R., Jensen L. J., Pasculescu A., Olhovsky M., Colwill K., Bork P., Yaffe M. B., Pawson T. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 29.Martin D. M., Miranda-Saavedra D., Barton G. J. Kinomer v.1.0: a database of systematically classified eukaryotic protein kinases. Nucleic Acids Res. 2009;37:D244–D250. doi: 10.1093/nar/gkn834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harthill J. E., Meek S. E., Morrice N., Peggie M. W., Borch J., Wong B. H., MacKintosh C. Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J. 2006;47:211–223. doi: 10.1111/j.1365-313X.2006.02780.x. [DOI] [PubMed] [Google Scholar]
- 31.Bronisz A., Sharma S. M., Hu R., Godlewski J., Tzivion G., Mansky K. C., Ostrowski M. C. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol. Biol. Cell. 2006;17:3897–3906. doi: 10.1091/mbc.E06-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller M. L., Jensen L. J., Diella F., Jorgensen C., Tinti M., Li L., Hsiung M., Parker S. A., Bordeaux J., Sicheritz-Ponten T., et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogre L., Okresz L., Henriques R., Anthony R. G. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 2003;8:424–431. doi: 10.1016/S1360-1385(03)00188-2. [DOI] [PubMed] [Google Scholar]
- 34.Champion A., Kreis M., Mockaitis K., Picaud A., Henry Y. Arabidopsis kinome: after the casting. Funct. Integr. Genomics. 2004;4:163–187. doi: 10.1007/s10142-003-0096-4. [DOI] [PubMed] [Google Scholar]
- 35.Miranda-Saavedra D., Barton G. J. Classification and functional annotation of eukaryotic protein kinases. Proteins. 2007;68:893–914. doi: 10.1002/prot.21444. [DOI] [PubMed] [Google Scholar]
- 36.Basu S., Totty N. F., Irwin M. S., Sudol M., Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 37.Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coblitz B., Shikano S., Wu M., Gabelli S. B., Cockrell L. M., Spieker M., Hanyu Y., Fu H., Amzel L. M., Li M. C-terminal recognition by 14-3-3 proteins for surface expression of membrane receptors. J. Biol. Chem. 2005;280:36263–36272. doi: 10.1074/jbc.M507559200. [DOI] [PubMed] [Google Scholar]
- 40.Coblitz B., Wu M., Shikano S., Li M. C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 2006;580:1531–1535. doi: 10.1016/j.febslet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 41.Sliva D., Gu M., Zhu Y. X., Chen J., Tsai S., Du X., Yang Y. C. 14-3-3ζ interacts with the α-chain of human interleukin 9 receptor. Biochem. J. 2000;345:741–747. [PMC free article] [PubMed] [Google Scholar]
- 42.O'Kelly I., Butler M. H., Zilberberg N., Goldstein S. A. Forward transport. 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell. 2002;111:577–588. doi: 10.1016/s0092-8674(02)01040-1. [DOI] [PubMed] [Google Scholar]
- 43.Bermudez I., Moroni M. Phosphorylation and function of α4β2 receptor. J. Mol. Neurosci. 2006;30:97–98. doi: 10.1385/JMN:30:1:97. [DOI] [PubMed] [Google Scholar]
- 44.Rong J., Li S., Sheng G., Wu M., Coblitz B., Li M., Fu H., Li X. J. 14-3-3 protein interacts with Huntingtin-associated protein 1 and regulates its trafficking. J. Biol. Chem. 2007;282:4748–4756. doi: 10.1074/jbc.M609057200. [DOI] [PubMed] [Google Scholar]
- 45.Ganguly S., Weller J. L., Ho A., Chemineau P., Malpaux B., Klein D. C. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1222–1227. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu H., Coburn J., Collier R. J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. U.S.A. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ottmann C., Yasmin L., Weyand M., Veesenmeyer J. L., Diaz M. H., Palmer R. H., Francis M. S., Hauser A. R., Wittinghofer A., Hallberg B. Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: from structure to pathogenesis. EMBO J. 2007;26:902–913. doi: 10.1038/sj.emboj.7601530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriuchi H., Okamoto C., Nishihama R., Yamashita I., Machida Y., Tanaka N. Nuclear localization and interaction of RolB with plant 14-3-3 proteins correlates with induction of adventitious roots by the oncogene rolB. Plant J. 2004;38:260–275. doi: 10.1111/j.1365-313X.2004.02041.x. [DOI] [PubMed] [Google Scholar]
- 49.Seimiya H., Sawada H., Muramatsu Y., Shimizu M., Ohko K., Yamane K., Tsuruo T. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–2661. doi: 10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sumioka A., Nagaishi S., Yoshida T., Lin A. N., Miura M., Suzuki T. Role of 14-3-3γ in FE65-dependent gene transactivation mediated by the amyloid beta-protein precursor cytoplasmic fragment. J. Biol. Chem. 2005;280:42364–42374. doi: 10.1074/jbc.M504278200. [DOI] [PubMed] [Google Scholar]
