Abstract
SPP (signal peptide peptidase) is an aspartyl intramembrane cleaving protease, which processes a subset of signal peptides, and is linked to the quality control of ER (endoplasmic reticulum) membrane proteins. We analysed SPP interactions with signal peptides and other membrane proteins by co-immunoprecipitation assays. We found that SPP interacts specifically and tightly with a large range of newly synthesized membrane proteins, including signal peptides, preproteins and misfolded membrane proteins, but not with all co-expressed type II membrane proteins. Signal peptides are trapped by the catalytically inactive SPP mutant SPPD/A. Preproteins and misfolded membrane proteins interact with both SPP and the SPPD/A mutant, and are not substrates for SPP-mediated intramembrane proteolysis. Proteins interacting with SPP are found in distinct complexes of different sizes. A signal peptide is mainly trapped in a 200 kDa SPP complex, whereas a preprotein is predominantly found in a 600 kDa SPP complex. A misfolded membrane protein is detected in 200, 400 and 600 kDa SPP complexes. We conclude that SPP not only processes signal peptides, but also collects preproteins and misfolded membrane proteins that are destined for disposal.
Keywords: endoplasmic reticulum quality control, intramembrane proteolysis, oligomeric membrane protein complex, signal peptide peptidase (SPP)
Abbreviations: BN-PAGE, blue native PAGE; 2D-BN-SDS/PAGE, two-dimensional blue native SDS/PAGE; DDM, n-dodecyl-β-D-maltoside; DTT, dithiothreitol; ER, endoplasmic reticulum; ERAD, ER-associated degradation; HA, haemagglutinin; HCMV, human cytomegalovirus; HEK, human embryonic kidney; Ii, MHC class II-associated invariant chain; pPrl, preprolactin; RAMP4, ribosome-associated membrane protein 4; RAMP4op, RAMP4 with an opsin tag encoding a glycosylation site appended to its C-terminus; SPPrl, signal peptide of preprolactin; SPFPrl, signal peptide fragment of preprolactin; SPP, signal peptide peptidase; SPPL, SPP-like; TBS-T, Tris-buffered saline with Triton X-100
INTRODUCTION
SPP (signal peptide peptidase) is an intramembrane cleaving protease of the GxGD-type aspartyl protease family [1]. These proteases are polytopic membrane proteins with their active sites located in two adjacent transmembrane domains. They promote peptide bond hydrolysis of transmembrane proteins in the plane of cellular membranes [2].
SPP is located in the ER (endoplasmic reticulum) membrane, where it functions in the processing of signal sequence-derived signal peptides [3]. Signal sequences are N-terminal extensions of nascent secretory and membrane proteins, and mediate protein targeting to the ER membrane and insertion into the membrane [4]. They typically have a tripartite structure consisting of an N-terminal region, a central hydrophobic h-region, and a C-terminal region including the cleavage site for signal peptidase [5]. Following insertion of the nascent polypeptide, signal peptidase usually cleaves off the signal sequence from the parent preprotein [6]. SPP processes some of the resulting signal peptides within their membrane-spanning h-region and thereby generates fragments that are liberated from the ER membrane [3,7]. Some of them act as bioactive peptides, for example those derived from the signal sequences of MHC class I molecules, the hepatitis C virus polyprotein, the HIV-1 Env glycoprotein or pPrl (preprolactin) [8,9]. In these cases, SPP-mediated processing affects diverse pathways such as immune surveillance, virus maturation or cellular signalling. All SPP substrates identified to date are signal peptides [10,11]. They span the ER membrane in a type II topology, exposing the N-terminus to the cytosol and the C-terminus to the exoplasm [12]. It is therefore assumed that SPP processes only type II oriented peptides, whereas type I oriented peptides are processed by other intramembrane proteases [11]. Signal peptides need to be released from their parent preproteins by signal peptidase cleavage to become SPP substrates, a process called ectodomain shedding [10,13].
Besides processing signal peptides, SPP may also contribute to protein quality control at the ER membrane. Such a function was proposed from the identification of SPP as a cross-linking partner of misfolded variants of the polytopic membrane protein opsin [14]. Furthermore, SPP is implicated in the US2-dependent dislocation of MHC class I heavy chains from the ER membrane. US2 is a HCMV (human cytomegalovirus)-encoded protein and forms a complex with SPP. Both proteins are required for the dislocation of full-length MHC class I heavy chains to the cytosol [15].
SPP can be isolated as a monomer and as an SDS-stable dimer [16,17]. For proteolytic processing, SPP does not require additional cofactors, since purified recombinant SPP is able to process a substrate [18]. This is in contrast with presenilin-dependent intramembrane processing. The aspartyl protease presenilin is the catalytic core unit of the γ-secretase complex consisting of at least four subunits. All components are essential for complex assembly, substrate selection and protease function [19].
Considering the processing of signal peptides and the proposed role of SPP in the dislocation of other membrane proteins [14,15], the question arises of how SPP selects its substrates for intramembrane processing or ER membrane dislocation and degradation. In the present study, we characterized SPP interactions with potential substrates and other proteins bearing a type II oriented transmembrane domain by co-immunoprecipitation. We found that SPP specifically interacts with a large range of newly synthesized membrane proteins including a signal peptide, a preprotein and a misfolded polytopic membrane protein. Using BN-PAGE (blue native PAGE), we identified distinct SPP complexes and discovered that a substrate and a preprotein, as well as a misfolded membrane protein, assemble with SPP complexes of different sizes.
EXPERIMENTAL
Plasmids
SPP–myc and SPPD/A (SPP-D265A)–myc were generated by PCR using the primers 5′-CCCGGGTCATTTCTCTTTCTTCAAGTCCTCTTCAGAAATGAGCTTTTGCTCCTCCAGCCCCTTCGATGC-3′ and 5′-ATTTAGGTGACACTATAG-3′ with pSV-Sport1-SPP and pSV-Sport1-SPP-D265A [17] as templates. Thereby, a myc tag is introduced N-terminal to the KKEK (Lys-Lys-Glu-Lys) ER retention signal. PCR products were cloned into pRK5rs. pRK5rs and pRK5rs-pPrl have been described previously [20]. Starting with pRK5rs-pPrl, pRK5rs-pPrlPP29 was generated by site-directed mutagenesis using the primers 5′-CTACTCTTGTGCCAGGGTCCGCCTTCCACCCCCGTCTGTCCC-3′ and 5′-GGGACAGACGGGGGTGGAAGGCGGACCCTGGCACAAGAGTAG-3′. C-terminal HA (haemagglutinin)-tagged forms of pPrl and pPrlPP29 were generated by PCR on pRK5rs-pPrl and pRK5rs-pPrlPP29 using primers 5′-CGCTCGAGTTAAGCGTAATCTGGAACATCGTATGGGTAGCAGTTGTTGTTGTAGATGATTCTGC-3′ and 5′-CGATTGAATTCCCCGGGGATCCTCTAGAGTCGACCCAGCTTG-3′ and cloned into pRK5rs. pcDNA3.1-RAMP4op is derived from pGEM4Z-RAMP4/3′UTR/op [21] by PCR using the primers 5′-GAATTCTCCACCATGGTCGCCAAGCAGAGG-3′ and 5′-GAATTCAAGCTTCAGCCCGTCTTGTTGG-3′. pSV51-Ii is described in [22]. pRK5rs-OP91H* was generated by PCR on pZEO-OP91H+(cys56) (compare with [14]) using the primers 5′-TAATACGACTCACTATAGGG-3′ and 5′-TAGAAGGCACAGTCGAGG-3′ and cloning into pRK5rs.
Antibodies
Anti-myc and anti-opsin antibodies were prepared from 9E10 or R2-15 hybridoma cell supernatants respectively, using standard procedures. SPP-specific rabbit antibodies (C18) recognize an epitope in amino acids 353–377 of human SPP [23]. Anti-HA and anti-α-tubulin antibodies were purchased from Covance and Sigma ImmunoChemicals respectively. Anti-pPrl, anti-SPPrl (signal peptide of pPrl)[24], anti-invariant chain and anti-RAMP4 (ribosome-associated membrane protein 4) antibodies [22] have been described previously. Anti-(mouse IgG) and anti-(rabbit IgG) antibodies conjugated with horseradish peroxidase were obtained from Sigma.
Cell culture and transient transfection
HEK (human embryonic kidney) -293 cells (European Collection of Animal Cell Cultures, 85120602) were grown in Dulbecco's modified Eagle's medium/Ham's F12 (Invitrogen) supplemented with 10% (v/v) fetal bovine serum and 2 mM L-glutamine. Cells were regularly controlled for the absence of mycoplasma using standard PCR. Cells (3×105) were seeded in six-well slots, used for calcium phosphate transfection as described in [20], and harvested 44 h later.
