Abstract
Sirtuin 1 (SirT1) is the largest of the seven members of the sirtuin family of class III nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylases, whose activation is beneficial for metabolic, neurodegenerative, inflammatory and neoplastic diseases, and augments life span in model organisms (Finkel et al., 2009; Lavu et al., 2008). In vitro studies show that SirT1 protects genome integrity and is involved in circadian physiological rhythms (Asher et al., 2008; Nakahata et al., 2008; Oberdoerffer et al., 2008). In the last few years, a fundamental role for SirT1 in the metabolism and differentiation of skeletal muscle cells has been uncovered (Fulco et al., 2003), and the use of specific transgenic or knockout SirT1 mouse models implicates it in the protection of heart muscle from oxidative and hypertrophic stresses (Alcendor et al., 2007). In this Perspective, we review the recent exciting findings that have established a key role for the ’longevity’ protein SirT1 in skeletal and heart muscle physiology and disease. Furthermore, given the multiple biological functions of SirT1, we discuss the unique opportunities that SirT1 mouse models can offer to improve our integrated understanding of the metabolism, as well as the regeneration and aging-associated changes in the circadian function, of skeletal and heart muscle.
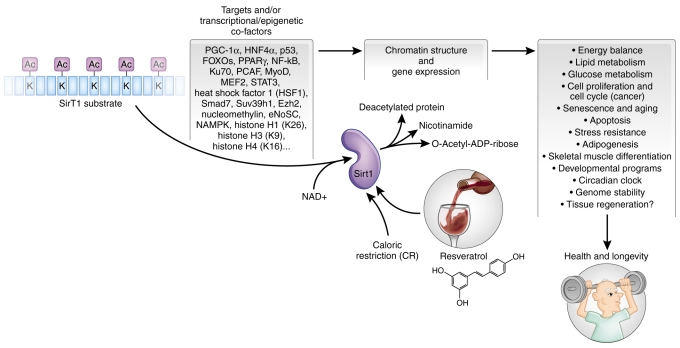
Why is SIRT1 considered to be a ‘longevity’ gene? SirT1 is the mammalian ortholog of yeast Sir2, an enzyme that is involved in protein deacetylation, which was first characterized as an important regulator of life span in this organism, and subsequently in higher eukaryotes (Longo and Kennedy, 2006). However, whether SirT1 is associated with an extension of the life span of human cells is a matter of some debate (Michishita et al., 2005). SirT1 substrates and transcriptional/epigenetic co-factors make up an impressive and constantly growing list, including, among others, PGC-1α, HNF4α, p53, FOXOs, PPARγ, NF-κB, Ku70, PCAF, MyoD, MEF2, STAT3, HSF1, Smad7, Suv39h1, Ezh2, nucleomethylin, eNoSC and various histones (Nemoto et al., 2004; Nemoto et al., 2005; Rodgers et al., 2005; Kume et al., 2007; Grummt and Ladurner, 2008; Finkel et al., 2009; Nie et al., 2009; Vaquero and Reinberg, 2009; Westerheide et al., 2009). SirT1 influences numerous processes that are crucial to cell viability, such as gene silencing or activation, apoptosis, stress resistance, senescence, energy balance, and lipid and glucose metabolism (Fig. 1). Recent elegant work on SirT1 knockout mouse embryonic fibroblasts (MEFs) and embryonic stem cells showed that SirT1 activity impacts functionally on the circadian clock (Asher et al., 2008; Nakahata et al., 2008) and on genome (chromatin) stability (Oberdoerffer et al., 2008; Wang et al., 2008), and an integrated picture of SirT1-dependent anti-cancer and anti-aging effects is just emerging (Fig. 1) (Jung-Hynes and Ahmad, 2009; Liu et al., 2009).
Fig. 1.
