Summary
HIV-1 employs strand transfer for recombination between the two viral genomes. We previously provided evidence that strand transfer proceeds by an invasion-mediated mechanism, in which a DNA segment on the original RNA template is invaded by a second RNA template at a gap site. The initial RNA-DNA hybrid then expands until the DNA is fully transferred. Ribonuclease H (RNase H) cleavages and nucleocapsid protein (NC) were required for long distance propagation of the hybrid. The evaluation was performed on a unique substrate with a short gap serving as a pre-created invasion site (PCIS). In our current work, this substrate provided the opportunity to test what factors influence a specific invasion site to support transfer, and distinguish factors that influence invasion site creation from those that impact later steps. RNase H can act in a polymerization-dependent or -independent mode. Polymerization-dependent and -independent RNase H were found to be important to create efficiently-used invasion sites in the primer-donor complex, with or without NC. Propagation and terminus transfer steps, emanating from a PCIS in the presence of NC, were stimulated by polymerization-dependent but not -independent RNase H. RNase H can carry out primary and secondary cleavages during synthesis. While both modes of cleavage promoted invasion, only primary cleavage promoted propagation in the presence of NC in our system. These observations suggest that once invasion is initiated at a short gap, it can propagate through an adjacent region interrupted only by nicks, with help by NC. We considered the possibility that propagation solely by strand exchange was a significant contributor to transfers. However, it did not promote transfer, even if synthetic progress of the RT was intentionally slowed, which is consistent with strand exchange by random walk in which rate declines precipitously with distance.
Keywords: HIV-1, reverse transcriptase, RNase H, nucleocapsid protein, recombination
Introduction
HIV-1 employs strand transfer recombination between its two co-packaged genomes during reverse transcription as a means of combining advantageous traits. Coordination of the polymerase and ribonuclease H (RNase H) activities of reverse transcriptase (RT) culminates in transfer of the cDNA from a donor RNA template to an acceptor RNA template. RNase H activity initiates an invasion-mediated strand transfer mechanism by degrading the donor RNA under the extended DNA primer exposing gaps of ssDNA.1; 2; 3 The acceptor RNA anneals with the DNA at these invasion sites, and the primer-acceptor hybrid then propagates out from the invasion site eventually completing transfer of the primer terminus.2; 3; 4; 5; 6 After transfer, the RT resumes DNA synthesis on the acceptor template.
Hybrid propagation can proceed through either branch migration or proximity mechanisms.7 Branch migration is the successive replacement of primer-donor base pairs with primer-acceptor base pairs beginning at the invasion site and progressing continuously until completion of strand transfer. For propagation by proximity, acceptor annealing to the primer at an invasion site increases the local concentration of the acceptor around the DNA, which facilitates other downstream interactions between the acceptor and primer.
We recently provided additional evidence for the invasion-mediated transfer mechanism by using a substrate designed to model a strand transfer intermediate that is predicted to form just after creation of the invasion site.8 The pre-created invasion site (PCIS) substrate consisted of a DNA primer with a 20 nt 5′-overhang annealed to the 3′-terminus of a donor RNA. The acceptor RNA used in the reactions shared homology with the donor, and had an additional 20 nt at the 3′-terminus that were complementary with the PCIS. This substrate allowed us to examine the effectiveness of a single invasion site during invasion-mediated strand transfer. We reported that efficient strand transfer from a specific invasion site required the RNase H activity of RT and the HIV-1 RNA chaperone properties of the viral nucleocapsid protein (NC).8 Other groups have also demonstrated that NC promotes efficient hybrid propagation from sites of acceptor invasion.9; 10 Moreover, strand transfer with the PCIS substrates and WT RT was more efficient than the combined actions of RNase H-deficient E478Q RT and E. coli RNase H, which suggests that linking polymerization and cleavage in the same protein provided an additional stimulation to strand transfer.8 These results imply that RNase H has a dual function during strand transfer – (1) creating invasion sites, and (2) facilitating the downstream steps of hybrid propagation and ultimate transfer of the DNA terminus.
RT endonucleolytically hydrolyzes the RNA template during minus strand synthesis in a process termed “polymerase-dependent” (pol-dep) cleavage.11; 12 During DNA polymerization, the RNase H active site makes pol-dep cuts periodically in the RNA template, approximately an order of magnitude slower than the rate of polymerization.13; 14 Non-polymerizing RTs bind the nucleic acid substrate at a nicked site. The polymerase active site is positioned over the RNA 5′ terminus, and RT cleaves the remaining RNA fragments in a “polymerase-independent” (pol-ind) mode.11; 15; 16; 17 Biochemical studies show that RNA fragments annealed to longer DNA strands are cut by a non-polymerizing RT approximately 18 bp upstream of the RNA 5′-terminus, the distance separating the polymerase and RNase H active sites.11; 12; 18; 19; 20 Whether RT binding is directed by pol-dep or -ind positioning, it makes a “primary” cut 15–20 bp upstream of the polymerase active site, followed by a “secondary” cut 8–10 bp toward the DNA 3′-terminus.11; 12; 15; 16; 21; 22; 23; 24; 25
There is an excess of non-polymerizing RT in the HIV-1 virion.26 This suggests that pol-ind cleavage contributes to efficient strand transfer by degrading the donor RNA beyond what is made by the pol-dep cleavage specificity. Hwang et al. evaluated how pol-ind cleavage affects template switching using trans-complementation assays in vivo in which the RNase H-deficient mutant D524N RT was introduced in trans with the polymerase-deficient mutant D150E RT virus. 27 They observed an increase in template switching approximately 1.6-fold upon the addition of pol-ind cleavage. Their results imply that pol-ind cleavage only mildly stimulates strand transfer.
Results from various laboratories suggest that RT pausing during synthesis allows time for primary and secondary RNase H cleavage to initiate invasion strand transfer.3; 5; 13; 28; 29; 30; 31; 32; 33 Mutations at H539 in HIV-1 RT render the enzyme defective in secondary cleavage activity.22; 23; 34; 35 Biochemical analyses comparing WT RT and H539-mutants indicate that pol-dep secondary cuts leave only a small overlap of RNA and DNA base pairing, causing premature separation of the strands.23 It was proposed that while DNA 3′-directed secondary cleavage is injurious to HIV replication, RNA 5′-directed secondary cleavage enhances replication by clearing donor RNA fragments. Significantly, template inactivation by pol-dep secondary cleavage was prevented by addition of NC because it reduced RT pausing during DNA synthesis.
NC promotes strand transfer despite a seemingly contradictory role in reducing RT pausing.23; 36; 37; 38; 39; 40 In its mature form, NC is a 55 amino acid proteolytic product of the HIV-1 gag polyprotein.41; 42 NC binding promotes nucleic acid annealing and aggregation by destabilizing nearby weak secondary structures.43 The annealing and aggregating properties of NC are consistent with its ability to prevent template inactivation caused by pol-dep secondary cleavage, and still potently stimulate strand exchange.
We previously described the strand exchange properties of NC in relation to distance limitations for hybrid propagation originating from a specific invasion site.8 Efficient strand exchange required a hybrid propagation length, the distance separating the invasion site and primer terminus, of less than 17–24 bp. NC and RNase H activity extended the effective hybrid propagation distance to approximately 32 bp. However, creation of new invasion sites was required when distances separating the PCIS and primer terminus were longer than approximately 64 bp. Propagation efficiency was measured by a strand exchange assay in which completed exchange was reported by incorporation of a radioactive nucleotide onto the 3′-terminus of the exchanged strand. This proved to be a sensitive and accurate method of strand exchange detection as nucleotide addition was significantly faster than the PCIS-mediated hybrid propagation reaction. Results incidentally demonstrated that hybrid propagation by branch migration is slower than primer extension in our strand transfer system, although the relative rates could not be determined.
Branch migration of a complex potentially moves forward, moves backward or remains static at the nucleotide level. Such movements are determined by the probability of individual base pair formation or separation. For example, to move forward an A-T primer-acceptor base pair would be formed at the same location a primer-donor A-T base pair would separate. Equivalent replacement of base pairs during branch migration would suggest an energy neutral movement and a rapid process. Previous results imply that branch migration is likely a relatively slow step in the strand transfer mechanism.8 Random walk, a mathematical model, provides the computational means to model branch migration kinetics. A one-dimensional random walk determines the number of probability roles, or steps, predicted to traverse a particular distance. For application to branch migration, steps are related to a length of time and the appropriate probability applied for progression. Much like branch migration, movement by random walk is determined by the probability of making a step forward, backward, or remaining static. Similarities shared by branch migration and random walk make the predictive properties of random walk useful in examining the rate of strand exchange and explain why strand exchange of long primer-donor complexes is slow.