Metabolic labelling and immunoprecipitation
Cells were depleted and metabolically labelled essentially as described in [20] and in the Figure legends. They were lysed in Triton X-100-containing lysis buffer [50 mM Hepes (pH 7.5), 150 mM NaCl, 1% (v/v) Triton X-100, 10% (v/v) glycerol, 1.5 mM MgCl2, 1 mM EGTA, 1 mM PMSF and Complete™ protease inhibitor cocktail (Roche)] or in digitonin-containing lysis buffer [1% (w/v) digitonin, 20 mM Tris/HCl (pH 7.5), 1.5 mM MgCl2, 1 mM EGTA, 50 mM NaCl, 10% (v/v) glycerol and protease inhibitors as described above]. Non-solubilized material was removed by centrifugation at 16000 g for 10 min at 4 °C. When indicated, 1/10 of the lysate was mixed directly with SDS-sample buffer [for Tris/Tricine gels: 4% (w/v) SDS, 75 mM Tris/HCl (pH 6.8), 12% (v/v) glycerol, 0.01% Serva Blue G and 50 mM DTT (dithiothreitol); for Tris/glycine gels: 2% (w/v) SDS, 50 mM Tris/HCl (pH 6.8), 10% (v/v) glycerol, 0.01% Bromophenol Blue and 50 mM DTT]. For immunoprecipitation, the cell lysate was diluted in HNTG low-salt buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 0.1% Triton X-100 and 10% (v/v) glycerol] or in TNDG buffer [20 mM Tris/HCl (pH 7.5), 50 mM NaCl, 0.1% digitonin and 10% (v/v) glycerol]. Antibodies and either Protein A–Sepharose or Protein G–Sepharose beads (GE Healthcare) were added for 3 h at 4 °C. Beads were washed five times with HNTG high-salt buffer (HNTG buffer with 500 mM NaCl instead of 150 mM) or with TNDG buffer. Samples were heated at 65 °C for 10 min and separated by Tris/Tricine or Tris/glycine SDS/PAGE using the methods of Schägger and von Jagow [25] and Laemmli [26] respectively. Dried gels were analysed by PhosphoImaging (Fuji BAS 1500).
Western blotting
Cells were lysed and proteins separated as described above. Then, proteins were transferred on to membranes in a semi-dry blotting unit. Blocking and first antibody incubation was done in 0.25% gelatin in TBS-T [Tris-buffered saline with Triton X-100: 50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA and 0.05% Triton X-100], washing and secondary antibody incubation was carried out in TBS-T. For immunodetection, BM chemiluminescence blotting substrate (POD, Roche Applied Science) was used. In the case of re-probing the membranes with anti-tubulin antibodies, the membranes were stripped with 62.5 mM Tris/HCl (pH 6.7), 2% (w/v) SDS and freshly added 0.5% 2-mercaptoethanol for 30 min at 50 °C.
BN-PAGE
Cells were lysed in BN-lysis buffer (digitonin-containing lysis buffer, as above, but containing 1% of detergents as indicated in the Figure legends). Non-solubilized material was removed by centrifugation at 16000 g for 10 min at 4 °C. A 1/40 volume of BN-sample buffer [500 mM 6-aminohexanoic acid, 100 mM Bis-Tris (pH 7) and 5% (w/v) Coomassie Blue G250] was added to the lysate before subjection to BN-PAGE [27]. Samples were loaded on to 5–9%, 6–13% or 6–16% gradient gels [with 67 mM 6-aminohexanoic acid and 50 mM Bis-Tris (pH 7)] and run at 4 °C for 1 h at 15 mA with cathode buffer B [50 mM Tricine (pH 7), 15 mM Bis-Tris and 0.02% Coomassie Blue G250]. Then, the cathode buffer was exchanged for cathode buffer B/10 (buffer B but containing only 0.002% Coomassie Blue G250) and gels were run at 100 V [anode buffer: 50 mM Bis-Tris (pH 7)]. As molecular markers, the high-molecular-mass kit for native electrophoresis (GE Healthcare) was used. For Western blotting, BN gels were soaked in Tris/glycine-SDS-electrophoresis buffer [26], then in BN-transfer buffer [25 mM Tris, 150 mM glycine, 0.02% SDS and 20% (v/v) methanol] and proteins were transferred via semi-dry blotting on to PVDF membranes (Millipore). Membranes were destained with methanol and immunodetection was performed as described above.
2D-BN-SDS/PAGE
Lanes from the BN gel were excised, incubated in SDS-sample buffer for 30 min and analysed using SDS/PAGE, and the following Western blots were performed according to standard protocols as described above.
Affinity purification of SPP complexes and MS
To purify SPPD/A–myc containing complexes, anti-myc antibodies were coupled to CNBr–Sepharose beads (GE Healthcare) according to the manufacturer's instructions. Cells transfected with pRK5rs-SPPD/A-myc and pRK5rs-Prl or mock-transfected cells were lysed in BN-lysis buffer containing 1% (w/v) digitonin. Beads were equilibrated in the same buffer, mixed with the lysates and incubated for 16 h at 4 °C. Non-bound material was removed by washing the beads four times with BN-lysis buffer, and bound material was eluted by competition with synthetic myc peptide (Sigma) for 2 h at 4 °C. Fractions of each step were mixed with BN-sample buffer and subjected to BN-PAGE. The gel was silver-stained [28]. For LC–MS/MS (liquid chromatography–tandem MS) analysis, the band containing the 200 kDa complex was excised from the gel together with a reference region from a lysate of mock-transfected cells.
Densitometric analyses of protein bands
Densitometric analysis was carried out with ImageJ. Each signal, represented by a defined area, was corrected for background by subtracting a signal from an identical area within the same lane. For statistical analyses, relative amounts were calculated, i.e. signals from each time point were adjusted to 100%. Relative amounts are given as mean±S.D. percentages for three independent experiments.
RESULTS
The SPP substrate SPPrl tightly interacts with SPPD/A
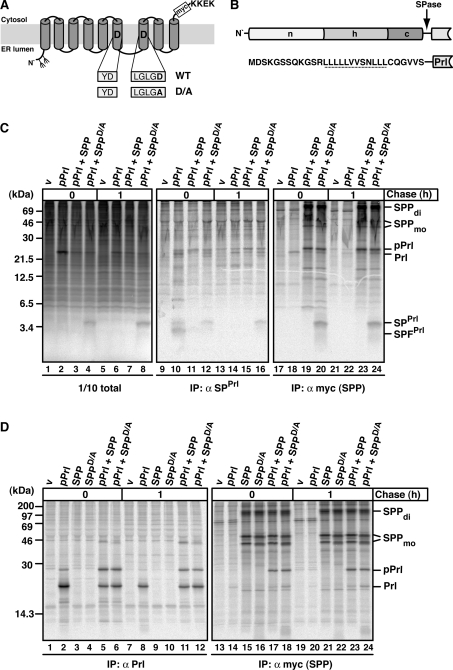
To characterize the interaction of SPP with potential substrates and with other integral membrane proteins, we employed co-immunoprecipitation of metabolically labelled proteins. As a model substrate, we used the pPrl-derived signal peptide, SPPrl (Figure 1B), which was shown previously to be processed by SPP in an in vitro translation/translocation system [3,7]. To generate SPPrl in cells, we overexpressed pPrl. Additionally, we expressed myc-tagged SPP or the catalytically inactive SPP mutant SPPD/A, which is known to lack proteolytic activity (Figure 1A [1,29]).
Figure 1. The effect of SPP and SPPD/A on SPPrl and pPrl processing and binding.
(A) Schematic representation of SPP in the ER membrane with amino acids of the active-site motif in one-letter code. The D→A mutation in SPPD/A and the position of the myc tag are indicated. KKEK, ER retrieval motif. Fork-like structures indicate N-linked glycosylation sites. (B) Outline and amino acid sequence of the signal sequence of pPrl with its N-terminal (n), hydrophobic (h) and C-terminal (c) region. The cleavage site for signal peptidase (SPase) is indicated, and the h-region is underlined. (C) SPPrl is trapped by the dominant-negative mutant SPPD/A. HEK-293 cells transiently expressing pPrl, SPP–myc or SPPD/A–myc were metabolically pulse labelled for 30 min and chased for 1 h where indicated. Cells were lysed in the presence of 1% Triton X-100. A 1/10 sample of the total lysate was analysed directly using SDS/PAGE and the rest was used for immunoprecipitation (IP) using either anti-SPPrl antibodies or anti-myc antibodies to identify myc-tagged SPP/SPPD/A. Samples were subjected to Tris/Tricine SDS/PAGE and labelled proteins were visualized by autoradiography. v, empty vector control; SPFPrl, signal peptide fragment of SPPrl; Prl, mature prolactin. SPP–myc dimer (di) and monomer (mo) are indicated. Note that the monomer is detected as a double-band, representing non-glycosylated and glycosylated SPP (results not shown). Molecular masses are indicated in kDa. (D) pPrl co-immunoprecipitates with SPP and SPPD/A. HEK-293 cells were transfected and pulse–chase labelled as described in (C). To detect pPrl and mature prolactin (Prl), anti-Prl antibodies were used for immunoprecipitation (IP). Anti-myc antibodies were used for the identification of myc-tagged SPP/SPPD/A and for co-immunoprecipitation. Proteins were separated by Tris/glycine SDS/PAGE and visualized as described in (C). Molecular masses are indicated in kDa.