The enzymatic reaction carried out by SirT1, its targets, including transcriptional co-factors, and dependent biological processes. SirT1 protein substrate(s) is represented as a string of blue rectangles, with acetylated (Ac) lysine (K) residues. Using NAD+ as a co-factor, SirT1 can deacetylate histones, and nuclear and cytoplasmic proteins on specific K residues. This reaction generates a deacetylated protein, nicotinamide and O-acetyl-ADP-ribose (OAADPR). SirT1 activity can be enhanced by caloric restriction (CR) and by the polyphenol resveratrol, affecting multiple developmental, physiological and pathological processes, and ultimately favoring health and increasing longevity.
Several mechanisms that are capable of activating sirtuin enzymatic activity have been shown to increase life span. Classical activators of SirT1 include the polyphenol resveratrol (contained in red grapes and green tea) (Howitz et al., 2003), as well as a regimen of caloric restriction (CR) (Cohen et al., 2004). CR, defined in mice as a reduction in food intake of 30%–50% compared with animals fed ad libitum, is a very well-known intervention that enhances longevity in laboratory animals (Fig. 1). CR may actually increase life span by triggering a complex interplay of signaling molecules, including not only SirT1, but also AMP-activated protein kinase (AMPK), forkhead box O transcription factors (FOXOs), mammalian target of rapamycin (mTOR), and the ratio of NAD+ to NADH (Cantó and Auwerx, 2009b). Similarly, resveratrol impacts on additional cellular pathways, probably owing to its chemical nature as a protein-binding polyphenol. Owing to the pleiotropic positive effects of SirT1 on the health of organisms, the pharmaceutical industry have shown a growing interest in developing compounds that are able to modulate SirT1 activity (Lavu et al., 2008).
In this review, we will narrow our focus on the role of SirT1 activity in two striated muscle tissues of embryonic mesodermic origin, skeletal and heart muscle, which govern fundamental processes such as glucose and lipid metabolism, physical activity, and propulsion of blood around the circulatory system, with a particular attention to relevant SirT1-specific mouse models. Readers who are interested in other sirtuins (SirT2–SirT7) or in other tissue-specific SirT1 mice models are referred to the recent excellent reviews by Finkel et al. and Guarente (Guarente, 2007; Finkel et al., 2009).
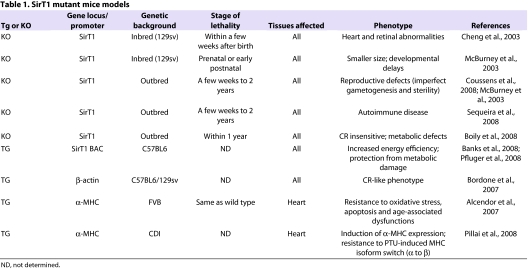
Given the technologies available to manipulate the mouse genome (van der Weyden et al., 2003), and the high degree of homology between murine and human genomes, the mouse is considered the premier organism for modeling human pathologies. Using the mouse as a model organism provides the possibility of generating loss-of-function and gain-of-function mutants of disease-candidate proteins, even in a conditional (tissue-specific) and/or inducible manner. In this respect, we learned in 2003 that, when generated in an inbred genetic background, whole-body SirT1 knockout (KO) mice carrying two null alleles of Sirt1 die prenatally or during the early postnatal period, with neurological and cardiac malformations (Cheng et al., 2003; McBurney et al., 2003). This points to a crucial role for active SirT1 in homeostasis (Table 1). However, in outbred backgrounds, whole-body SirT1 KO produces viable mice with diverse phenotypes such as imperfect gametogenesis and sterility (McBurney et al., 2003; Coussens et al., 2008); an autoimmune-like condition (Sequeira et al., 2008); and an impairment in obtaining benefits from the positive CR-induced metabolic effects (Table 1) (Boily et al., 2008). These findings highlight the importance of considering the impact of genetic background variability when analyzing murine phenotypes. Conversely, whole-body bacterial artificial chromosome (BAC)-driven transgenic (Tg) overexpression of SirT1 in mice, even at moderate levels (∼ twofold to threefold), has been unequivocally proven to be beneficial, inducing an increase in energy efficiency and preventing metabolic damage (Banks et al., 2008; Pfluger et al., 2008). SirT1 overexpression is thus thought to resemble closely the beneficial phenotype induced by CR (Table 1) (Bordone et al., 2007). Given that CR is a very efficient strategy to reverse both the clinical features of metabolic syndromes such as obesity and insulin resistance in humans (Opie, 2009), and the CR-like phenotypes of SirT1-overexpressing mice, this evidence suggests that new SirT1-activating compounds could be useful for the future management of patients suffering from metabolic disturbances.