In the current study, we revisited the unique PCIS substrates to more closely examine the coordination of NC with various modes of RNase H cleavage by RT during a strand transfer reaction. We demonstrate that pol-dep, pol-ind, primary and secondary RNase H cleavage modes create invasion sites. Moreover, we show that pol-dep, primary cleavages and NC coordinate to promote hybrid propagation. We also employed conditions that reduce the RT polymerization rate in the expectation that at a specific reduced level, hybrid propagation by branch migration could catch the extending primer terminus. This allowed a quantitative assessment of the branch migration rate. We describe a surprising outcome suggesting that migration rate steeply declines with distance, as also suggested by random walk calculations. To describe the kinetics of branch migration a random walk model was designed to represent the base pairing events of branch migration. Random walk predicted a steep increase in time for the process to reach longer distances, suggesting a precipitous decline in branch migration over increasing distances.
Results
The Contributions of Polymerase-Dependent and -Independent RNase H Cleavages to the Efficiency of Strand Transfer
RT exhibits different modes of RNase H cleavage, each of which is likely to contribute its unique characteristics to the mechanism of strand transfer. The significance of RNase H cleavage specificity is highlighted by previous results showing that PCIS-mediated transfer efficiency was higher in reactions with WT RT compared to those with E478Q RT and E. coli RNase H.8 These results suggest that the coordinated action of polymerization and RNase H is significant to prepare the substrates for transfer.
To assess the contribution of pol-ind cleavage on transfer efficiency with respect to a specific invasion site, RT concentration was reduced from 37 nM to the concentration of the primer, 0.64 nM. This represents a reduction from a large excess of RT over substrate, to a level of RT similar to that of the substrate. RT was preincubated with the substrate before the start of the reaction so that at the lower RT concentration, available RTs would be encouraged to bind 3′ termini for synthesis, and presumably less would participate in pol-ind cleavage.
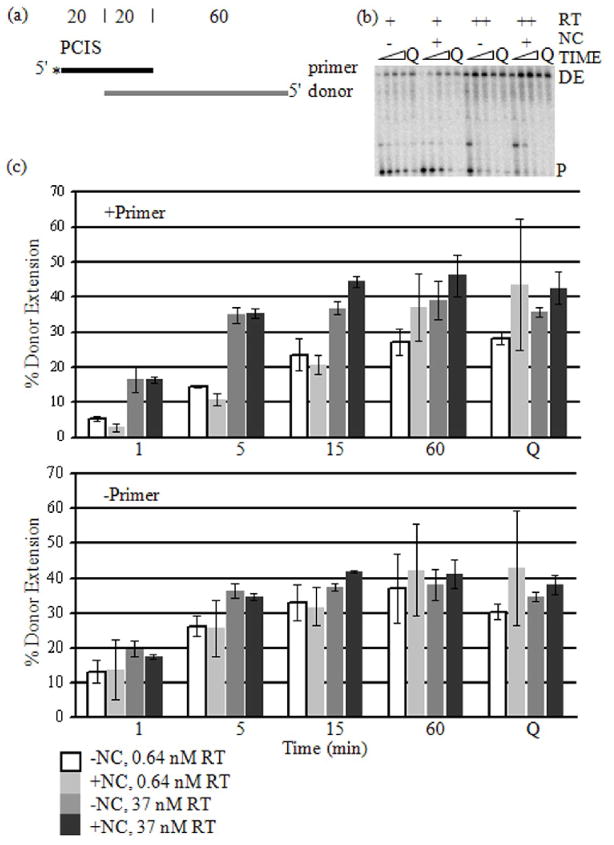
The proper design of this experiment requires that the RTs engaged in DNA synthesis at low concentration polymerize with similar efficiency as those operating at the higher concentration. This was examined by determining the percentage of radiolabeled DNA primers that were extended by RT along the full length of the RNA donor template (DE). The substrate was the RNA donor annealed to a DNA primer having or lacking the PCIS (Fig. 1a). RT concentrations tested were 37 nM and 0.64 nM WT RT (Fig. 1b and c). Significantly, we calculated this value in two ways, by including (+primer) or excluding (−primer) unextended primer in the denominator of the quotient (Fig. 1c) (see Materials and Methods). Excluding unextended primer from the calculation assessed the efficiency of polymerization on those primers that sustained synthesis. Results indicated a similar degree of primer extension at either 37 nM or 0.64 nM RT concentrations. This shows that the RT was sufficiently active and processive on those primers that were being extended that synthesis occurred with similar efficiency to that at the higher RT concentration. Moreover, supplementing the reaction with 37 nM E478Q RT, which should have augmented synthesis of any primers from which the WT RT dissociated during the reaction, had little effect on the −primer results.
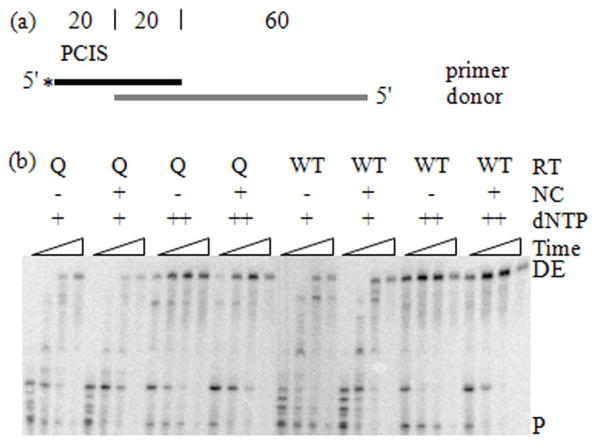
Figure 1. Primer Extension on Donor RNA at High and Low RT Concentrations.
(a) The substrates used in the reaction were a 5′-radiolabeled (asterisk) DNA primer (black) annealed to an RNA donor (gray) as shown in the schematic. The numbers above the templates indicate the length of segments of the template in nucleotides. The pre-created invasion site (PCIS) is indicated above the primer. (b) Extension reactions were performed with 0.64 nM (+) and 37 nM (++) RT concentrations, with or without NC and sampled at 1, 5, 15 and 60 min. E478Q RT (Q) was added with additional dNTPs and incubated for another 5 min. The reaction samples were run in denaturing PAGE and visualized. Unextended primers (P) and donor extension products (DE) are labeled. (c) Donor extension (DE) efficiencies (see “Materials and Methods”) versus time (min) were graphed. DE efficiency after addition of 34 nM E478Q RT (Q) is also included. The graph labeled “+Primer” represents DE efficiencies calculated with the unextended primers included. The graph labeled “−Primer” represents DE efficiencies excluding unextended primers.
Including unextended primer (+primer) in the denominator of the quotient allowed an assessment of primer utilization by the RTs present in the reaction. In this case there was a significantly lower fraction of fully extended primers in the reaction with 0.64 nM compared to 37 nM RT. This demonstrates a lower efficiency of initiation of synthesis in the reactions with lower RT. This encouraging result shows that the primer termini and any sites for pol-ind cleavage are in considerable excess over the RT, a situation in which we expect the rate of pol-ind cleavage to be reduced. At 0.64 nM RT, the primer and RT concentrations were about the same. However, the number of active polymerizing RT molecules was likely less than the number of primers, consistent with the incomplete utilization of primers.