- 51.Yang X., Lee W. H., Sobott F., Papagrigoriou E., Robinson C. V., Grossmann J. G., Sundstrom M., Doyle D. A., Elkins J. M. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bustos D. M., Iglesias A. A. Intrinsic disorder is a key characteristic in partners that bind 14-3-3 proteins. Proteins. 2006;63:35–42. doi: 10.1002/prot.20888. [DOI] [PubMed] [Google Scholar]
- 53.Oldfield C. J., Meng J., Yang J. Y., Yang M. Q., Uversky V. N., Dunker A. K. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9:S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Obsilova V., Nedbalkova E., Silhan J., Boura E., Herman P., Vecer J., Sulc M., Teisinger J., Dyda F., Obsil T. The 14-3-3 protein affects the conformation of the regulatory domain of human tyrosine hydroxylase. Biochemistry. 2008;47:1768–1777. doi: 10.1021/bi7019468. [DOI] [PubMed] [Google Scholar]
- 55.Silhan J., Vacha P., Strnadova P., Vecer J., Herman P., Sulc M., Teisinger J., Obsilova V., Obsil T. 14-3-3 protein masks the DNA binding interface of forkhead transcription factor FOXO4. J. Biol. Chem. 2009;284:19349–19360. doi: 10.1074/jbc.M109.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S., Murphy J., Toth R., Campbell D. G., Morrice N. A., MacKintosh C. Complementary regulation of TBC1D1 and AS160 by growth factors, insulin and AMPK activators. Biochem. J. 2008;409:449–459. doi: 10.1042/BJ20071114. [DOI] [PubMed] [Google Scholar]
- 57.Demmel L., Beck M., Klose C., Schlaitz A. L., Gloor Y., Hsu P. P., Havlis J., Shevchenko A., Krause E., Kalaidzidis Y., Walch-Solimena C. Nucleocytoplasmic shuttling of the Golgi phosphatidylinositol 4-kinase Pik1 is regulated by 14-3-3 proteins and coordinates Golgi function with cell growth. Mol. Biol. Cell. 2008;19:1046–1061. doi: 10.1091/mbc.E07-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yaffe M. B. How do 14-3-3 proteins work?. Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 59.She Q. B., Solit D. B., Ye Q., O'Reilly K. E., Lobo J., Rosen N. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell. 2005;8:287–297. doi: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watkins J. L., Lewandowski K. T., Meek S. E., Storz P., Toker A., Piwnica-Worms H. Phosphorylation of the Par-1 polarity kinase by protein kinase D regulates 14-3-3 binding and membrane association. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18378–18383. doi: 10.1073/pnas.0809661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giacometti S., Camoni L., Albumi C., Visconti S., De Michelis M. I., Aducci P. Tyrosine phosphorylation inhibits the interaction of 14-3-3 proteins with the plant plasma membrane H+-ATPase. Plant Biol. 2004;6:422–431. doi: 10.1055/s-2004-820933. [DOI] [PubMed] [Google Scholar]
- 62.Waterman M. J., Stavridi E. S., Waterman J. L., Halazonetis T. D. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 63.Winter S., Simboeck E., Fischle W., Zupkovitz G., Dohnal I., Mechtler K., Ammerer G., Seiser C. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2008;27:88–99. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sorrell D. A., Marchbank A. M., Chrimes D. A., Dickinson J. R., Rogers H. J., Francis D., Grierson C. S., Halford N. G. The Arabidopsis 14-3-3 protein, GF14omega, binds to the Schizosaccharomyces pombe Cdc25 phosphatase and rescues checkpoint defects in the rad24-mutant. Planta. 2003;218:50–57. doi: 10.1007/s00425-003-1083-7. [DOI] [PubMed] [Google Scholar]
- 65.Kulma A., Villadsen D., Campbell D. G., Meek S. E., Harthill J. E., Nielsen T. H., MacKintosh C. Phosphorylation and 14-3-3 binding of Arabidopsis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Plant J. 2004;37:654–667. doi: 10.1111/j.1365-313x.2003.01992.x. [DOI] [PubMed] [Google Scholar]
- 66.Pozuelo Rubio M., Peggie M., Wong B. H., Morrice N., MacKintosh C. 14-3-3s regulate fructose-2,6-bisphosphate levels by binding to PKB-phosphorylated cardiac fructose-2,6-bisphosphate kinase/phosphatase. EMBO J. 2003;22:3514–3523. doi: 10.1093/emboj/cdg363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGee S. L., van Denderen B. J., Howlett K. F., Mollica J., Schertzer J. D., Kemp B. E., Hargreaves M. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 68.McCart A. E., Mahony D., Rothnagel J. A. Alternatively spliced products of the human kinesin light chain 1 (KNS2) gene. Traffic. 2003;4:576–580. doi: 10.1034/j.1600-0854.2003.00113.x. [DOI] [PubMed] [Google Scholar]