Table 1.
SirT1 mutant mice models
What happens if SirT1 is artificially manipulated in mouse skeletal or heart muscle cells? Skeletal muscle-specific SirT1 Tg or KO mice models have not yet been reported, but the effects of SirT1 have been studied extensively in skeletal muscle cells. An original report using cultured murine myotubes and human primary skeletal muscle cells demonstrated that SirT1 overexpression represses the muscle transcriptional regulator MyoD (Fulco et al., 2003). As a consequence, the production of several transcripts including those encoding myogenin and muscle contractile proteins was blocked, and muscle differentiation, which was monitored as a reduced fusion of myoblasts into myotubes, was severely inhibited (Fulco et al., 2003). The ratio of NAD+ to NADH and the redox state are intimately linked to nutrient availability in muscle cells, and an elegant follow-up of this work places SirT1 at the crossroads between the two. If cultured myoblasts are exposed to glucose restriction, SirT1 activity is enhanced through AMPK-dependent regulation of NAM phosphoribosyltransferase (NAMPT), the rate-limiting enzyme that is responsible for NAD+ turnover, and this blocks differentiation into myotubes (Fulco et al., 2008). Moreover, in cultured myotubes, the presence of SirT1 was shown to be necessary for the cell-autonomous switch from glucose utilization to fatty acid oxidation in the presence of a low glucose concentration; this flexible metabolic response occurs during CR and is generally impaired during metabolic diseases (Gerhart-Hines et al., 2007). These and other seminal in vitro studies thus uncovered a key role for SirT1 in shaping muscle cellular metabolism and differentiation by functional interaction with other regulators of cellular energy stores, such as AMPK and peroxisome proliferator-activated receptor gamma (PPARγ) coactivator-1 alpha (PGC-1α) (Nemoto et al., 2005; Lagouge et al., 2006; Gerhart-Hines et al., 2007; Amat et al., 2009; Cantó and Auwerx, 2009a). The field now awaits skeletal muscle-specific SirT1 Tg or KO mice models, which would confirm, in vivo, the role of SirT1 in the differentiation, metabolism and contractile function of this tissue at the level of a whole organism.
By contrast, two cardiac-restricted Tg SirT1 mouse models, obtained by using the tissue specific α-myosin heavy chain (α-MHC) promoter, have been described (Alcendor et al., 2007; Pillai et al., 2008). In one study, low to moderate (about threefold to eightfold) SirT1 overexpression efficiently protected mice from paraquat-induced cardiac stress and apoptosis, and delayed the onset of age-dependent heart dysfunctions (Table 1) (Alcendor et al., 2007). Conversely, greater increases in SirT1 levels (about 13-fold) induced oxidative stress and apoptosis, ultimately leading to cardiomyopathy and decreased survival (Table 1) (Alcendor et al., 2007). This was the first report to introduce the ‘hormesis’ concept, meaning that SirT1 activation – depending on its extent – can be either beneficial or deleterious in the heart. A second study showed that Tg heart-restricted SirT1 overexpression upregulated α-MHC levels and protected against the switch in cardiac MHC isoform expression (α to β) that is induced by 6-propyl-2-thiouracil (PTU), a potent antithyroid drug (Table 1) (Pillai et al., 2008).