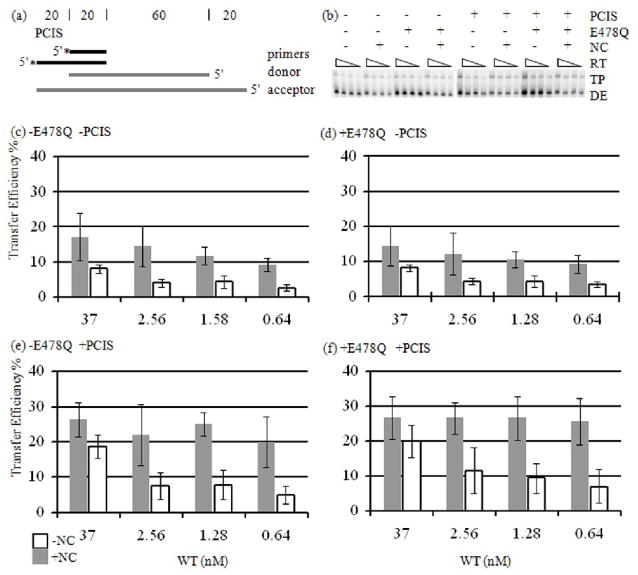
To parse pol-dep and pol-ind contributions to the creation of new invasion sites, strand transfer reactions were first performed without the PCIS (Fig. 2a). Strand transfer without the PCIS requires creation of new, alternate invasion sites by RNase H.8 We measured strand transfer efficiency (TE) at 37, 2.56, 1.28 and 0.64 nM WT RT (see Materials and Methods for the definition of TE) (Fig. 2b–f). As the RT concentration was lowered in reactions lacking NC, the transfer efficiency dropped from 8.0 % with 37 nM RT to 2.5 % with 0.64 nM RT (Fig. 2c). Upon the addition of NC, transfer efficiency dropped with lowered RT from 17.1 % to 9.2 %. The reactions were repeated with addition of E478Q RT (activity equivalent to 37 nM WT RT) for 5 min at the end of the initial incubation period. This assured that even at the low RT level, transfers that initiated on the acceptor RNA would be fully extended. Results were similar with the 5 min incubation with E478Q RT. Transfer efficiency decreased from 8.2 % to 3.3 % without NC, and from 14.4 % to 9.3 % with NC, indicating that transfer efficiency at low RT was not being influenced by a limitation in synthesis (Fig. 2d). These results indicate that pol-ind cleavage is important to achieve maximal transfer efficiency. They also imply that gap creation in the primer-donor transfer intermediate, the most likely consequence of pol-ind cleavage, is critical for producing the structure needed for proper invasion, and possibly for hybrid propagation.
Figure 2. Strand Transfer Reactions with RT Titration.
(a) The substrates used are the same as in Figure 1, except that reactions were run either with or without the PCIS. Also, acceptor RNA (gray) was added at the start of the reaction. (b) Reactions with −PCIS or +PCIS substrates were incubated with or without NC for 60 min, and samples were taken. E478Q RT and dNTP were added to the reaction, and incubated for 5 min. RT concentrations were 37, 2.56, 1.28 and 0.64 nM. Donor extension (DE) and strand transfer (TP) products are indicated. (c–f) Average transfer efficiencies determined from strand transfer reactions are represented in the graphs.
We next performed reactions with a substrate having a PCIS to determine how pol-dep and pol-ind cleavages influence strand transfer initiated at a specific invasion site (Fig. 2a). Transfer efficiency with the PCIS, but no NC, declined with decreasing RT from 18.7 % at 37 nM RT to 4.9 % at 0.64 nM before E478Q RT addition, and 20.1 % to 7.0 % after addition (Fig. 2e and f). The drop in transfer efficiency with the decline in RT concentration indicated that removal of pol-ind cleavage reduced strand transfer efficiency in the presence of the PCIS.
Interestingly, the impact of pol-ind cleavage was eliminated when the PCIS and NC were combined. Transfer efficiency in reactions with the PCIS and NC together remained relatively unaltered with decreasing RT concentrations, which varied from 26.4 % for 37 nM RT to 20 % for 0.64 nM RT before E478Q RT addition, and 26.6 % to 25.6 % after (Fig. 2e–f). When strand transfer initiated from the PCIS, pol-dep cleavage and NC appeared to be all that was necessary for high efficiency. Results indicate that both pol-dep and pol-ind contributed to invasion site formation. Of the two modes of cleavage, pol-dep cleavage was more influential in the presence of NC. The most significant conclusion possible to draw from these results with a PCIS substrate is that only pol-dep cleavage stimulated hybrid propagation in the presence of NC.
The Contributions of Primary and Secondary Cleavage to Strand Transfer Efficiency
RT pausing during minus strand synthesis allows time for RT to make a concentration of RNase H cuts in the RNA template. Recent work indicates that DNA 3′-directed secondary cuts, cleave the RNA donor close to the DNA 3′-terminus promoting separation of the DNA and RNA donor hybrid.23 Although allowing pol-dep primary cleavage before initiating DNA synthesis did not perturb subsequent synthesis, preliminary pol-dep secondary cleavage did inhibit DNA synthesis. This suggests that secondary cleavage during natural synthesis is an uncommon event. However, secondary cleavage during the pol-ind positioning of RT at the 5′ ends of RNA segments could create gaps relevant to invasion site creation or efficient hybrid propagation.
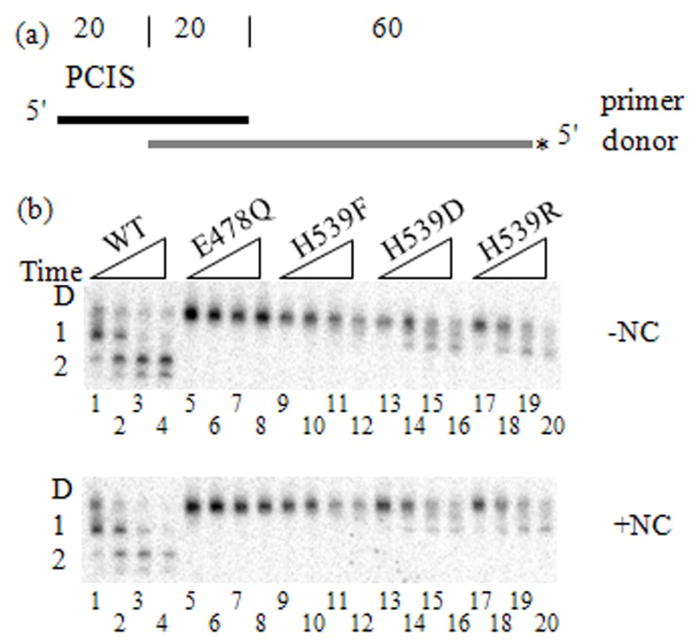
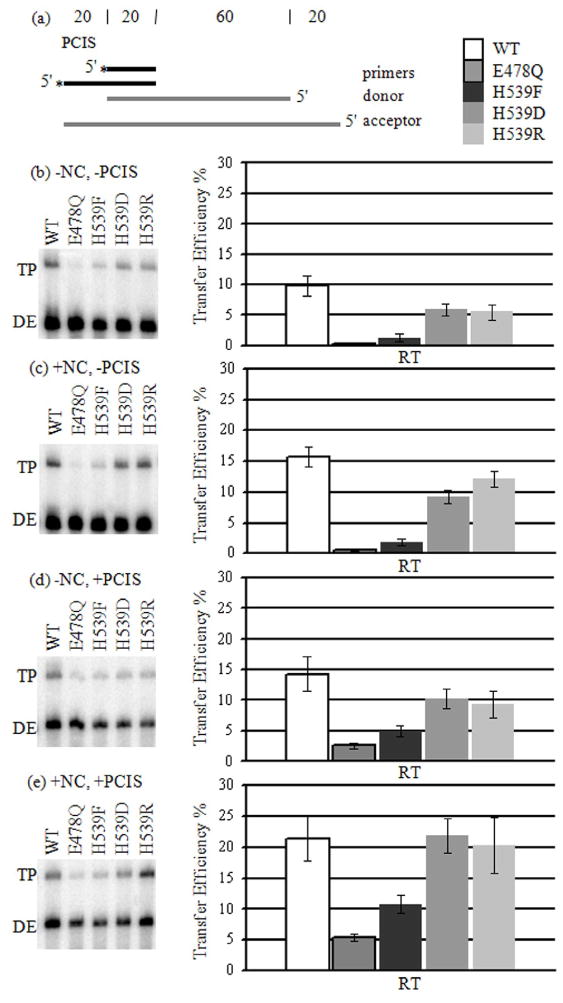
We evaluated the influence of primary and secondary cleavages on the creation of an invasion site. PCIS substrates were used in donor extension and strand transfer reactions comparing WT, E478Q and H539 RT mutants (Fig. 3a, 4a). H539F, H539D and H539R RTs are defective in secondary cleavage without and with NC (Fig. 3b).22; 23; 35 H539F RT also has diminished primary cleavage activity as shown by the slight fade of the donor RNA band into smaller cleavage products (Fig. 3b).22 A similar, somewhat more intense, spreading of bands can be seen in lanes 1, 2 and 14–20 of both −NC and +NC gel images (Fig. 3b). E478Q RT represented a lack of both primary and secondary cleavage. Strand transfer reactions with E478Q RT yielded the lowest transfer efficiency with 0.3 % (Fig. 4b). E478Q RT presumably produced very little strand transfer because there was no PCIS to initiate hybrid propagation, no RNase H activity to create alternate invasion sites, and no NC to promote strand exchange (Fig. 3b). This implies that a relative increase in strand transfer with WT or H539 mutant RTs is a result of invasion site formation by RNase H activity. As compared to E478Q RT, the H539F mutant generated slightly higher transfer efficiency of 1.3 %. H539D and H539R RTs, capable of only primary cuts, yielded 6.0 % and 5.5 %, respectively. WT RT had the highest transfer efficiency of 9.8 %. RNase H in these strand transfer assays lacking a PCIS could create invasion sites or enhance hybrid propagation. Presumably, the foremost effect of both primary and secondary cleavages was creating invasion sites since effective invasion was needed for all subsequent steps.