HEK-293 cells transiently expressing pPrl alone or together with either SPP or SPPD/A were metabolically labelled and then lysed either directly or chased for 1 h. To detect SPPrl by immunoprecipitation, we used an antibody recognizing an epitope in the N-terminal region of the pPrl signal sequence (anti-SPPrl), which had been characterized previously [24]. Aliquots of total lysates (Figure 1C, lanes 1–8) and the immunoprecipitated proteins (Figure 1C, lanes 9–16) were separated by Tris/Tricine SDS/PAGE, which allows a high resolution of small proteins. Finally, labelled proteins were visualized by autoradiography. When pPrl alone was expressed, the 3 kDa SPPrl and its smaller fragment, SPFPrl, were detected after pulse labelling in the immunoprecipitate (Figure 1C, lane 10), but not after 1 h of chase (Figure 1C, lane 14). This indicates that SPPrl in cells is processed and that the resulting fragment SPFPrl has a rather short half-life. After co-expression with SPP, no SPPrl and only small amounts of SPFPrl were found (Figure 1C, lanes 11 and 15), suggesting that overexpressed SPP increases the rate of processing and leads to a rapid degradation of SPFPrl. When SPPD/A was co-expressed with pPrl, SPPrl accumulated (Figure 1C, lanes 4 and 12) and was stabilized over time (Figure 1C, lanes 8 and 16). Thus SPPD/A can efficiently prevent SPPrl processing and degradation. The anti-SPPrl antibody did not efficiently immunoprecipitate SPPrl under these native conditions (Figure 1C, lanes 12 and 16 compared with lanes 4 and 8). This may indicate that, under these conditions, SPPrl is not accessible for the antibody.
To see whether SPPrl interacts with SPPD/A, we performed co-immunoprecipitation using the anti-myc antibody. This antibody recognizes myc-tagged SPP and SPPD/A, which are detected in the gel as monomers and dimers (Figure 1C, lanes 17–24). Similar amounts of SPP and SPPD/A were immunoprecipitated (Figure 1C, lanes 19, 20, 23 and 24) and could be detected after Western blotting (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/427/bj4270523add.htm), demonstrating similar expression levels of both proteins. SPPrl was efficiently co-immunoprecipitated with SPPD/A (Figure 1, lanes 20 and 24, and see Supplementary Figure S2 at http://www.BiochemJ.org/bj/427/bj4270523add.htm).
In order to test whether SPPrl also interacts with an SPP-like (SPPL) protease, pPrl was co-expressed with the ER-localized SPPL3 [30] and its catalytically inactive mutant SPPL3-D271A, here termed SPPL3D/A (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/427/bj4270523add.htm). No SPPrl accumulated in the presence of SPPL3D/A and no co-immunoprecipitation of SPPrl and SPPL3D/A was detected. This suggests that SPPD/A cannot be functionally replaced by SPPL3D/A. Taken together, these results show that SPPrl is processed by SPP in cells and that the catalytically inactive mutant SPPD/A is an efficient dominant-negative mutant that interacts tightly with SPPrl.
pPrl interacts with SPP and is no substrate for SPP-mediated processing
In addition to SPPrl, we observed the specific co-immunoprecipitation of a 25 kDa protein with SPP and SPPD/A (Figure 1C, right-hand panel and Supplementary Figure S2). As this protein could be immunoprecipitated with an anti-prolactin antibody [24], but is approx. 3 kDa larger than prolactin, it must be preprolactin (Figure 1D). When pPrl alone was expressed, mainly prolactin of approx. 22 kDa was immunoprecipitated and only a small amount of pPrl accumulated (Figure 1D, lanes 2 and 8), indicating that pPrl is efficiently inserted into ER membranes where its signal sequence is cleaved off. In contrast, higher amounts of pPrl accumulated in the presence of co-expressed SPP or SPPD/A (Figure 1D, lanes 5, 6, 11 and 12) and were co-immunoprecipitated (Figure 1D, lanes 17, 18, 23 and 24). This suggests that overexpression of SPP increases the amount of the preprotein pPrl at the expense of processed prolactin. pPrl interacts with SPPD/A and catalytically active SPP, accumulates and is not processed with time. This strongly indicates that it is not a substrate for SPP. This is consistent with the earlier finding that cleavage by signal peptidase is required for intramembrane proteolysis by SPP [10].
SPP does not interact with the type II oriented membrane proteins Ii (MHC class II-associated invariant chain) and RAMP4
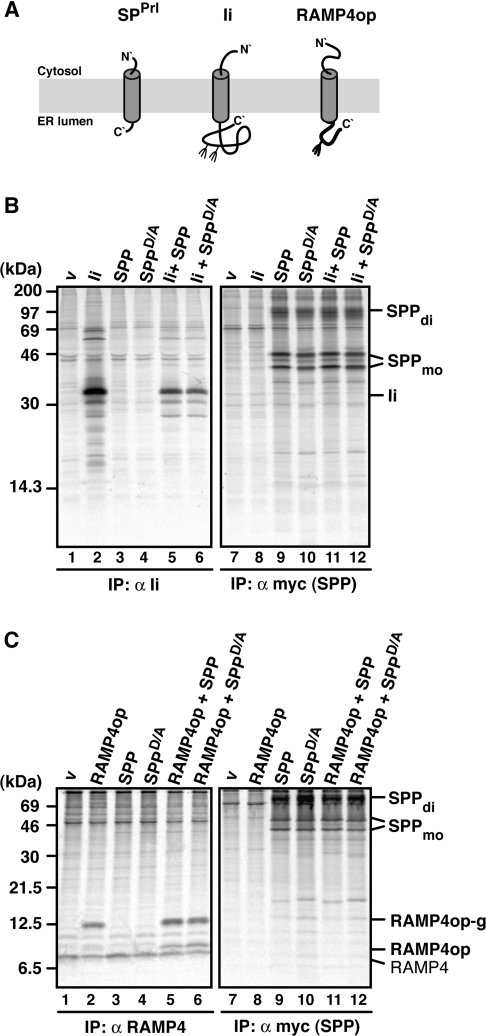
Signal peptides, like their preproteins, span the ER membrane in a type II orientation, exposing the N-terminus towards the cytosolic side of the membrane [12]. In order to investigate whether other type II oriented membrane proteins that comprise non-cleaved signal sequences also interact with SPP or SPPD/A, we tested Ii and RAMP4. Ii is a signal-anchor glycoprotein with an N-terminal transmembrane domain [31], whereas RAMP4 is a tail-anchored membrane protein, which is inserted into the ER membrane via its C-terminal transmembrane domain [21,32]. To detect RAMP4 membrane insertion, an opsin tag encoding a glycosylation site was appended to its C-terminus (RAMP4op) [21] (Figure 2A).
Figure 2. SPP does not associate with the type II membrane proteins Ii and RAMP4op.
(A) Membrane disposition of a signal peptide (SPPrl), Ii and RAMP4op. Fork-like structures indicate N-linked glycosylation sites. (B and C) Ii (B) and RAMP4op (C) were transiently expressed alone or together with SPP–myc or SPPD/A–myc, pulse labelled for 30 min, and proteins immunoprecipitated (IP) with anti-Ii (B), anti-RAMP4 (C) or anti-myc antibodies (B and C). Proteins were separated by Tris/glycine SDS/PAGE (B) or Tris/Tricine SDS/PAGE (C) and visualized by autoradiography. v, empty vector control; RAMP4, endogenous RAMP4; RAMP4op, non-glycosylated RAMP4op; RAMP4op-g, glycosylated RAMP4op. SPP dimer (di) and monomer (mo) are indicated. Molecular masses are indicated in kDa.
In transfected cells, Ii was immunoprecipitated mainly as a bi-glycosylated protein of 33 kDa (Figure 2B, lane 2). Similar amounts of Ii were detected upon co-expression with SPP or SPPD/A, but Ii was not co-immunoprecipitated with SPP or SPPD/A (Figure 2B, lanes 11 and 12). As no smaller forms of Ii accumulated in the presence of SPP or SPPD/A, this indicates that Ii is neither an SPP substrate nor a binding partner (Figure 2B, lanes 5 and 6).
RAMP4op was immunoprecipitated as a 12 kDa glycoprotein as characterized previously [21] (Figure 2C, lane 2). In addition, endogenous RAMP4 of 8 kDa was immunoprecipitated by the anti-RAMP4 antibody (compare lanes 1 and 2 in Figure 2C). The co-expression of SPP and SPPD/A did not affect the amount of either RAMP4op or endogenous RAMP4. No co-immunoprecipitation of RAMP4op with overexpressed SPP or SPPD/A was found (Figure 2C, lanes 11 and 12). Together, these data indicate that Ii and RAMP4 neither are SPP substrates nor interact with co-expressed SPPs. Furthermore, we conclude that not all type II oriented membrane proteins interact with SPP/SPPD/A.