In parallel, in vitro findings from cultured or primary cardiomyocyte models expanded our understanding of the cardioprotective effects of the longevity protein SirT1 and increased NAD+ availability, including the increased resistance to ischemia/reperfusion-induced oxidative stress (Hsu et al., 2009; Rane et al., 2009), angiotensin II-dependent hypertrophy (Pillai et al., 2006) and apoptosis (Alcendor et al., 2004; Pillai et al., 2005), thus strengthening the view that pharmacological SirT1 activation might be beneficial for the treatment of cardiac diseases (Hsu et al., 2008; Lavu et al., 2008; Borradaile et al., 2009).
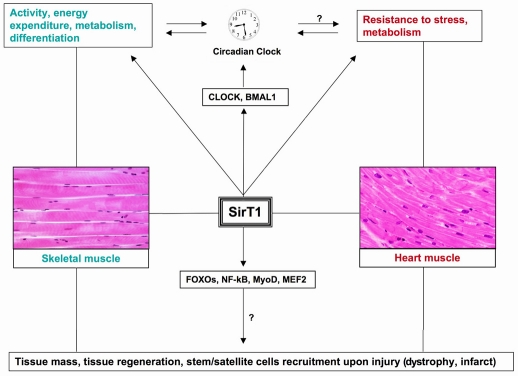
Therapeutic areas that will probably benefit from SirT1 research are (1) the control of reduced muscle mass and (2) muscle regeneration upon injury. First, muscle mass is reduced during a condition of atrophy (such as in response to starvation, immobilization or treatment with glucocorticoids) or cachexia, which leads to muscle wasting owing to increased protein catabolism (McKinnell and Rudnicki, 2004; Mourkioti and Rosenthal, 2005). Muscle-specific RING finger protein 1 (MuRF1) and MAFbx/atrogin-1, which are E3 ubiquitin ligases involved in proteasome-mediated proteolysis of muscle proteins, are transcriptionally controlled by NF-κB and FOXOs, respectively (Glass, 2005). Since SirT1 modulates both NF-κB and FOXOs, in concert with regulating MyoD and myocyte enhancer factor-2 (MEF2), it may also control muscle mass during injury (Fig. 2). In addition, SirT1 controls angiogenesis and vasculogenesis during development (Potente et al., 2007; Potente and Dimmeler, 2008); however, its regenerative potential in other tissues is unknown. This is an active field of investigation in skeletal and heart muscle diseases since these tissues were long considered ‘post-mitotic’ and therefore have a limited regenerative capacity upon aging, damage, skeletal muscle dystrophy or cardiac infarct.
Fig. 2.
Role of SirT1 in skeletal and heart muscle physiology, disease and regeneration upon damage. SirT1 modulates the activity of circadian clock molecular components and regulates the activity, energy expenditure, metabolism and differentiation of skeletal muscle, as well as the stress resistance and metabolism of heart muscle. Hypothetical connections that are not yet supported by experimental evidence are accompanied by a question mark.