Figure 3. Donor Cleavage Profiles for RT mutants and WT.
(a) The substrates used are the same as in Figure 1, except that the RNA donor is 5′-radiolabeled, not the DNA primer. (b) Intact donor RNA (D) and primary (1) or secondary (2) cleavage products are generated over time (1, 5, 15 30 min) in the absence (−NC) and presence (+NC) of NC. RNA cleavage products were visualized in denaturing polyacrylamide gel electrophoresis. Gel lanes are numbered 1–20. Lanes 1 - 5 WT RT, 5 - 8 E478Q RT, 9 - 12 H539F RT, 13 - 16 H539D RT, 17 - 20 H539R RT.
Figure 4. Strand Transfer Reactions with WT and mutant RTs.
(a) See Figure 2(a). (b)–(e) Strand transfer reactions with WT and mutant RTs were performed and transfer efficiencies were calculated (see Materials and Methods). Transfer efficiency averages are shown. Outlined, white boxes represent results from reactions with WT RT, and outlined, gray boxes represent E478Q RT. Progressively lighter shades of gray represent H539F, H539D and H539R RTs.
NC-stimulation of transfer efficiency with WT and mutant RTs varied only slightly, ranging from 1.5 to 2.2-fold. E478Q RT transfer was stimulated by NC 1.7-fold to 0.5 % (Fig. 4c). H539F RT transfer was stimulated 1.5-fold to 1.9 %. With H539D and H539R RTs, transfer efficiency increased 1.5-fold to 9.2 %, and 2.2-fold to 12.1%, respectively. NC stimulated WT RT strand transfer efficiency 1.5-fold to 15.7 %. With H539F RT, transfer efficiency remained far less than with H539D and H539R RTs suggesting that NC does not augment strand transfer when primary cleavage activity is reduced. Moreover, transfer efficiencies with H539D and H539R RTs were still less than with WT, indicating that NC does not improve strand transfer through compensating for a lack of secondary cleavages. Because NC affected transfer with all RTs similarly, it is likely that the primary effect of NC is to promote interaction between the DNA primer and RNA acceptor at the PCIS, rather than facilitating RNase H-catalyzed creation of invasion sites.
Unlike NC, the PCIS impacted strand transfer differently with WT and the mutant RTs. Because there was no possibility for the creation of alternate invasion sites with E478Q RT, strand exchange originating at the PCIS was the only available strand transfer mechanism. The addition of the PCIS increased transfer efficiency in the E478Q RT reactions 8.7-fold to 2.6 % (Fig. 4d). Transfer efficiency with H539F RT increased 3.8-fold to 5.0 %, and 1.7-fold for both RT mutants H539D and H539R to 10.2 % and 9.3 %, respectively. WT RT was affected the least by the PCIS, increasing 1.5-fold to 14.3 %. Because transfer efficiency with H539F RT was less than with H539D and H539R RTs it is likely that increasing amounts of primary cuts improve the efficiency of strand transfer. Also, because strand transfer efficiencies with H539D and H539R RTs were less than with WT RT, secondary cleavage is seemingly important for effective strand transfer in reactions with the PCIS, but lacking NC.
The combination of NC and PCIS had the most striking influence on strand transfer stimulating E478Q RT 18-fold to 5.4 %, and increasing H539F RT transfer efficiency 8.3-fold to 10.8 % (Fig. 4e). H539D RT transfer efficiency was increased 3.6-fold to 21.8 %, and H539R RT 3.7-fold to 20.3 %. Transfer efficiency with WT RT was increased only 2.2-fold to 21.4 %. Significantly, H539D and H539R RTs yielded transfer efficiencies lower than WT RT in all experimental permutations except when NC and the PCIS were combined. However, even with NC and PCIS combined, the H539F RT mutant was still unable to attain transfer efficiency similar to WT.
Comparison of the effects of secondary cleavage on substrates with and without the PCIS allows differentiation of secondary cleavage contributions in invasion site creation versus hybrid propagation. For instance, in reactions lacking the PCIS and NC, secondary cuts stimulated strand transfer 1.6-fold from 6.0 % with H539D RT to 9.8 % with WT RT (Fig. 4b). Reactions with the PCIS showed a reduced effect on strand transfer by secondary cleavage increasing transfer efficiency 1.4-fold from 10.2 % with H539D RT to 14.3% with WT RT (Fig. 4d). Moreover, similar transfer efficiencies for WT and H539D RTs when both NC and the PCIS were present together suggest that contributions from secondary cuts were rendered negligible (Fig. 4e). This significant observation indicates that the propagation steps of transfer are not significantly promoted by secondary cuts.
These results with the PCIS substrates indicate that secondary cleavage has an impact on transfers without a PCIS, but that impact is diminished by the addition of the PCIS. Apparently, secondary cleavage and gap formation are not critical for efficient hybrid propagation, and the primary role for secondary cleavage in strand transfer is in creating gaps for efficient invasion. Further, primary cleavage in combination with NC and the PCIS appear to be all that is required for efficient hybrid propagation.
Limited Effective Distance of Hybrid Propagation Solely by Branch Migration
We previously reported that strand transfer from a PCIS mediated by E478Q RT occurs at only approximately 34% of the efficiency of transfer with WT RT.8 Moreover, NC-promoted strand exchange by branch migration, in the absence of RNase H, is undetectable after a distance of approximately 32 nt. This suggests that the rate of polymerization is sufficiently faster than that of branch migration that the expanding hybrid rarely catches up to the primer terminus. However, branch migration could be the dominant method of hybrid propagation for transfer involving an RT paused a short distance beyond the invasion site. Since some transfer was measured with E478Q RT we considered the possibility that branch migration is only slightly slower than polymerization, at least up to about 32 nucleotides. To test this hypothesis we slowed the progress of primer extension by limiting dNTP concentration, to promote the branch migration mechanism of transfer.
We used a PCIS substrate with a primer-donor hybrid length of 20 bp, which allowed for 12 nt of synthesis until the approximate 32 bp hybrid propagation distance limitation was reached (Fig. 5a, 6a). Five of the first seven template nucleotides downstream of the unextended primer terminus were A or G (71%), and seven of the first twelve were A or G (58%), requiring either dCTP or dTTP for extension. The dNTP concentration used in the standard reaction is 50 μM. To reduce primer extension rate, the dATP and dGTP concentrations were lowered to 5 μM, and dTTP and dCTP lowered to 50 nM (Fig. 5–6).