SPP interacts with misfolded opsin
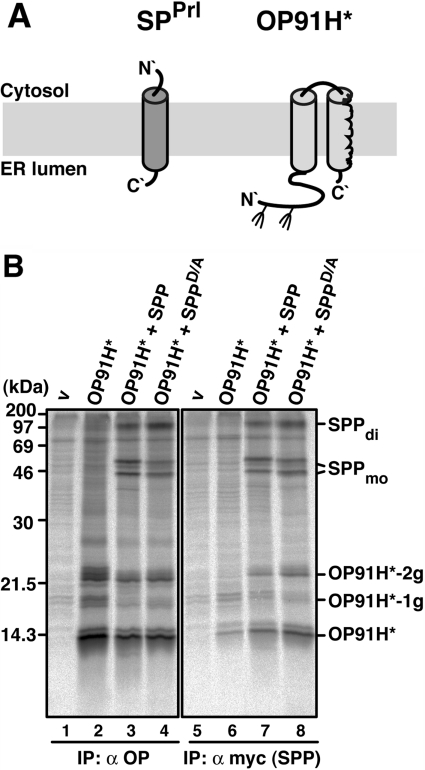
Previously, Crawshaw et al. [14] have shown that SPP can be cross-linked to a misassembled transmembrane domain of a truncated mutant of polytopic opsin. This protein–protein interaction could be prevented by addition of an SPP inhibitor [14]. In the present study, we characterized this interaction in our cell-based assay using co-immunoprecipitation. We used OP91H*, a truncated opsin mutant that spans the membrane twice (type I and type II oriented transmembrane domains) and can be bi-glycosylated in the N-terminal region [14] (Figure 3A and Figure 3B, lanes 2–4).
Figure 3. SPP interacts with misfolded opsin.
(A) Membrane disposition of OP91H* in comparison with a signal peptide (SPPrl). Fork-like structures indicate N-linked glycosylation sites. (B) OP91H* co-immunoprecipitates with SPP and SPPD/A. HEK-293 cells transiently expressing OP91H*, SPP–myc or SPPD/A–myc were pulse labelled for 30 min, lysed with 1% digitonin and immunoprecipitated (IP) with anti-opsin antibodies (lanes 1–4), or anti-myc antibodies (lanes 5–8). Proteins were separated by Tris/glycine SDS/PAGE and visualized by autoradiography. v, empty vector control. -1g and -2g indicate mono- and bi-glycosylated opsin forms respectively. SPP dimer (di) and monomer (mo) are indicated. Molecular masses are indicated in kDa.
OP91H* was immunoprecipitated in its non-, mono- and bi-glycosylated forms after pulse labelling as characterized previously [14] (Figure 3B, lane 2). Similar amounts of OP91H* were immunoprecipitated upon co-expression with SPP or SPPD/A. Additionally, SPP and SPPD/A co-immunoprecipitated with OP91H* (Figure 3B, lanes 3 and 4). Employing the anti-myc antibody for immunoprecipitation revealed that a small portion of OP91H* was non-specifically immunoprecipitated in the absence of myc-tagged SPP (Figure 3B, lane 6). However, increased amounts OP91H* specifically co-immunoprecipitated with co-expressed SPP and SPPD/A (Figure 3B, lanes 7 and 8). Taken together, these data indicate that both SPP and SPPD/A interact tightly with a newly synthesized misfolded membrane protein.
SPP impedes the degradation of misfolded opsin
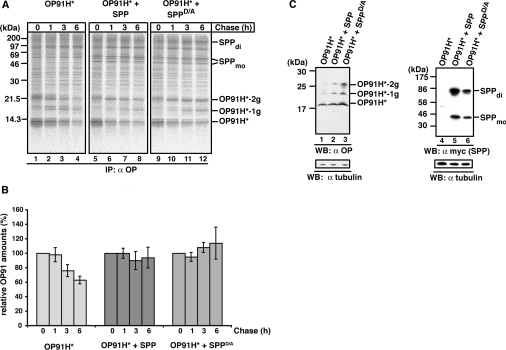
Truncated and misfolded opsin mutants are retained in the ER membrane where they do not assemble correctly, but can aggregate or undergo ERAD (ER-associated degradation) [33,34]. To test whether co-expression of SPP affects the degradation of OP91H*, we pulse–chase labelled cells expressing either OP91H* alone or together with SPP or SPPD/A.
OP91H* was substantially degraded after 6 h, when expressed alone (Figure 4A, lanes 1–4 and quantification in Figure 4B). In contrast, the co-expression of either SPP or SPPD/A resulted in a longer half-life of OP91H*, indicating that its degradation was significantly impeded (Figure 4A, lanes 5–6 and 9–12, and quantifications in Figure 4B). Accumulation was found in particular for the bi-glycosylated OP91H*, suggesting that SPP-bound OP91H* was still competent for glycosylation. The stabilizing effect of co-expressed SPP/SPPD/A on OP91H* could also be confirmed under steady-state conditions as depicted by the Western blot in Figure 4(C). Together, these results show that the misfolded membrane protein is stabilized in the presence of high SPP amounts. Since co-expression of both SPP and SPPD/A leads to similar observations, we conclude that the stabilization is independent of the catalytic SPP activity.
Figure 4. SPP impedes the degradation of misfolded opsin.
(A) Pulse-chase analysis of OP91H*. HEK-293 cells transiently expressing OP91H* in the absence or presence of SPP–myc or SPPD/A–myc were pulse labelled for 10 min and chased for the indicated times. Digitonin lysates were used for immunoprecipitation (IP) with the anti-opsin antibody. -1g and -2g indicate mono- and bi-glycosylated opsin forms respectively. SPP dimer (di) and monomer (mo) are indicated. Molecular masses are indicated in kDa. (B) Quantification of OP91H* accumulation. Three independent experiments as shown in (A) were used for the quantification of OP91H* accumulation in the absence (left-hand panel) or in presence of SPP–myc (middle panel) or SPPD/A–myc (right-hand panel) respectively. The OP91H* amount after the pulse (0 h chase) was set to 100%. Results are relative means±S.D. for three independent experiments. (C) Western blotting (WB) shows OP91H* accumulation in the presence of SPP. HEK-293 cells transiently expressing OP91H* and SPP–myc or SPPD/A–myc as indicated were lysed with 1% digitonin, and aliquots were separated by Tris/glycine SDS/PAGE. OP91H* and SPP/SPPD/A–myc proteins were identified using Western blotting using anti-opsin (left-hand panel) and anti-myc antibodies (right-hand panel) respectively. Membranes were stripped and re-probed with anti-tubulin antibodies as a loading control (lower panels). -1g and -2g indicate mono- and bi-glycosylated opsin forms respectively. SPP dimer (di) and monomer (mo) are indicated. Molecular masses are indicated in kDa.
SPP is found in distinct high-molecular-mass complexes
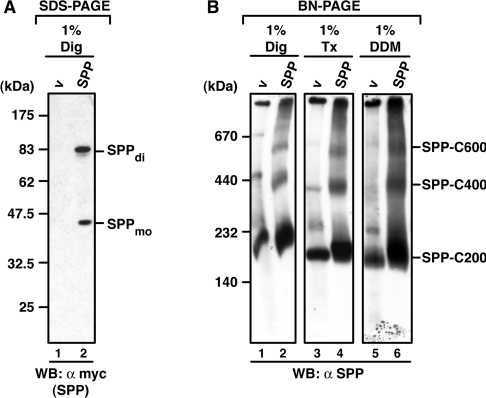
Our co-immunoprecipitation data revealed that SPP interacts with a range of membrane proteins, including substrates as well as preproteins and misfolded opsins that are not SPP substrates. In order to characterize further the SPP complexes, we analysed them using BN-PAGE. First, we tested whether SPP, which has been characterized under denaturing conditions as a monomer of 40 kDa and as an SDS-stable dimer of 90 kDa [16] (Figure 5A) is detectable under native conditions in higher-molecular-mass complexes. Mock-transfected cells expressing endogenous SPP and transfected cells expressing SPP–myc were solubilized by three different detergents [1% digitonin, 1% Triton X-100, and 1% DDM (n-dodecyl-β-D-maltoside)], and proteins were separated by BN-PAGE (Figure 5B). Using an SPP-specific antibody we detected endogenous (Figure 5B, lanes 1, 3 and 6) and overexpressed (Figure 5B, lanes 2, 4 and 6) SPP complexes after Western blotting. Both migrated as a major complex of approx. 200 kDa (C200). Additionally, complexes of approx. 400 kDa (C400) and 600 kDa (C600) were detected. Endogenous SPP was detected in one further complex of approx. 250 kDa. All of these complexes were detected after solubilization with the three detergents, indicating that they are rather stable.
Figure 5. Endogenous and overexpressed SPP is found in distinct high-molecular-mass complexes after BN-PAGE.