Recently, many efforts have relied on the characterization of skeletal or heart muscle-specific stem and/or satellite cells that, once recruited, may contribute to repairing injured tissues. Since it has been proposed that SirT1 may also influence the lineage/cell-fate decisions of stem cells by sensing redox status (Machida and Booth, 2004; Mantel and Broxmeyer, 2008), it is conceivable that SirT1 may also influence the regenerative potential of skeletal muscle and the heart, by affecting the activation, proliferation and differentiation of their respective adult stem cells (Fig. 2). Two lines of evidence support this hypothesis. First, class I and II histone deacetylase inhibitors proved to be beneficial for muscular functional recovery in dystrophic mouse models (Minetti et al., 2006; Colussi et al., 2008). However, we ignore whether modulation of SirT1 activity in dystrophic skeletal muscle may ameliorate the disease. Pharmacological activation/inhibition of SirtT1 together with combined genetic studies using conditional satellite cell-specific SirT1 Tg/KO and dystrophic (such as MDX) mice models would undoubtedly shed light on this issue. Second, locally acting isoforms of insulin growth factor-1 (IGF-1) have great repair and regenerative effects in injured skeletal and heart muscle tissue (Musaro et al., 1999; Musaro et al., 2001; Barton et al., 2002; Winn et al., 2002; Musarò et al., 2004; Mourkioti and Rosenthal, 2005; Schulze et al., 2005; Pelosi et al., 2007). Interestingly, IGF-1 and SirT1 share downstream signaling targets, such as FOXOs, in skeletal myocytes and cardiomyocytes. It has been reported that circulating IGF-1 counteracted SirT1 activity (Cohen et al., 2004). Moreover, the levels of circulating IGF-1 are lowered upon caloric restriction (Huffman et al., 2008). Hence, SirT1 and circulating IGF-1 play opposite biological roles in longevity (Longo, 2009), but it is unknown how muscle and cardiac SirT1 activity impacts on the regenerative potential of separate IGF-1 isoforms, acting locally or systemically. We postulate that significant advances in this research field will be achieved by the combined use of IGF-1 and SirT1 Tg and KO genetic mice models.
Based on these considerations, and the established role of SirT1 in skeletal muscle cell proliferation and differentiation (Fulco et al., 2003), we predict that SirT1 agonists and antagonists may be useful in treating muscle damage. In the initial phases of muscle injury, SirT1 agonists may help in amplifying the expansion of the satellite cell pool, and later on, administration of SirT1 antagonists may favor the differentiation of this expanded satellite cell population.
In both skeletal and heart muscle, gene expression is regulated in a circadian fashion (24-hour cycles), and the molecular architecture of this phenomenon relies on a complex transcription-translation feedback loop in which a heterodimeric transcription factor, CLOCK/BMAL1, regulates the expression of other clock genes [such as Bmal1 (also known as Arntl), Per2, Dbp, Rora and Cry1]. This molecular circadian clock machine governs all of the physiological rhythms that are present in living beings, including sleep-wake cycles and feeding. Twenty-four-hour rhythms are endogenously generated but can also be entrained by external cues, such as light and food availability, which act on the hypothalamic suprachiasmatic nucleus (SCN) (Crosio et al., 2000). The SCN clock is believed to set the phases of peripheral tissues, such as skeletal and heart muscle (Lamia et al., 2008). Whereas in the skeletal muscle the circadian clock is crucial for activity and body weight (Zambon et al., 2003; McDearmon et al., 2006), the cardiac clock enables the heart to anticipate environmental stimuli, ensuring an appropriate response (Esser and Young, 2009). In fact, diurnal changes in myocardial contractions are well known, both in mouse models and in humans. Clock gene expression patterns are altered in animal models of hypertension, myocardial infarction or ischemia (Esser and Young, 2009), and in humans, myocardial infarction more often occurs early in the morning and in shift workers (with altered sleep-wake cycles) (Esser and Young, 2009). However, the cause-effect relationships of this phenomenon are not understood.
SirT1 deacetylates CLOCK and BMAL1 in a circadian fashion in MEFs and is a core component of the circadian clock (Asher et al., 2008; Nakahata et al., 2008). Genetic mouse models have uncovered unequivocal links between metabolic intracellular activity and circadian rhythms. Mice that are deficient for CLOCK and BMAL1 display metabolic phenotypes with altered glucose and fat homeostasis (Rudic et al., 2004; Turek et al., 2008). Nonetheless, our understanding of the connections between SirT1-dependent metabolism and the circadian clock is just beginning. What is clear is that NAD+ is central for both metabolism and circadian rhythm. By activating SirT1, NAD+ can control the production of NAMPT, through CLOCK and BMAL1, in a circadian fashion (Nakahata et al., 2009; Ramsey et al., 2009). Therefore, the circadian clock governs intracellular NAD+ levels through an interlocked and transcriptional feedback loop. These in vitro findings, mainly obtained in MEFs, will undoubtedly be the basis for in vivo studies to assess the functional relevance of the interaction between the circadian clock and SirT1 in skeletal and heart muscle physiopathology. A functional relationship between other energy metabolism regulators, which are in turn intimately bound to SirT1 function, such as AMPK (Fulco et al., 2008; Cantó et al., 2009a) and PGC-1α (Nemoto et al., 2005; Rodgers et al., 2005), and the mammalian clock has been found in skeletal muscle (Liu et al., 2007; Vieira et al., 2008), whereas in the heart such links have not yet been explored.