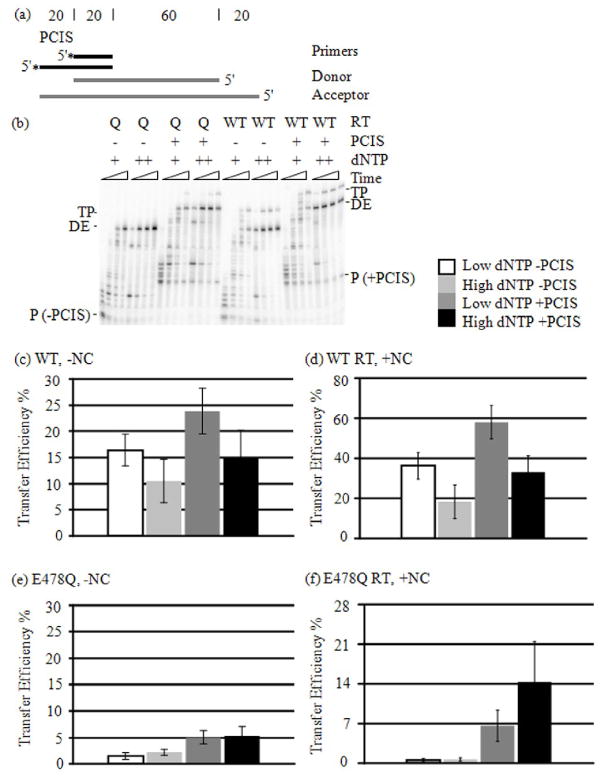
Figure 5. Primer Extension on Donor RNA with Low dNTP Concentrations.
(a) See Figure 1(a). (b) Extension reactions were performed with low (+) and high (++) dNTP concentrations (see “Materials and Methods”), with or without NC, with E478Q (Q) or WT RTs and sampled at 1, 5, 15 and 60 min. The reaction samples were run in denaturing PAGE and visualized.
Figure 6. Strand Transfer Reactions with Low dNTP Concentration.
(a) Please see Figure 2(a). (b) Strand transfer reactions were performed with low (+) and high (++) dNTP concentrations (see “Materials and Methods”), with or without the PCIS, with E478Q (Q) or WT RTs and sampled at 1, 5, 15 and 60 min. Products were visualized by denaturing PAGE. Unextended primers (P), Donor extension (DE) and strand transfer (TP) products are indicated. Strand transfer products shown are from reactions with NC, but other reactions were also done without NC (not shown). (c–f) Strand transfer efficiencies at 60 min only are represented. (c) WT RT without NC; (d) WT RT with NC; (e) E478Q RT without NC; (f) E478Q with NC.
We measured DNA primer extension on the donor RNA template in high and low dNTP concentrations, with and without NC (Fig. 5). For both E478Q and WT RTs, the formation of full-length donor extension products (DE) in low dNTP concentrations required considerably more time than in high dNTP concentrations. DE products appeared within the first minute of synthesis in high dNTPs, but only began to emerge between five and fifteen minutes in low dNTPs for reactions with and without NC (Fig. 5b). Complete extension of nearly all donor primers was achieved after 60 min incubation for all reaction component permutations (Fig. 5b). There is a general decrease in total counts per lane for all reactions (data not shown). The decrease in counts is particularly noticeable at the 60 min time points with WT RT (Fig. 5b). In all of the reactions the products evacuated the wells and migrated into the gel. Also, no fold-back products were formed. The general loss in substrate is likely due to moderate degradation, aggregation, precipitation or tube adherence of substrate in the reaction tubes sustained over the long incubation times.
For WT RT, lowering the dNTP concentration increased the transfer efficiency. In strand transfer reactions for 60 min without both the PCIS and NC the transfer efficiency with high dNTPs was 10.5 %, and 16.4 % for low dNTPs (Fig. 6c). For reactions with just the PCIS, but not NC, the transfer efficiency was 15.1 % with high dNTPs, and increased to 23.8 % with low dNTPs. The transfer efficiency for WT RT in the presence of NC, on a substrate lacking the PCIS, was 18.3 % with high dNTPs, and increased to 36.3 % with low dNTPs (Fig. 6d). WT RT with the PCIS and NC yielded 33.0 % transfer efficiency with high dNTPs, and 58.0 % with low.
Comparing transfer efficiency results with high and low dNTPs for the PCIS substrate with NC suggests that the effectiveness of the PCIS in promoting transfer was only slightly altered by dNTP reduction. In reactions with WT RT lacking the PCIS, NC increased transfer efficiency 2.2-fold from 16.4 % to 36.3 % at low dNTPs, and 1.7-fold from 10.5 % to 18.3 % at high dNTPs (Fig. 6c and d). With both WT RT and the PCIS, NC stimulated transfer efficiency 2.4-fold from 23.8 % to 58 % at low dNTPs, and 2.2-fold from 15.1 % to 33 % at high dNTPs. The PCIS without NC increased transfer efficiency 1.5-fold from 16.4 % to 23.8 % at low dNTPs, and 1.4-fold from 10.5 % to 15.1 % at high dNTPs (Fig. 6c). With NC, the PCIS increased transfer efficiency 1.6-fold from 36.3 % to 58.0 % with low dNTPs, and 1.8-fold from 18.3 % to 33.0 % with high dNTPs (Fig. 6d). Lowering dNTPs slightly augmented the stimulatory effect NC had on strand transfer, suggesting that more closely spaced pol-dep primary cuts facilitate NC in promoting hybrid propagation.
Since the reduction of dNTPs can promote transfers through RNase H mechanisms, the effects of dNTP reduction on the relative rates of branch migration and primer extension could only be determined with E478Q RT. Unexpectedly, lowering dNTPs with E478Q RT slightly reduced, rather than increased, transfer efficiency in all cases (Fig. 6e and f). Transfer efficiencies using the substrate with no PCIS and without NC yielded 2.2 % transfer efficiency with high dNTPs, and 1.5 % with low dNTPs (Fig. 6e). With the PCIS, but no NC, transfer efficiency was 5.3 % with high dNTPs, and 5.1 % with low dNTPs. In reactions with NC and without the PCIS, transfer efficiency at 60 min went from 0.7 % in high dNTPs to 0.5 % in low (Fig. 6f). The PCIS and NC combined produced 14.3 % transfer efficiency with high dNTPs, and 6.6 % with low. The lack of stimulation by dNTP reduction demonstrated an inability of the acceptor-primer hybrid to branch migrate quickly enough to catch the extending primer terminus, even when RT polymerization was slowed. It is likely that synthesis was so much faster than branch migration that the residual transfer previously observed with E478Q RT happened on a subset of substrate molecules in which the RT had stalled for some long period.
The Characteristics of Branch Migration in this System are Consistent with a Random Walk Mechanism
Our previous observation that low efficiency transfers are possible in the absence of RNase H, but that essentially no branch migration could be detected beyond a distance of 32 nt suggested that the rate of branch migration declines precipitously with distance.8 The current results showing that reduction of the rate of primer extension does not allow the spreading hybrid to catch the primer terminus are consistent with this conclusion.
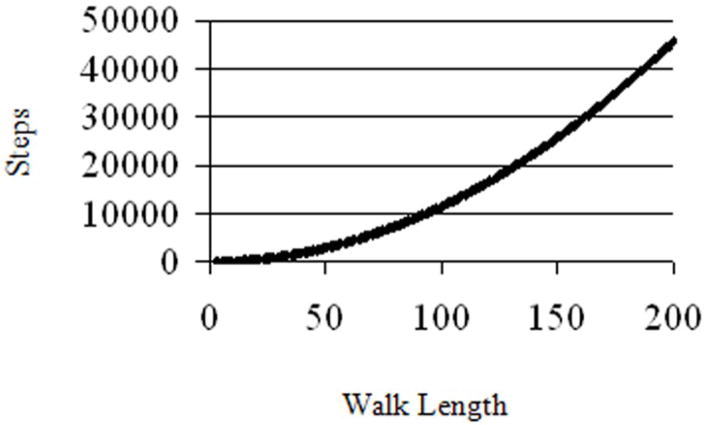
A mathematical model was used to analyze a defined probability of movement over a predetermined distance in a given time period, such as a primer-acceptor hybrid expanding through a primer-donor hybrid. The principles of a one-dimensional Random Walk were applied to study a walk progressing from one to 200 units (Fig. 7). The number of steps were calculated to reach a given length with specified probability. Each step represented the equal possibilities of movement forward, backward or no progression. Steps could be potentially expressed as increments of time. A unit of walking distance could be defined as any measurement of progress, such as a primer-acceptor hybrid progressing towards the primer 3′-terminus. Results indicate that there is a parabolic increase in the average number of steps required to progress from one to 200 units. A total of 110 steps were needed for half of simulations to walk a length of 10 units, 1053 steps were needed for half to walk a length of 30, and 11503 steps were needed for a length of 100 (Fig. 7). Consistent with this model are results in this study demonstrating that slowing polymerization does not allow more efficient branch migration. This model suggests a rapidly declining rate of branch migration that is dependent on the distance the hybrid has progressed. This is because the process is stochastic; complete displacement of the primer occurs by chance. This chance decreases with length because it requires more steps forward than backward. This is in keeping with results previously reported showing that strand exchange efficiency decreased with increasing hybrid propagation distance.8; 44
Figure 7. Steps Needed for Half of 105 Simulations to First Reach Specific Distances.