(A) After SDS/PAGE, SPP is detected as a monomer and SDS-stable dimer. HEK-293 cells transiently expressing SPP–myc were lysed with digitonin (Dig). Proteins were separated by SDS/PAGE, and SPP monomers (mo) and dimers (di) were identified by Western blotting (WB) using anti-myc antibodies. v, empty vector control. (B) Detection of SPP complexes after BN-PAGE. HEK-293 cells transiently expressing SPP–myc or transfected with an empty vector (v), were lysed with digitonin (Dig), Triton X-100 (Tx) or DDM. Total lysates were subjected to BN-PAGE (5–9%). Endogenous and overexpressed SPP complexes of 200 kDa (C200), 400 kDa (C400) and 600 kDa (C600) were then identified by Western blotting (WB) using an SPP-specific antibody. Molecular masses are indicated in kDa.
A signal peptide is trapped in a 200 kDa SPP complex
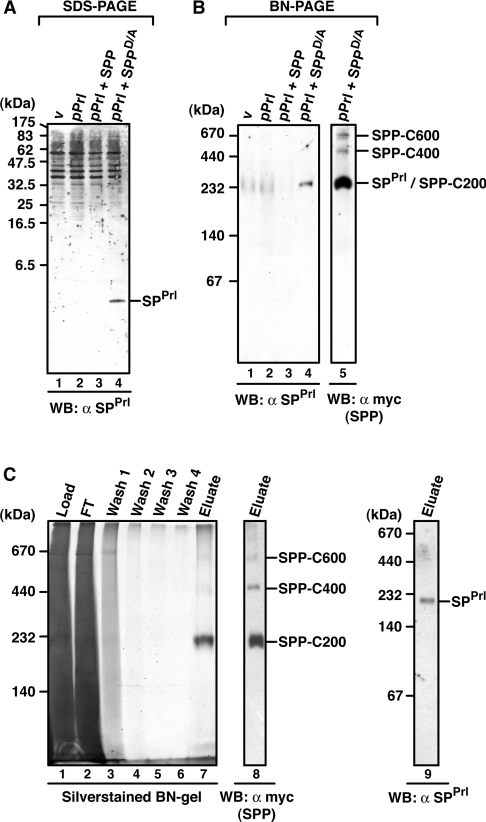
Since SPPrl accumulates in the presence of SPPD/A and can be identified by Western blotting (Figure 6A, lane 4), we next used BN-PAGE to identify SPPrl/SPPD/A complexes. Upon Western blotting, the anti-SPPrl antibody recognized its epitope in a 200 kDa complex, when pPrl was co-expressed with SPPD/A (Figure 6B, lanes 1–4). SPPD/A complexes were identified using the anti-myc antibody, which recognized the above-described complexes C200, C400 and C600 (Figure 6B, lane 5). The 200 kDa SPPrl complex co-migrates with the SPPD/A-C200, suggesting that SPPrl is trapped in this SPP complex. SPPrl could also be detected in this complex, when proteins were solubilized in the presence of Triton X-100 or DDM, indicating that the SPPrl/SPPD/A-C200 is stable under different detergent conditions (see Supplementary Figure S4 at http://www.BiochemJ.org/bj/427/bj4270523add.htm).
Figure 6. Characterization of SPPrl/SPP complexes.
(A) Steady-state accumulation of SPPrl in the presence of SPPD/A. HEK-293 cells transiently expressing pPrl alone or together with SPP–myc or SPPD/A–myc were lysed in 1% Triton X-100 buffer, and proteins were separated by Tris/Tricine SDS/PAGE. SPPrl was identified upon SPPD/A–myc co-expression by Western blotting (WB) using the anti-SPPrl antibody. (B) SPPrl is trapped in a 200 kDa SPP complex. Digitonin lysates of HEK-293 cells expressing pPrl, SPP–myc or SPPD/A–myc as indicated were separated by BN-PAGE (6–13%). SPPrl-containing complexes were identified by Western blotting (WB) using an anti-SPPrl antibody. SPPD/A–myc complexes were detected with an anti-myc antibody (lane 5). The position of the 200 kDa SPPrl/SPPD/A-C200 complex and higher-molecular-mass SPP complexes are indicated. (C) Affinity purification of SPPrl/SPP complexes. HEK-293 cells co-expressing pPrl and SPPD/A–myc were lysed with 1% digitonin, and SPPD/A–myc complexes were affinity-purified on anti-myc beads. Aliquots of the total lysate, flow-through (FT), wash fraction (wash) and eluted fractions (eluate) were characterized by BN-PAGE (5–9%) and silver staining (lanes 1–7) or by Western blotting (WB) with an anti-myc antibody (lane 8) and an anti-SPPrl antibody (lane 9). Molecular masses are indicated in kDa.
To identify the protein components of SPPrl/SPPD/A-C200, we affinity-purified SPPD/A complexes on anti-myc antibodies. Figure 6(C) shows the purification profile and elution of the three SPP complexes on a silver-stained blue native gel and after Western blotting (lanes 1–8). SPPrl could be identified as a component of purified SPPD/A-C200 (Figure 6C, lane 9). Next, MS was used to identify polypeptides within the three SPP complexes. In the C200 complex, three different peptide sequences derived from SPP were obtained. Other peptide sequences were non-specifically found in the SPP complexes since they were also found in samples obtained after a mock-purification. Together, these data suggest that SPP is the main, possibly the sole, constituent of the C200 complex. We also attempted to identify the protein constituents of the SPP-C400 and -C600 complexes. However, no peptide sequences were obtained by MS analysis, probably due to the small amounts.
pPrl is found in a 600 kDa SPP complex
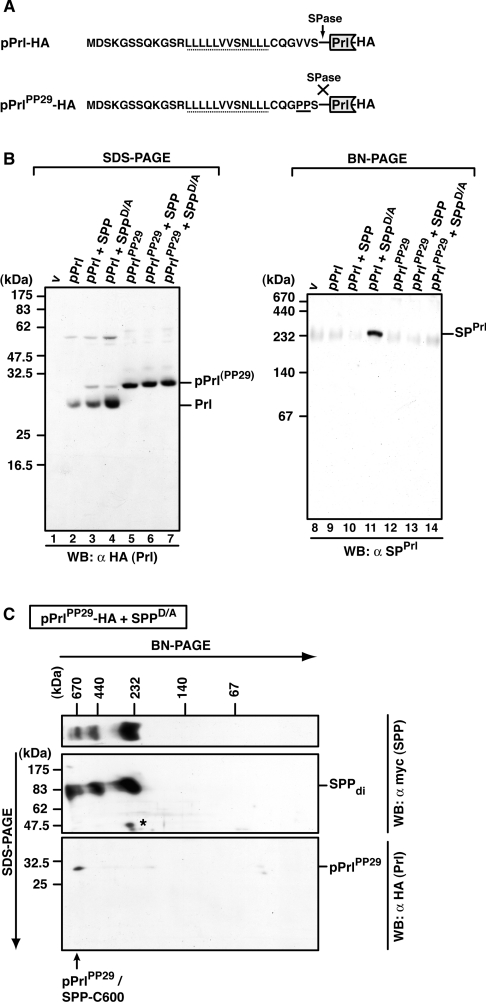
In addition to SPPrl that is trapped by SPPD/A, pPrl interacts tightly with SPPD/A and SPP (compare with Figure 1). Therefore we also identified pPrl/SPP complexes using BN-PAGE. Since only small amounts of pPrl accumulated in cells when pPrl was co-expressed with SPP or SPPD/A (Figure 7B, lanes 1–4), we generated the pPrl mutant pPrlPP29, which is not cleaved by signal peptidase and thus leads to the accumulation of pPrl [10,13] (Figure 7A, and Figure 7B, lanes 5–7).
Figure 7. Identification of pPrl–SPP complexes.
(A) Amino acid sequence of the signal sequence of wild-type pPrl and the mutant pPrlPP29, which is not cleaved by signal peptidase (SPase) (both are C-terminally HA-tagged). The V→P mutation (PP29) is underlined. h-regions are indicated with a dotted line. (B) Detection of mature prolactin (Prl), pPrlPP29 and SPPrl after SDS/PAGE or BN-PAGE. HEK-293 cells transiently expressing pPrl–HA or pPrlPP29–HA alone or together with SPP–myc or SPPD/A–myc were lysed with 1% digitonin, and total lysates were subjected to Tris/glycine SDS/PAGE (lanes 1–7) or to BN-PAGE (6–16%; lanes 8–14). Mature prolactin (Prl–HA) and pPrl(PP29)–HA were identified after SDS/PAGE by Western blotting (WB) using anti-HA antibodies, and SPPrl was identified after BN-PAGE and Western blotting using anti-SPPrl antibodies. v, empty vector control. (C) Identification of pPrlPP29 in the 600 kDa SPP complex after 2D-BN-SDS/PAGE. HEK-293 cells transiently co-expressing pPrlPP29–HA and SPPD/A–myc were lysed with 1% digitonin, and the total lysate was first separated by BN-PAGE (6–13%) and then in the second dimension by SDS/PAGE (12.5%). SPPD/A–myc complexes and pPrlPP29–HA were identified by Western blotting (WB) using the anti-myc and the anti-HA antibody respectively. SPP dimers (di) and pPrlPP29/SPP-C600 are indicated. The asterisk indicates a cross-reacting band. Molecular masses are indicated in kDa.