New SirT1 mutant mouse models will allow us to decipher its role in regulating the activity and structure of many proteins that are at the core of muscle and heart muscle function, including the important connections to the observed circadian patterns of metabolic behavior and to pathophysiological events in muscle (dys)function (Fig. 2).
Acknowledgments
We would like to thank members of the Rosenthal lab for helpful discussions. N.R. acknowledges support by the European Union grants ‘Heart Repair’ ( LSHM-CT-2005-018630) and EUMODIC ( LSHG-CT-2006-037188), and V.S. acknowledges support from the Intramural Research Program of the National Institute of Arthritis, Musculoskeletal, and Skin Diseases (NIH-NIAMS) . M.V. is the recipient of an EIPOD fellowship from the EMBL. We apologize to colleagues whose research work we could not cite for length limits. Deposited in PMC for release after 12 months.
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Alcendor R, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. (2004). Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 95, 971–980 [DOI] [PubMed] [Google Scholar]
- Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. (2007). Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 100, 1512–1521 [DOI] [PubMed] [Google Scholar]
- Amat R, Planavila A, Chen SL, Iglesias R, Giralt M, Villarroya F. (2009). SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma Co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J Biol Chem. 284, 21872–21880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. (2008). SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 [DOI] [PubMed] [Google Scholar]
- Banks A, Kon N, Knight C, Matsumoto M, Gutiérrez-Juárez R, Rossetti L, Gu W, Accili D. (2008). SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 8, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton E, Morris L, Musaro A, Rosenthal N, Sweeney HL. (2002). Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 157, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boily G, Seifer EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, et al. (2008). SirT1 regulates energy metabolism and response to caloric restriction in mice. PloS One 3, e1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, et al. (2007). SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6, 759–767 [DOI] [PubMed] [Google Scholar]
- Borradaile N, Pickering JG. (2009). NAD(+), sirtuins, and cardiovascular disease. Curr Pharm Des. 15, 110–117 [DOI] [PubMed] [Google Scholar]
- Cantó C, Auwerx J. (2009a). PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 20, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Auwerx J. (2009b). Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 20, 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Mostoslavsky R, Saito S, Manis J, Gu Y, Patel P, Bronson R, Appella E, Alt F, Chua K. (2003). Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. (2004). Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- Colussi C, Mozzetta C, Gurtner A, Illi B, Rosati J, Straino S, Ragone G, Pescatori M, Zaccagnini G, Antonini A, et al. (2008). HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc Natl Acad Sci USA 105, 19183–19187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. (2008). Sirt1 deficiency attenuates spermatogenesis and germ cell function. PloS One 3, e1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. (2000). Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 3, 1241–1247 [DOI] [PubMed] [Google Scholar]
- Esser K, Young ME. (2009). The role of clock genes in cardiometabolic disease. J Appl Physiol. 107, 1316–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. (2009). Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Sartorelli V. (2008). Comparing and contrasting the roles of AMPK and SIRT1 in metabolic tissues. Cell Cycle 7, 3669–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. (2003). Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell 12, 51–62 [DOI] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. (2008). Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell 14, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. (2007). Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass D. (2005). A signaling role for dystrophin: inhibiting skeletal muscle atrophy pathways. Cancer Cells. 8, 351–352 [DOI] [PubMed] [Google Scholar]