A random walk calculation is represented graphically in number of steps taken for half of all simulations to complete versus the length of the walk for completion.
Discussion
Previous work indicates that retroviral strand transfer proceeds by a three-step process: (1) acceptor RNA invasion of the donor RNA-DNA complex, (2) propagation of the acceptor RNA-DNA hybrid, and (3) transfer of the DNA 3′-terminus to the acceptor RNA.2 The viral proteins RT and NC enable these steps.
We considered that different modes of RNase H cleavage by RT have distinct contributions to strand transfer because of temporal and spatial differences in their specificities. For instance, results suggest that although pol-ind cleavage only moderately stimulates template switching, pol-dep cleavage is critical for efficient recombination.27 In addition, we recently demonstrated that spatially separating RNase H activity from polymerization reduced strand transfer efficiency.8 These observations suggest that pol-dep and pol-ind cleavages impact strand transfer very differently. Further, when RT pauses, it concentrates primary and secondary cuts in the RNA template. These cleavages generate gaps for acceptor invasion. Purohit et al. showed that secondary cleavage can occur in a pol-dep manner, but it can break up the primer-template causing disruption of DNA synthesis.23 However, NC suppresses template inactivation caused by pol-dep secondary cleavage. These observations imply that pol-dep, pol-ind, primary and secondary cleavages have differing contributions to strand transfer, and that NC may alter their effectiveness in promoting the transfer event.
In the current study we have employed the unique properties of the PCIS substrate to evaluate in vitro how RT and NC, and features of template structure, affect invasion strand transfer. Specifically, we assessed the effects of pol-dep, pol-ind, primary and secondary RNase H cleavages on invasion site creation and hybrid propagation. Moreover, we determined the influence of NC on the different modes of RNase H cleavage and their impacts on strand transfer.
We demonstrated that all RNase H specificities tested contribute to invasion site formation. It is probable that all cleavage specificities contribute to creation of longer gaps, which have been shown to increase transfer efficiency.9 In addition, rapid invasion site formation by contributions from all RNase H specificities would likely enhance strand transfer since transfer efficiency decreases as the RT extends the DNA 3′-terminus further downstream.8
In contrast, we found that pol-ind and secondary cleavages are not needed for NC-assisted hybrid propagation. The distance separating the RNase H and polymerase active sites is approximately 18 bp; this distance has probably evolved so that the primer-template will not fall apart as a consequence of pol-dep, primary cuts.11; 12; 18; 19; 20 The length of template occupied by an RT molecule is approximately 23 bp.20 Based on these distances, a non-polymerizing RT positioned immediately next to a polymerizing RT engaged at the DNA 3′-terminus can cleave the RNA template a minimum of 41 bp upstream of the terminus. This distance is already beyond the maximum for branch migration (32 bp) in the presence of RT and NC, implying that pol-ind cleavage is not capable of contributing significantly to branch migration. This is consistent with results presented here indicating that pol-ind cleavage does not stimulate hybrid propagation.
Interestingly, pol-dep, primary cleavages were sufficient to promote propagation in the presence of NC. The rate of pol-dep cleavage compared to RT polymerization suggests that RT makes only nicks in the RNA template during DNA synthesis.13; 14 Evidently, the properties of NC allow it to promote an exchange of the nicked donor for the intact acceptor. We envision a progressive mechanism in which acceptor RNA interaction with the DNA at a gap in the donor RNA promotes interaction at downstream nicks or small gaps generated by pol-dep activity that would otherwise be inaccessible. This has previously been described as the proximity mechanism of strand transfer.7
An alternative mechanism of invasion-mediated strand transfer is branch migration. Expansion of the acceptor-cDNA hybrid by branch migration on the PCIS substrates proceeds unaided through 17 bp, and, as mentioned above, no more than approximately 32 bp with the assistance of NC and RNase H cleavages. Pol-dep, primary RNase H makes only periodic cuts in the RNA substrate rather than completely degrading it. This suggests that a typical strand transfer intermediate is likely comprised of RNA fragments hybridized to the DNA. Hybrid propagation by branch migration could take advantage of such a fragmented template by displacement of donor RNA segments 15–20 nucleotides long. In this way, branch migration could function as an alternative to the proximity mechanism in hybrid propagation through an RNase H-nicked region.
We considered the possible contribution of branch migration through regions that have sustained little or no pol-dep, primary cleavages. If branch migration were only slightly slower than polymerization, it could be an effective means of propagation through uncut template RNA in cases where the RT naturally paused a distance after the invasion site. Therefore, a local environment favoring reduced polymerization rate may lead to increased transfer efficiency by branch migration.
To evaluate this possibility we slowed RT polymerization in a strand transfer reaction by decreasing the dNTP concentration. Transfer increased with reduced WT RT polymerization. This could have been caused by more effective branch migration, but also by promotion of invasion by the increased frequency of RNase H cuts when the RT was slowed. To distinguish these possibilities we employed E478Q RT, lacking RNase H, in lieu of the WT RT, and saw no increase in transfer. This suggests three characteristics of the branch migration in our assays - (1) the rate of branch migration is far less than polymerization; (2) branch migration over long distances is too inefficient to complete strand transfer; (3) efficient branch migration likely requires RT to stall a very short distance from the invasion site. These conclusions are consistent with results implying that RNase H activity is required for efficient hybrid propagation.
We originally viewed branch migration as an energy-neutral process, in which each base pair of the donor-primer was successively replaced by the equivalent base pair of the acceptor-primer. Any structure that stabilized folding of the single-stranded acceptor would have to be disrupted. Yet, a new, identical structure of the same negative free energy would form in the single-stranded donor once it was free of the DNA primer. By this view, steps in migration should have occurred freely, and the migration process could have been very dynamic, with rapidly shifting strands. However, our results are consistent with only a slow exchange, and one in which the migration in one direction slows considerably with distance. Calculations based on the principle of random walk indicate a reduction in the probability of forward movement with increasing distance for a given length of time. One can infer from the random walk predictions and branch migration data shown herein that increasing branch migration distance inhibits the progression of an acceptor-primer hybrid.
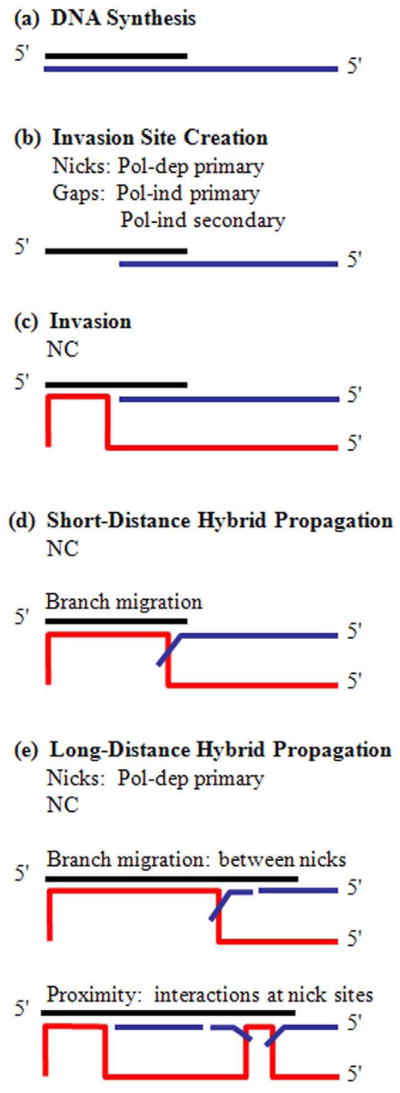
We conclude that all RNase H specificities have evolved to coordinate in generation of accessible invasion sites (Fig. 8). NC then facilitates acceptor invasion, followed by primer-acceptor hybrid propagation. Pol-dep, primary cleavages are the dominant RNase H activities in promoting hybrid propagation, while pol-ind and secondary cleavages contribute to transfer almost exclusively during invasion site creation. In addition NC and RNase H work together to overcome the inefficiencies of branch migration. There is a precipitous decrease in the efficiency of hybrid propagation with each additional base incorporated by RT. Therefore, simple branch migration can contribute to transfer only if the polymerizing RT stalls a very short distance from the invasion site.