After BN-PAGE and Western blotting, the anti-SPPrl antibody recognized its epitope in a 200 kDa complex when wild-type pPrl was co-expressed with SPPD/A (Figure 7B, lanes 8–14). However, we could not detect pPrlPP29 complexes after BN-PAGE and Western blotting. Most likely, the corresponding epitope was not available after blotting from the blue native gel. To circumvent this problem, we separated the complexes in a second dimension using SDS/PAGE, termed 2D-BN-SDS/PAGE (Figure 7C). When HA-tagged pPrlPP29 was co-expressed with SPPD/A, it was detected as a 30 kDa protein after the second-dimension SDS/PAGE (Figure 7C, bottom panel). SPP migrated as 90 kDa dimers (Figure 7C, middle panel), indicating an even-numbered stoichiometry of SPP in the complexes. pPrlPP29 co-migrates with the large SPP-C600 complex, strongly indicating that it is present in this complex. As a control, a similar analysis applying wild-type pPrl showed that mature prolactin is not incorporated into higher-molecular-mass complexes, but migrated in a range below 60 kDa according to the first-dimension BN-PAGE (results not shown). We conclude that SPPrl and its preprotein assemble into SPP complexes of different sizes. While SPPrl is trapped in C200, the preprotein predominantly accumulates in C600.
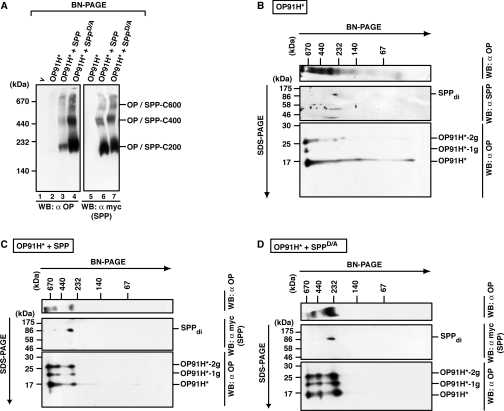
Misfolded opsin accumulates in three distinct SPP complexes
As misfolded opsin OP91H* also interacts with SPP and SPPD/A (Figure 3), we next analysed OP91H*–SPP complexes by BN-PAGE. Lysates of cells expressing OP91H* alone or together with SPP or SPPD/A were analysed by BN-PAGE and Western blotting (Figure 8A). Upon co-expression with SPP and SPPD/A, OP91H* is found to co-localize with the three SPP complexes C200, C400 and C600 (Figure 8A, compare lanes 3 and 4 with lanes 6 and 7). Very low amounts of the OP91H*–SPP complexes were detected in lysates from cells expressing OP91H* alone (Figure 8A, lane 2 and compare with Supplementary Figure S5 at http://www.BiochemJ.org/bj/427/bj4270523add.htm for long exposure). This again corroborates the strong stabilizing effect of SPP on OP91H* as found after pulse–chase labelling and immunoprecipitation (compare with Figure 4).
Figure 8. Misfolded opsin accumulates in several SPP complexes.
(A) Identification of OP91H*–SPP complexes after BN-PAGE. HEK-293 cells transiently expressing OP91H* and SPP–myc or SPPD/A–myc as indicated were lysed with 1% digitonin, and aliquots were separated by BN-PAGE (5–9%). OP91H* and SPP/SPPD/A–myc complexes were identified by Western blotting (WB) using anti-opsin and anti-myc antibodies respectively. SPP complexes of 200 kDa (SPP-C200), 400 kDa (SPP-C400) and 600 kDa (SPP-C600) co-migrating with OP91H* are indicated. (B–D) Glycosylated and non-glycosylated opsin is found in complexes of the same size. Digitonin lysates of HEK-293 cells transiently expressing OP91H* (B) or co-expressing SPP–myc (C) or SPPD/A–myc (D) were first separated by BN-PAGE (6–13%) and then separated in the second dimension by SDS/PAGE (15%). Top panels show Western blots (WB) of the BN gels using anti-opsin antibodies. Middle panels show Western blots of the upper part of the second-dimension SDS gels using anti-SPP antibodies in (B) and anti-myc antibodies in (C) and (D). Bottom panels show Western blots of the lower part of the second-dimension gel using anti-opsin antibodies. SPP dimers (di) and non-, mono- (−1g) and bi- (−2g) glycosylated forms of OP91H* are indicated. Molecular masses are indicated in kDa.
OP91H* has two N-glycosylation sites and is detected as a non-, mono- or bi-glycosylated form after SDS/PAGE (Figures 3 and 4). In order to test whether the glycosylation status determines the assembly into one particular SPP complex, we analysed OP91H*–SPP and OP91H*–SPPD/A complexes using 2D-BN-SDS/PAGE (Figures 8B–8D). When OP91H* was expressed alone, glycosylated and non-glycosylated forms of OP91H* were mainly detected in high-molecular-mass complexes of approx. 600 kDa after second-dimension SDS/PAGE (Figure 8B). When co-expressed with SPP or SPPD/A, glycosylated and non-glycosylated OP91H* forms were detected to similar extents in the 200, 400 and 600 kDa complexes (Figures 8C and 8D), indicating that the glycosylation status does not determine OP91H* assembly into one particular SPP complex.
DISCUSSION
SPP interacts tightly with a signal peptide and a range of membrane proteins
We have investigated SPP interactions with a set of membrane proteins with type II oriented transmembrane regions. The pPrl-derived signal peptide SPPrl is an SPP substrate and interacts tightly with the catalytically inactive SPP mutant SPPD/A. The parent preprotein, pPrl, as well as the misfolded opsin OP91H* also interact with SPPD/A and furthermore with catalytically active SPP. Since pPrl and OP91H* accumulate in the presence of catalytically active SPP and no smaller processing fragments were detected, these proteins are not substrates for SPP-mediated processing. They bind to SPP, but are not processed and are in the following section termed SPP client proteins. No interaction of the type II membrane proteins Ii and RAMP4op with SPP was detected. We conclude that SPP interacts with a range of membrane proteins that is not restricted to substrates, but it does not interact with all type II membrane proteins.
The question arises as to what the features are that distinguish proteins that bind to SPP from those that do not. A preprotein, such as pPrl, accumulates because its signal sequence has not been cleaved off by signal peptidase. It remains anchored in the ER membrane via its signal sequence, resulting in a type II oriented membrane protein. SPP co-expression enhances the accumulation of pPrl and both also interact tightly with each other. The most likely explanation is that SPP is in close proximity to the translocation site in the ER membrane and optimally placed to compete with signal peptidase for binding to the signal sequence in the newly synthesized preprotein. Such an SPP binding may then prevent signal peptidase from cleavage. Consistent with this model, SPP is known to interact with the translocon-associated protein TRAM1 (translocation-associated membrane protein 1) [35]. A signal peptide and its parent preprotein are only distinguished by the absence or presence of the luminal ectodomain of the preprotein. Since mature prolactin did not interact with SPP, it must be the signal sequence that mediates the interaction with SPP. Although the preceding signal sequence cleavage is required to allow SPP-mediated processing of the signal peptide, as demonstrated previously [10], a luminal ectodomain does not generally prevent an interaction with SPP. Similarly, additional transmembrane domains, as in OP91H*, do not prevent an SPP interaction.
In its overall topology, Ii resembles a preprotein that spans the membrane with its signal anchor sequence. However, no interaction between SPP and Ii has been detected. Since a luminal domain, in principle, does not abolish an SPP interaction, there may be intrinsic features in the transmembrane domain required for SPP interaction. RAMP4 and a signal peptide are similar in their overall topology. Both expose their N-terminal domains in the cytosol, span the membrane with their hydrophobic domains and have a small C-terminal domain in the ER lumen. In contrast with a signal peptide, RAMP4 does not interact with SPP. Taken together, a type II topology and a small C-terminal domain in the ER lumen are not sufficient for an interaction with SPP, but a large luminal domain, as well as additional transmembrane domains, do not prevent an SPP interaction. It appears that not one single and simple feature determines whether a protein interacts with SPP. Since all proteins that interact with SPP have a transmembrane domain, there might be intrinsic features in this domain that are essential for an SPP interaction. Possibly the structural arrangement of the transmembrane domains determines the accessibility to SPP. Truncated opsin proteins were shown to have a less compact structure and a reduced α-helical content compared with the correctly assembled wild-type opsin [36]. Thus misfolded proteins that are not correctly assembled may expose their hydrophobic domains that are in turn accessible to SPP. Similarly, preproteins are usually generated as by-products of an inefficient process, so that one can assume them to remain unassembled and destined for disposal. In contrast, Ii and RAMP4 are known to oligomerize with their physiological interaction partners. Ii rapidly assembles into a homo-oligomeric complex upon membrane insertion [37], whereas RAMP4 associates with the Sec61 translocon and ribosomes [38]. Consequently, both proteins are presumably not accessible to SPP and therefore do not interact. In conclusion, we propose that the oligomeric state of type II oriented membrane proteins may be one critical determinant for their accessibility to SPP.