- Grummt I, Ladurner AG. (2008). A metabolic throttle regulates the epigenetic state of rDNA. Cell 133, 577–580 [DOI] [PubMed] [Google Scholar]
- Guarente L. (2007). Sirtuins in aging and disease. Cold Spring Harb. Symp. Quant Biol. 72, 483–488 [DOI] [PubMed] [Google Scholar]
- Howitz K, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. (2003). Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- Hsu C, Odewale I, Alcendor R, Sadoshima J. (2008). Sirt1 protects the heart from aging and stress. J Biol Chem. 389, 221–231 [DOI] [PubMed] [Google Scholar]
- Hsu C, Oka S, Shao D, Hariharan N, Sadoshima J. (2009). Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ Res. 105, 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman DM, Moellering DR, Grizzle WE, Stockard CR, Johnson MS, Nagy TR. (2008). Effect of exercise and calorie restriction on biomarkers of aging in mice. Am J Physiol Regul Integr Comp Physiol 294, R1618–R1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung-Hynes B, Ahmad N. (2009). SIRT1 controls circadian clock circuitry and promotes cell survival: a connection with age-related neoplasms. FASEB J. 23, 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isshiki K, Isono M, Uzu T, Guarente L, Kashiwagi A, et al. (2007). SIRT1 inhibits transforming growth factor beta-induced apoptosis in glomerular mesangial cells via Smad7 deacetylation. J Biol Chem. 282, 151–158 [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. (2006). Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- Lamia K, Storch KF, Weitz CJ. (2008). Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA 105, 15172–15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavu S, Boss O, Elliott PJ, Lambert PD. (2008). Sirtuins-novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 7, 841–853 [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. (2007). Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature 447, 477–481 [DOI] [PubMed] [Google Scholar]
- Liu T, Liu PY, Marshall GM. (2009). The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 69, 1702–1705 [DOI] [PubMed] [Google Scholar]
- Longo V. (2009). Linking sirtuins, IGF-I signaling, and starvation. Exp Gerontol. 44, 70–74 [DOI] [PubMed] [Google Scholar]
- Longo V, Kennedy BK. (2006). Sirtuins in aging and age-related disease. Cell 126, 257–268 [DOI] [PubMed] [Google Scholar]
- Machida S, Booth FW. (2004). Increased nuclear proteins in muscle satellite cells in aged animals as compared to young growing animals. Exp Gerontol. 39, 1521–1525 [DOI] [PubMed] [Google Scholar]
- Mantel C, Broxmeyer HE. (2008). Sirtuin 1, stem cells, aging, and stem cell aging. Curr Opin Hematol. 15, 326–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney M, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. (2003). The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 23, 38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDearmon E, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, et al. (2006). Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 314, 1304–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell I, Rudnicki MA. (2004). Molecular mechanisms of muscle atrophy. Cell 119, 907–910 [DOI] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. (2005). Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti G, Colussi C, Adami R, Serra C, Mozzetta C, Parente V, Fortuni S, Straino S, Sampaolesi M, Di Padova M, et al. (2006). Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat Med. 12, 1147–1150 [DOI] [PubMed] [Google Scholar]
- Mourkioti F, Rosenthal N. (2005). IGF-1, inflammation and stem cells: interactions during muscle regeneration. Trends Immunol. 26, 535–542 [DOI] [PubMed] [Google Scholar]
- Musaro A, McCullagh KJ, Naya FJ, Olson EN, Rosenthal N. (1999). IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 400, 581–585 [DOI] [PubMed] [Google Scholar]
- Musaro A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, Barton ER, Sweeney HL, Rosenthal N. (2001). Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet. 27, 195–200 [DOI] [PubMed] [Google Scholar]
- Musarò A, Giacinti C, Borsellino G, Dobrowolny G, Pelosi L, Cairns L, Ottolenghi S, Cossu G, Bernardi G, Battistini L, et al. (2004). Stem cell-mediated muscle regeneration is enhanced by local isoform of insulin-like growth factor 1. Proc Natl Acad Sci USA 101, 1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. (2008). The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. (2004). Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. (2005). SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. (2009). STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 11, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. (2008). SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opie L. (2009). Caloric restriction models reverse metabolic syndrome. J Am Coll Cardiol. 53, 899–900 [DOI] [PubMed] [Google Scholar]