Figure 8. The Collaboration of Factors That Influence Strand Transfer.
(a) The strand transfer substrate is composed of a nascent DNA (black) synthesized by RT on the RNA donor (blue). (b) Strand transfer is initiated by multiple RNase H modes creating an invasion site. (c) NC promotes acceptor RNA (red) invasion at gaps. (d) Hybrid propagation over short distances is promoted by NC and proceeds by branch migration. (e) Hybrid propagation over long distances is promoted by NC and pol-dep, primary cleavage. Propagation by branch migration likely progresses through short RNA donor fragments. Propagation by proximity progresses by interaction of acceptor RNA and DNA at nick sites.
Materials and Methods
Materials
Donor and acceptor RNA strands were synthesized and annotated throughout the text as previously described.8 Integrated DNA Technologies synthesized the DNA oligonucleotides. Polynucleotide kinase was obtained from Roche Applied Science. The radionucleoside triphosphate, [γ-32P] ATP (6000 Ci/mmol), was acquired from PerkinElmer Life Sciences. HIV-1 nucleocapsid protein (NCp9) was expressed and purified as previously described.40; 45 The HIV-1 reverse transcriptase (RT) WT and H539 mutants were purified as previously described.32; 46 Dr. Stuart F. J. Le Grice generously provided HIV-1 E478Q RT. Units of polymerase specific activity of WT and mutant RTs were determined as previously described (1 Unit = incorporation of 1 nmol radioactive nt/10 min). 8; 32;40; 45; 46 All other materials were the best commercially available.
DNA Primer Extension on Donor RNA
Radiolabeled DNA primer (0.64 nM), −PCIS or +PCIS, and the donor RNA template (4 nM) were heat annealed at 95° C for 5 min, and then slow-cooled to 37° C in RT reaction buffer (50 mM Tris, pH 8.0, 50 mM KCl, 1 mM DTT, and 1 mM EDTA, pH 8.0). When indicated, the primer-donor complex was incubated with NC for 3 minutes, coating the nucleic acid 100 % (1 NC binds 7 nt). RT (37 nM; 2 U) was incubated in the reaction mixture for an additional 2 min at 37° C. To start the reaction, MgCl2 (6 mM) and dNTP (50 μM) were simultaneously introduced to the reaction mixture. The reactions were incubated at 37° C for various times up to 60 min, and were subsequently terminated with a buffer comprised of 90 % formamide, 10 mM EDTA, pH 8.0, 0.1 % xylene cyanole, and 0.1 % bromophenol blue.
Strand Transfer Assays with HIV-1 RT
Reactions were prepared and performed exactly as primer extension assays, except that acceptor RNA (8 nM) was added with MgCl2 and dNTPs at the start of the reaction.
Variations on Primer Extension and Strand Transfer Reactions
Primer extension reactions and strand transfer assays were altered for various assays. For mutant RT assays, E478Q, H539F, H539R or H539D RTs were used in lieu of WT RT. For low RT concentration assays, RT was titrated from 37 nM to 2.56, 1.28 and 0.64 nM. Also, for low RT assays, as indicated, after the 60 min time-point E478Q RT (34 nM; 2 U) and dNTPs (40 μM) were added, and the reactions were incubated at 37° C for an additional 5 min. For low dNTP concentration assays, the dNTP concentration was changed from 50 μM each to a combination of dATP and dGTP at 5 μM, and dTTP and dCTP at 50 nM.
Detection and Analysis
All reaction products were denatured by heating at 95° C for 5 min, and separated by electrophoresis in a 6 % polyacrylamide gel (7 M urea). The gel image was taken with a Storm PhosphorImager, and reaction products were quantitated with ImageQuant software, version 1.2. Polymerization efficiency (PE) was calculated by PE = DE/Total Lane, in which “DE” is the pixel intensity count of the DNA band that extended the full length of the donor RNA template, and “Total Lane” is the count for all products in the gel lane. PE was determined with (+primer) and without (−primer) unextended primer included in the “Total Lane” value. Transfer efficiencies (TE) were determined with the equation TE = (TP/(DE + TP)), in which “TP” represents pixel counts for transfer products. Fold-stimulation of transfer efficiency by NC (SNC) was calculated with the equation SNC = TE+NC/TE−NC, in which the transfer efficiency with NC (TE+NC) was divided by the transfer efficiency without NC (TE−NC). Similarly, the stimulation of transfer efficiency by the PCIS was calculated by SPCIS = TE+PCIS/TE−PCIS. All error bars represent standard deviation for a minimum of three experiments.
Random Walk Determination
To investigate the behavior of a random walk with a process with equal probabilities for either moving forward, backward, or remaining in the same location, a C++ program was written to perform simulations that roll an independent sequence of random numbers for each of a set number of simulations. Random numbers were generated using the ran2 algorithm.47 The probability for moving forward, moving backward, and staying in the same location was set to 1/3. Rolls for each simulation are repeated, and the position of the simulations are updated depending on the roll. The origin (position 0) is a reflective boundary; there are no negative positions. The position where the simulations are considered to be complete is iterated from 1 to the final maximum moves for completion. This value is an absorbing boundary and represents the point at which the RNA template dissociates from the donor. The number of rolls it takes for half of the total simulations to complete to a given length is plotted on the y-axis in Fig. 7. For these calculations, the average number of steps for the completion of 100,000 independent simulations was determined. The X-axis is the total walk length to completion.