SPP assembles with substrates and client proteins into complexes of different sizes
Using BN-PAGE, we identified endogenous and overexpressed SPPs in three distinct high-molecular-mass complexes of 200, 400 and 600 kDa (C200, C400 and C600). A substrate, i.e. a signal peptide, is trapped in SPP-C200 when SPPD/A was co-expressed, while its preprotein is predominantly found in SPP-C600. In contrast, a misfolded membrane protein was identified in all three SPP complexes.
Since the substrate is only detected in the SPP-C200 complex, we suggest that this is the catalytically active SPP complex. Besides SPP, we did not find any other protein in this complex, consistent with the fact that SPP alone is sufficient to process signal peptides [1,18]. The 200 kDa complex is probably a dimer of SPP-dimers, since we detect dimers after 2D-BN-SDS/PAGE and since one SPP dimer has a molecular mass of approx. 90 kDa [16]. In contrast, SPP client proteins accumulate predominantly in the large SPP complexes C400 and C600. The presence of these complexes correlates with the finding that the interacting proteins are not substrates. As the amount of these complexes was small after affinity purification, we could not identify their protein constituents using MS. However, we found SPP dimers using 2D-BN-SDS/PAGE and Western blotting, indicating that this is the minimal SPP core in the large complexes. The higher mass suggests that either SPP dimers homo-oligomerize into larger complexes or that SPP dimers associate with further proteins.
Taken together, our findings indicate that substrates and client proteins can interact with SPP and are segregated by SPP. Both SPP substrates and its clients may bind initially to a docking site in SPP. Substrates may then enter the active site and are processed, whereas other proteins may be excluded from this site owing to steric hindrance by their ectodomains. This model could apply for a signal peptide and its preprotein. As the amino acid sequences of the signal peptide and the signal sequence are identical, it must be the large luminal ectodomain of the preprotein that prevents processing [10,39]. Similarly, also misfolded polytopic membrane proteins might be sterically hindered to enter the active site as they contain more than one transmembrane domain. The model of distinct docking and active sites has previously been deduced from the finding that a helical peptide inhibitor binds to another SPP site than a transition-state analogue [40]. Moreover, this concept also implies that a signal peptide, which entered the SPP active site, may be buried inside the protease. In contrast, when a preprotein or a misfolded polytopic membrane protein is bound to SPP, their additional domains, which are the luminal domain and the transmembrane domain respectively, may stick out of the SPP molecule, enabling further interactions and accumulation in larger SPP complexes. In these complexes, SPP itself or additional cofactors could contribute accessory binding sites. Since we identified all SPP complexes under steady-state conditions after Western blotting, we do not know how they are generated. It may be a dynamic process in which initially all SPP interacting proteins bind the SPP-C200 homo-oligomer and then, if not processed, mature into larger complexes. Alternatively, all SPP complexes could pre-exist and recruit preferentially signal peptides or unassembled membrane proteins with type II oriented transmembrane domains.
Also for the SPP-like proteases SPPL2a and SPPL2b, an interaction with substrates and the corresponding precursors has been shown [41–43]. As observed for SPP, the precursors interact with active SPPLs and accumulate. In contrast with this, no interaction of precursors with presenilin as the catalytic core of the γ-secretase complex has been observed so far [44,45]. One explanation is that γ-secretase is a multi-protein complex that consists of four subunits [19]. The catalytic core subunit is presenilin, an aspartyl intramembrane protease related to SPP, whereas a further subunit pre-selects peptide substrates and transfers them to the docking site in presenilin [46]. The characterization of γ-secretase by BN-PAGE identified heterogeneous complexes ranging in size from 200 to 900 kDa, but, in contrast with SPP, always included the subunits mentioned [47,48].
In contrast with γ-secretase, SPP functions may be characterized by a broad range of clients. Besides processing of signal peptides, SPP may collect disposal-prone membrane proteins. The interaction of SPP with clients that are prone to disposal is consistent with the proposed role of SPP in ER quality control of membrane proteins [14]. In such a scenario, SPP may shield unassembled exposed transmembrane regions. The SPP complexes we detect are possibly disposal-intermediates that represent part of a platform at which proteins are dislocated from the ER membrane [49]. In fact, SPP-dependent protein dislocation of full-length MHC class I heavy chains from the ER membrane has been demonstrated previously in cells expressing the HCMV-encoded immunoevasin US2 [15]. Furthermore, a recent study demonstrates that SPP co-operates with the ubiquitin E3 ligase TRC8 in the dislocation of MHC class I molecules from the ER membrane [50].
ERAD includes substrate recognition, targeting to the dislocation-machinery, dislocation from the ER membrane and degradation of the substrate by the proteasome [51]. Misfolded opsin proteins can be retained in the ER and can undergo ER-associated degradation via the proteasome [34]. We have shown that OP91H* is substantially degraded when overexpressed in cells. An interaction of misfolded opsin with endogenous SPP has been demonstrated previously [14]. However, co-expression of SPP impedes OP91H* degradation, resulting in accumulation. This apparent discrepancy can be explained by the overexpression situation. It is conceivable that high SPP amounts result in a prolonged interaction with degradation substrates, thereby impeding their efficient degradation. We attempted to clarify whether reduced SPP levels in the cell may affect the degradation of misfolded proteins by employing RNA interference. Substantial reduction of SPP did not lead to impeded degradation of OP91H* (see Supplementary Figure S6 at http://www.BiochemJ.org/bj/427/bj4270523add.htm). This could mean that SPP is not necessary for the degradation of OP91H or that low SPP amounts in the cell are sufficient for SPP function.
In an alternative model, SPP may act in the collection of disposal-prone membrane proteins by co-aggregating with them. Misfolded membrane proteins that expose hydrophobic domains tend to aggregate in the cell in an uncontrolled manner and can even recruit correctly folded proteins into aggregates, which in turn may lead to cell toxicity [34]. SPP with its ability to tightly bind such membrane proteins and to assemble with them into large oligomeric complexes, possibly aggregates, may contribute to a controlled sequestering of toxic membrane protein species. Large protein aggregates may then be eliminated by autophagic mechanisms.
Online data
AUTHOR CONTRIBUTION
Bianca Schrul designed and performed most experiments, interpreted the data, and wrote the manuscript. Katja Kapp contributed to experimental design and data interpretation, and wrote the manuscript. Irmgard Sinning contributed to the overall project and manuscript design. Bernhard Dobberstein conceived and supervised the project, interpreted the data, and wrote the manuscript.
ACKNOWLEDGEMENTS
We thank Bruno Martoglio (Novartis Pharma AG, Basel, Switzerland), Chica Schaller (ZMNH, Heidelberg, Germany), Benedict Cross (University of Manchester, Manchester, U.K.) and Stephen High (University of Manchester, Manchester, U.K.) for providing plasmids and antibodies, Thomas Ruppert (ZMBH) and Armin Bosserhoff (ZMBH) for MS, and Klaus Meese (ZMBH) for excellent technical assistance. We are grateful to Marius Lemberg (ZMBH) and Vincenzo Favaloro (ZMBH) for critical comments during the manuscript preparation, as well as to members of our group for stimulating discussions.
FUNDING
This work was funded by a fellowship from the Deutsche Forschungsgemeinschaft [grant number GRK1188 (to B.S.)].