- Pelosi L, Giacinti C, Nardis C, Borsellino G, Rizzuto E, Nicoletti C, Wannenes F, Battistini L, Rosenthal N, Molinaro M, et al. (2007). Local expression of IGF-1 accelerates muscle regeneration by rapidly modulating inflammatory cytokines and chemokines. FASEB J. 21, 1393–1402 [DOI] [PubMed] [Google Scholar]
- Pfluger P, Herranz D, Velasco-Miguel S, Serrano M, Tschöp MH. (2008). Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA 105, 9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai J, Isbatan A, Imai S, Gupta MP. (2005). Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 280, 43121–43130 [DOI] [PubMed] [Google Scholar]
- Pillai JB, Gupta M, Rajamohan SB, Lang R, Raman J, Gupta MP. (2006). Poly(ADP-ribose) polymerase-1-deficient mice are protected from angiotensin II-induced cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 291, H1545–H1553 [DOI] [PubMed] [Google Scholar]
- Pillai J, Chen M, Rajamohan SB, Samant S, Pillai VB, Gupta M, Gupta MP. (2008). Activation of SIRT1, a class III histone deacetylase, contributes to fructose feeding-mediated induction of the alpha-myosin heavy chain expression. Am J Physiol Heart Circ Physiol. 294, H1388–H1397 [DOI] [PubMed] [Google Scholar]
- Potente M, Dimmeler S. (2008). Emerging roles of SIRT1 in vascular endothelial homeostasis. Cell Cycle 7, 2117–2122 [DOI] [PubMed] [Google Scholar]
- Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, et al. (2007). SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 21, 2644–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey K, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. (2009). Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. (2009). Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 104, 879–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. (2005). Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- Rudic R, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. (2004). BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PloS Biol. 2, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze P, Fang J, Kassik KA, Gannon J, Cupesi M, MacGillivray C, Lee RT, Rosenthal N. (2005). Transgenic overexpression of locally acting insulin-like growth factor-1 inhibits ubiquitin-mediated muscle atrophy in chronic left-ventricular dysfunction. Circ Res. 97, 418–426 [DOI] [PubMed] [Google Scholar]
- Sequeira J, Boily G, Bazinet S, Saliba S, He X, Jardine K, Kennedy C, Staines W, Rousseaux C, Mueller R, et al. (2008). sirt1-null mice develop an autoimmune-like condition. Exp Cell Res. 314, 3069–3074 [DOI] [PubMed] [Google Scholar]
- Turek F, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. (2008). Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L, Adams DJ, Bradley A. (2003). Tools for targeted manipulation of the mouse genome. Physiol Genomics 11, 133–164 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Reinberg D. (2009). Calorie restriction and the exercise of chromatin. Genes Dev. 23, 1849–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira E, Nilsson EC, Nerstedt A, Ormestad M, Long YC, Garcia-Roves PM, Zierath JR, Mahlapuu M. (2008). Relationship between AMPK and the transcriptional balance of clock-related genes in skeletal muscle. Am J Physiol Endocrinol Metab. 295, E1032–E1037 [DOI] [PubMed] [Google Scholar]
- Wang R, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. (2008). Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14, 312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide S, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. (2009). Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn N, Paul A, Musaro A, Rosenthal N. (2002). Insulin-like growth factor isoforms in skeletal muscle aging, regeneration, and disease. Cold Spring Harbor Symp Quant Biol. 67, 507–518 [DOI] [PubMed] [Google Scholar]
- Zambon A, McDearmon EL, Salomonis N, Vranizan KM, Johansen KL, Adey D, Takahashi JS, Schambelan M, Conklin BR. (2003). Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 4, R61. [DOI] [PMC free article] [PubMed] [Google Scholar]