Acknowledgments
This work was supported by grants from the National Institutes of Health: GM 049573 (R.A.B.), GM 076485 (D.H.M.), and Training in HIV Replication and Pathogenesis 5T32 AI 049815 (S.T.R.). This work was also funded in part by NIH contract N01-CO-12400 (R.J.G.). Contents do not necessarily reflect the views of NIH, nor does mention of products imply endorsement by the US government. We thank Dr. Vandana Basu-Purohit for the H539 RT mutants, and Dr. Stuart F. J. Le Grice for providing E478Q RT. We express our appreciation to Dr. Mark N. Hanson for development of the PCIS substrate. Thank you to Dr. Lisa M. Demeter, Dr. Baek Kim and the members of the Bambara laboratory for helpful discussions. We thank Cody Spencer for contributions to the early stages of this project. We also thank Dr. Jeffrey DeStefano, Dr. Lata Balakrishnan and Dr. Dorota Piekna-Przybylska for reviewing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanson MN, Balakrishnan M, Roques BP, Bambara RA. Evidence that creation of invasion sites determines the rate of strand transfer mediated by HIV-1 reverse transcriptase. J Mol Biol. 2006;363:878–90. doi: 10.1016/j.jmb.2006.08.068. [DOI] [PubMed] [Google Scholar]
- 2.Negroni M, Buc H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc Natl Acad Sci U S A. 2000;97:6385–90. doi: 10.1073/pnas.120520497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roda RH, Balakrishnan M, Kim JK, Roques BP, Fay PJ, Bambara RA. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J Biol Chem. 2002;277:46900–11. doi: 10.1074/jbc.M208638200. [DOI] [PubMed] [Google Scholar]
- 4.DeStefano JJ, Bambara RA, Fay PJ. The mechanism of human immunodeficiency virus reverse transcriptase-catalyzed strand transfer from internal regions of heteropolymeric RNA templates. J Biol Chem. 1994;269:161–8. [PubMed] [Google Scholar]
- 5.DeStefano JJ, Mallaber LM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J Virol. 1992;66:6370–8. doi: 10.1128/jvi.66.11.6370-6378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeStefano JJ, Roberts B, Shriner D. The mechanism of retroviral recombination: the role of sequences proximal to the point of strand transfer. Arch Virol. 1997;142:1797–812. doi: 10.1007/s007050050198. [DOI] [PubMed] [Google Scholar]
- 7.Song M, Basu VP, Hanson MN, Roques BP, Bambara RA. Proximity and branch migration mechanisms in HIV-1 minus strand strong stop DNA transfer. J Biol Chem. 2008;283:3141–50. doi: 10.1074/jbc.M707343200. [DOI] [PubMed] [Google Scholar]
- 8.Rigby ST, Rose AE, Hanson MN, Bambara RA. Mechanism analysis indicates that recombination events in HIV-1 initiate and complete over short distances, explaining why recombination frequencies are similar in different sections of the genome. J Mol Biol. 2009;388:30–47. doi: 10.1016/j.jmb.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath MJ, Destefano JJ. A complementary single-stranded docking site is required for enhancement of strand exchange by human immunodeficiency virus nucleocapsid protein on substrates that model viral recombination. Biochemistry. 2005;44:3915–25. doi: 10.1021/bi0477945. [DOI] [PubMed] [Google Scholar]
- 10.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–70. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furfine ES, Reardon JE. Reverse transcriptase. RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991;266:406–12. [PubMed] [Google Scholar]
- 12.Gopalakrishnan V, Peliska JA, Benkovic SJ. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1992;89:10763–7. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeStefano JJ, Buiser RG, Mallaber LM, Myers TW, Bambara RA, Fay PJ. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J Biol Chem. 1991;266:7423–31. [PubMed] [Google Scholar]
- 14.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–97. [PubMed] [Google Scholar]
- 15.DeStefano JJ, Mallaber LM, Fay PJ, Bambara RA. Determinants of the RNase H cleavage specificity of human immunodeficiency virus reverse transcriptase. Nucleic Acids Res. 1993;21:4330–8. doi: 10.1093/nar/21.18.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palaniappan C, Fuentes GM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA. Helix structure and ends of RNA/DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J Biol Chem. 1996;271:2063–70. [PubMed] [Google Scholar]
- 17.Schultz SJ, Zhang M, Champoux JJ. Recognition of internal cleavage sites by retroviral RNases H. J Mol Biol. 2004;344:635–52. doi: 10.1016/j.jmb.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 18.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–90. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 19.Pullen KA, Ishimoto LK, Champoux JJ. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992;66:367–73. doi: 10.1128/jvi.66.1.367-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA: DNA. EMBO J. 2001;20:1449–61. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeStefano JJ, Buiser RG, Mallaber LM, Bambara RA, Fay PJ. Human immunodeficiency virus reverse transcriptase displays a partially processive 3′ to 5′ endonuclease activity. J Biol Chem. 1991;266:24295–301. [PubMed] [Google Scholar]
- 22.Purohit V, Balakrishnan M, Kim B, Bambara RA. Evidence that HIV-1 reverse transcriptase employs the DNA 3′ end-directed primary/secondary RNase H cleavage mechanism during synthesis and strand transfer. J Biol Chem. 2005;280:40534–43. doi: 10.1074/jbc.M507839200. [DOI] [PubMed] [Google Scholar]
- 23.Purohit V, Roques BP, Kim B, Bambara RA. Mechanisms that prevent template inactivation by HIV-1 reverse transcriptase RNase H cleavages. J Biol Chem. 2007;282:12598–609. doi: 10.1074/jbc.M700043200. [DOI] [PubMed] [Google Scholar]
- 24.Wisniewski M, Balakrishnan M, Palaniappan C, Fay PJ, Bambara RA. Unique progressive cleavage mechanism of HIV reverse transcriptase RNase H. Proc Natl Acad Sci U S A. 2000;97:11978–83. doi: 10.1073/pnas.210392297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wisniewski M, Balakrishnan M, Palaniappan C, Fay PJ, Bambara RA. The sequential mechanism of HIV reverse transcriptase RNase H. J Biol Chem. 2000;275:37664–71. doi: 10.1074/jbc.M007381200. [DOI] [PubMed] [Google Scholar]
- 26.Garcia Lerma JG, Yamamoto S, Gomez-Cano M, Soriano V, Green TA, Busch MP, Folks TM, Heneine W. Measurement of human immunodeficiency virus type 1 plasma virus load based on reverse transcriptase (RT) activity: evidence of variabilities in levels of virion-associated RT. J Infect Dis. 1998;177:1221–9. doi: 10.1086/515272. [DOI] [PubMed] [Google Scholar]
- 27.Hwang CK, Svarovskaia ES, Pathak VK. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc Natl Acad Sci U S A. 2001;98:12209–14. doi: 10.1073/pnas.221289898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeStefano JJ. Kinetic analysis of the catalysis of strand transfer from internal regions of heteropolymeric RNA templates by human immunodeficiency virus reverse transcriptase. J Mol Biol. 1994;243:558–67. doi: 10.1016/0022-2836(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 29.Hanson MN, Balakrishnan M, Roques BP, Bambara RA. Effects of donor and acceptor RNA structures on the mechanism of strand transfer by HIV-1 reverse transcriptase. J Mol Biol. 2005;353:772–87. doi: 10.1016/j.jmb.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 30.Lanciault C, Champoux JJ. Pausing during reverse transcription increases the rate of retroviral recombination. J Virol. 2006;80:2483–94. doi: 10.1128/JVI.80.5.2483-2494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negroni M, Buc H. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J Mol Biol. 1999;286:15–31. doi: 10.1006/jmbi.1998.2460. [DOI] [PubMed] [Google Scholar]
- 32.Roda RH, Balakrishnan M, Hanson MN, Wohrl BM, Le Grice SF, Roques BP, Gorelick RJ, Bambara RA. Role of the Reverse Transcriptase, Nucleocapsid Protein, and Template Structure in the Two-step Transfer Mechanism in Retroviral Recombination. J Biol Chem. 2003;278:31536–46. doi: 10.1074/jbc.M304608200. [DOI] [PubMed] [Google Scholar]
- 33.Wu W, Blumberg BM, Fay PJ, Bambara RA. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J Biol Chem. 1995;270:325–32. doi: 10.1074/jbc.270.1.325. [DOI] [PubMed] [Google Scholar]
- 34.Schatz O, Cromme FV, Gruninger-Leitch F, Le Grice SF. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;257:311–4. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- 35.Wohrl BM, Volkmann S, Moelling K. Mutations of a conserved residue within HIV-1 ribonuclease H affect its exo- and endonuclease activities. J Mol Biol. 1991;220:801–18. doi: 10.1016/0022-2836(91)90119-q. [DOI] [PubMed] [Google Scholar]
- 36.Drummond JE, Mounts P, Gorelick RJ, Casas-Finet JR, Bosche WJ, Henderson LE, Waters DJ, Arthur LO. Wild-type and mutant HIV type 1 nucleocapsid proteins increase the proportion of long cDNA transcripts by viral reverse transcriptase. AIDS Res Hum Retroviruses. 1997;13:533–43. doi: 10.1089/aid.1997.13.533. [DOI] [PubMed] [Google Scholar]
- 37.Ji JP, Loeb LA. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992;31:954–8. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- 38.Klasens BI, Huthoff HT, Das AT, Jeeninga RE, Berkhout B. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim Biophys Acta. 1999;1444:355–70. doi: 10.1016/s0167-4781(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes GM, Bambara RA, Fay PJ. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–11. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 40.Wu W, Henderson LE, Copeland TD, Gorelick RJ, Bosche WJ, Rein A, Levin JG. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–42. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dib-Hajj F, Khan R, Giedroc DP. Retroviral nucleocapsid proteins possess potent nucleic acid strand renaturation activity. Protein Sci. 1993;2:231–43. doi: 10.1002/pro.5560020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.You JC, McHenry CS. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–27. [PubMed] [Google Scholar]
- 43.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–86. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 44.Landgraf R, Ramamurthi KS, Sigman DS. Kinetics of spontaneous displacement of RNA from heteroduplexes by DNA. Nucleic Acids Res. 1996;24:3246–52. doi: 10.1093/nar/24.16.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morcock DR, Thomas JA, Sowder RC, 2nd, Henderson LE, Crise BJ, Gorelick RJ. HIV-1 inactivation by 4-vinylpyridine is enhanced by dissociating Zn(2+) from nucleocapsid protein. Virology. 2008;375:148–58. doi: 10.1016/j.virol.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–79. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 47.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C: The Art of Scientific Computing. 2. Cambridge University Press; Cambridge, UK: 1992. [Google Scholar]