- 69.Boutros R., Bailey A. M., Wilson S. H., Byrne J. A. Alternative splicing as a mechanism for regulating 14-3-3 binding: interactions between hD53 (TPD52L1) and 14-3-3 proteins. J. Mol. Biol. 2003;332:675–687. doi: 10.1016/s0022-2836(03)00944-6. [DOI] [PubMed] [Google Scholar]
- 70.Toyo-oka K., Shionoya A., Gambello M. J., Cardoso C., Leventer R., Ward H. L., Ayala R., Tsai L. H., Dobyns W., Ledbetter D., et al. 14-3-3ε is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat. Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 71.Mori D., Yano Y., Toyo-oka K., Yoshida N., Yamada M., Muramatsu M., Zhang D., Saya H., Toyoshima Y. Y., Kinoshita K., et al. NDEL1 phosphorylation by Aurora-A kinase is essential for centrosomal maturation, separation, and TACC3 recruitment. Mol. Cell. Biol. 2007;27:352–367. doi: 10.1128/MCB.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang Y., Yu W., Li Y., Yu L., Zhang Q., Wang F., Yang Z., Du J., Huang Q., Yao X., Zhu X. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol. Biol. Cell. 2007;18:2656–2666. doi: 10.1091/mbc.E06-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taya S., Shinoda T., Tsuboi D., Asaki J., Nagai K., Hikita T., Kuroda S., Kuroda K., Shimizu M., Hirotsune S., et al. DISC1 regulates the transport of the NUDEL/LIS1/14-3-3ε complex through kinesin-1. J. Neurosci. 2007;27:15–26. doi: 10.1523/JNEUROSCI.3826-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hayashi M. A., Portaro F. C., Bastos M. F., Guerreiro J. R., Oliveira V., Gorrao S. S., Tambourgi D. V., Sant'Anna O. A., Whiting P. J., Camargo L. M., et al. Inhibition of NUDEL (nuclear distribution element-like)-oligopeptidase activity by disrupted-in-schizophrenia 1. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3828–3833. doi: 10.1073/pnas.0500330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hennah W., Tomppo L., Hiekkalinna T., Palo O. M., Kilpinen H., Ekelund J., Tuulio-Henriksson A., Silander K., Partonen T., Paunio T., et al. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum. Mol. Genet. 2007;16:453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- 76.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 77.Kim H. J., Park H. J., Jung K. H., Ban J. Y., Ra J., Kim J. W., Park J. K., Choe B. K., Yim S. V., Kwon Y. K., Chung J. H. Association study of polymorphisms between DISC1 and schizophrenia in a Korean population. Neurosci. Lett. 2008;430:60–63. doi: 10.1016/j.neulet.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 78.Millar J. K., Christie S., Anderson S., Lawson D., Hsiao-Wei Loh D., Devon R. S., Arveiler B., Muir W. J., Blackwood D. H., Porteous D. J. Genomic structure and localisation within a linkage hotspot of Disrupted In Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol. Psychiatry. 2001;6:173–178. doi: 10.1038/sj.mp.4000784. [DOI] [PubMed] [Google Scholar]
- 79.Song W., Li W., Feng J., Heston L. L., Scaringe W. A., Sommer S. S. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem. Biophys. Res. Commun. 2008;367:700–706. doi: 10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- 80.Szeszko P. R., Hodgkinson C. A., Robinson D. G., Derosse P., Bilder R. M., Lencz T., Burdick K. E., Napolitano B., Betensky J. D., Kane J. M., et al. DISC1 is associated with prefrontal cortical gray matter and positive symptoms in schizophrenia. Biol. Psychol. 2008;79:103–110. doi: 10.1016/j.biopsycho.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grover D., Verma R., Goes F. S., Mahon P. L., Gershon E. S., McMahon F. J., Potash J. B. Family-based association of YWHAH in psychotic bipolar disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:977–983. doi: 10.1002/ajmg.b.30927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schneider T. D., Stephens R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aslan J. E., You H., Williamson D. M., Endig J., Youker R. T., Thomas L., Shu H., Du Y., Milewski R. L., Brush M. H., et al. Akt and 14-3-3 control a PACS-2 homeostatic switch that integrates membrane traffic with TRAIL-induced apoptosis. Mol. Cell. 2009;34:497–509. doi: 10.1016/j.molcel.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.