References
- 1.Weihofen A., Binns K., Lemberg M. K., Ashman K., Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 2.Fluhrer R., Steiner H., Haass C. Intramembrane proteolysis by signal peptidepeptidases: a comparative discussion of GxGD -type aspartyl proteases. J. Biol. Chem. 2009;284:13975–13979. doi: 10.1074/jbc.R800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weihofen A., Lemberg M. K., Ploegh H. L., Bogyo M., Martoglio B. Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J. Biol. Chem. 2000;275:30951–30956. doi: 10.1074/jbc.M005980200. [DOI] [PubMed] [Google Scholar]
- 4.Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Heijne G. Signal sequences: the limits of variation. J. Mol. Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 6.Paetzel M., Karla A., Strynadka N. C., Dalbey R. E. Signal peptidases. Chem. Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 7.Lyko F., Martoglio B., Jungnickel B., Rapoport T. A., Dobberstein B. Signal sequence processing in rough microsomes. J. Biol. Chem. 1995;270:19873–19878. doi: 10.1074/jbc.270.34.19873. [DOI] [PubMed] [Google Scholar]
- 8.Martoglio B., Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 9.Kapp K., Schrempf S., Lemberg M. K., Dobberstein B. Post-targeting functions of signal peptides. In: Zimmermann R., editor. Protein Transport into the Endoplasmic Reticulum. Austin: Landes Bioscience; 2009. pp. 1–16. [Google Scholar]
- 10.Lemberg M. K., Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell. 2002;10:735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 11.Beel A. J., Sanders C. R. Substrate specificity of γ-secretase and other intramembrane proteases. Cell. Mol. Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw A. S., Rottier P. J., Rose J. K. Evidence for the loop model of signal-sequence insertion into the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 1988;85:7592–7596. doi: 10.1073/pnas.85.20.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dev K. K., Chatterjee S., Osinde M., Stauffer D., Morgan H., Kobialko M., Dengler U., Rueeger H., Martoglio B., Rovelli G. Signal peptide peptidase dependent cleavage of type II transmembrane substrates releases intracellular and extracellular signals. Eur. J. Pharmacol. 2006;540:10–17. doi: 10.1016/j.ejphar.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Crawshaw S. G., Martoglio B., Meacock S. L., High S. A misassembled transmembrane domain of a polytopic protein associates with signal peptide peptidase. Biochem. J. 2004;384:9–17. doi: 10.1042/BJ20041216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loureiro J., Lilley B. N., Spooner E., Noriega V., Tortorella D., Ploegh H. L. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature. 2006;441:894–897. doi: 10.1038/nature04830. [DOI] [PubMed] [Google Scholar]
- 16.Nyborg A. C., Kornilova A. Y., Jansen K., Ladd T. B., Wolfe M. S., Golde T. E. Signal peptide peptidase forms a homodimer that is labeled by an active site-directed γ-secretase inhibitor. J. Biol. Chem. 2004;279:15153–15160. doi: 10.1074/jbc.M309305200. [DOI] [PubMed] [Google Scholar]
- 17.Friedmann E., Lemberg M. K., Weihofen A., Dev K. K., Dengler U., Rovelli G., Martoglio B. Consensus analysis of signal peptide peptidase and homologous human aspartic proteases reveals opposite topology of catalytic domains compared with presenilins. J. Biol. Chem. 2004;279:50790–50798. doi: 10.1074/jbc.M407898200. [DOI] [PubMed] [Google Scholar]
- 18.Narayanan S., Sato T., Wolfe M. S. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J. Biol. Chem. 2007;282:20172–20179. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe M. S. The γ-secretase complex: membrane-embedded proteolytic ensemble. Biochemistry. 2006;45:7931–7939. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 20.Dultz E., Hildenbeutel M., Martoglio B., Hochman J., Dobberstein B., Kapp K. The signal peptide of the mouse mammary tumor virus Rem protein is released from the endoplasmic reticulum membrane and accumulates in nucleoli. J. Biol. Chem. 2008;283:9966–9976. doi: 10.1074/jbc.M705712200. [DOI] [PubMed] [Google Scholar]
- 21.Favaloro V., Spasic M., Schwappach B., Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroder K., Martoglio B., Hofmann M., Holscher C., Hartmann E., Prehn S., Rapoport T. A., Dobberstein B. Control of glycosylation of MHC class II-associated invariant chain by translocon-associated RAMP4. EMBO J. 1999;18:4804–4815. doi: 10.1093/emboj/18.17.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urny J., Hermans-Borgmeyer I., Gercken G., Schaller H. C. Expression of the presenilin-like signal peptide peptidase (SPP) in mouse adult brain and during development. Gene Expression Patterns. 2003;3:685–691. doi: 10.1016/s1567-133x(03)00094-2. [DOI] [PubMed] [Google Scholar]
- 24.Martoglio B., Graf R., Dobberstein B. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. EMBO J. 1997;16:6636–6645. doi: 10.1093/emboj/16.22.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schägger H., von Jagow G. Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 28.Mortz E., Krogh T. N., Vorum H., Gorg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2001;1:1359–1363. doi: 10.1002/1615-9861(200111)1:11<1359::AID-PROT1359>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 29.Heimann M., Roman-Sosa G., Martoglio B., Thiel H. J., Rumenapf T. Core protein of pestiviruses is processed at the C terminus by signal peptide peptidase. J. Virol. 2006;80:1915–1921. doi: 10.1128/JVI.80.4.1915-1921.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krawitz P., Haffner C., Fluhrer R., Steiner H., Schmid B., Haass C. Differential localization and identification of a critical aspartate suggest non-redundant proteolytic functions of the presenilin homologues SPPL2b and SPPL3. J. Biol. Chem. 2005;280:39515–39523. doi: 10.1074/jbc.M501645200. [DOI] [PubMed] [Google Scholar]
- 31.Lipp J., Dobberstein B. Signal recognition particle-dependent membrane insertion of mouse invariant chain: a membrane-spanning protein with a cytoplasmically exposed amino terminus. J. Cell Biol. 1986;102:2169–2175. doi: 10.1083/jcb.102.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefanovic S., Hegde R. S. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 33.Heymann J. A., Subramaniam S. Expression, stability, and membrane integration of truncation mutants of bovine rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4966–4971. doi: 10.1073/pnas.94.10.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saliba R. S., Munro P. M., Luthert P. J., Cheetham M. E. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J. Cell Sci. 2002;115:2907–2918. doi: 10.1242/jcs.115.14.2907. [DOI] [PubMed] [Google Scholar]
- 35.Oresic K., Ng C. L., Tortorella D. TRAM1 participates in human cytomegalovirus US2- and US11-mediated dislocation of an endoplasmic reticulum membrane glycoprotein. J. Biol. Chem. 2009;284:5905–5914. doi: 10.1074/jbc.M807568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X., Garriga P., Khorana H. G. Structure and function in rhodopsin: correct folding and misfolding in two point mutants in the intradiscal domain of rhodopsin identified in retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4554–4559. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roche P. A., Marks M. S., Cresswell P. Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature. 1991;354:392–394. doi: 10.1038/354392a0. [DOI] [PubMed] [Google Scholar]
- 38.Gorlich D., Rapoport T. A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 39.Lemberg M. K., Martoglio B. On the mechanism of SPP-catalysed intramembrane proteolysis: conformational control of peptide bond hydrolysis in the plane of the membrane. FEBS Lett. 2004;564:213–218. doi: 10.1016/S0014-5793(04)00192-9. [DOI] [PubMed] [Google Scholar]
- 40.Sato T., Nyborg A. C., Iwata N., Diehl T. S., Saido T. C., Golde T. E., Wolfe M. S. Signal peptide peptidase: biochemical properties and modulation by nonsteroidal antiinflammatory drugs. Biochemistry. 2006;45:8649–8656. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- 41.Martin L., Fluhrer R., Reiss K., Kremmer E., Saftig P., Haass C. Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J. Biol. Chem. 2008;283:1644–1652. doi: 10.1074/jbc.M706661200. [DOI] [PubMed] [Google Scholar]
- 42.Fluhrer R., Grammer G., Israel L., Condron M. M., Haffner C., Friedmann E., Bohland C., Imhof A., Martoglio B., Teplow D. B., Haass C. A γ-secretase-like intramembrane cleavage of TNFα by the GxGD aspartyl protease SPPL2b. Nat. Cell Biol. 2006;8:894–896. doi: 10.1038/ncb1450. [DOI] [PubMed] [Google Scholar]
- 43.Kirkin V., Cahuzac N., Guardiola-Serrano F., Huault S., Luckerath K., Friedmann E., Novac N., Wels W. S., Martoglio B., Hueber A. O., Zornig M. The Fas ligand intracellular domain is released by ADAM10 and SPPL2a cleavage in T-cells. Cell Death Differ. 2007;14:1678–1687. doi: 10.1038/sj.cdd.4402175. [DOI] [PubMed] [Google Scholar]
- 44.Esler W. P., Kimberly W. T., Ostaszewski B. L., Ye W., Diehl T. S., Selkoe D. J., Wolfe M. S. Activity-dependent isolation of the presenilin–γ-secretase complex reveals nicastrin and a γ substrate. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2720–2725. doi: 10.1073/pnas.052436599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thinakaran G., Regard J. B., Bouton C. M., Harris C. L., Price D. L., Borchelt D. R., Sisodia S. S. Stable association of presenilin derivatives and absence of presenilin interactions with APP. Neurobiol. Dis. 1998;4:438–453. doi: 10.1006/nbdi.1998.0171. [DOI] [PubMed] [Google Scholar]
- 46.Shah S., Lee S. F., Tabuchi K., Hao Y. H., Yu C., LaPlant Q., Ball H., Dann C. E., 3rd, Sudhof T., Yu G. Nicastrin functions as a γ-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Hebert S. S., Serneels L., Dejaegere T., Horre K., Dabrowski M., Baert V., Annaert W., Hartmann D., De Strooper B. Coordinated and widespread expression of γ-secretase in vivo: evidence for size and molecular heterogeneity. Neurobiol. Dis. 2004;17:260–272. doi: 10.1016/j.nbd.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Evin G., Canterford L. D., Hoke D. E., Sharples R. A., Culvenor J. G., Masters C. L. Transition-state analogue γ-secretase inhibitors stabilize a 900 kDa presenilin/nicastrin complex. Biochemistry. 2005;44:4332–4341. doi: 10.1021/bi0481702. [DOI] [PubMed] [Google Scholar]
- 49.Loureiro J., Ploegh H. L. Antigen presentation and the ubiquitin–proteasome system in host–pathogen interactions. Adv. Immunol. 2006;92:225–305. doi: 10.1016/S0065-2776(06)92006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stagg H. R., Thomas M., van den Boomen D., Wiertz E. J., Drabkin H. A., Gemmill R. M., Lehner P. J. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J. Cell Biol. 2009;186:685–692. doi: 10.1083/jcb.200906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai B., Ye Y., Rapoport T. A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.