Abstract
Some simplified adociaquinone B analogs and a series of 1,4-naphthoquinone derivatives were synthesized and tested against the three enzymes Cdc25B, MKP-1, and MKP-3. Cdc25B and MKP1 in particular are enzymes overexpressed in human cancer cells, and they represent potential molecular targets for novel cancer chemotherapeutic treatments. A number of analogs exhibited significant inhibitory activity against these enzymes, and the bioassay data in addition to structure–activity relationships of these compounds will be discussed.
1. Introduction
The cell cycle is a highly regulated process that controls eukaryotic growth and proliferation. Transition through the four phases of the cell cycle (G1, S, G2, M) is regulated by cyclin dependant kinase (CDK)-cyclin complexes, which are activated by a subclass of dual-specificity protein tyrosine phosphatases, namely Cdc25A, B, and C.1 Studies have linked the oncogenesis of numerous human tumors with the overexpression of Cdc25A and B, thus suggesting that the inhibition of these dual-specificity phosphatases may be a viable and attractive method of cancer treatment.1-4
Cdc25B was initially shown to primarily activate CDK1-cyclin A and CDK1-cyclin B at the G2-M transition of the cell cycle via dephosphorylation of Thr14 and Tyr15 residues,4-8 although more recent studies have revealed considerable functional overlap among the three Cdc25s in the G1-S and G2-M transitions.9-11 For example, Cdc25B can activate Cdk2-cyclin A. 12 These findings and others demonstrate the Cdc25 enzymes and their corresponding CDK-cyclin complexes have multiple cellular roles.13 Expression of Cdc25B is uniquely increased after DNA-damage induced by carcinogens, which may reflect a casual role the genetic instability associated with cancer.3,14
At a chemical level, promotion of the transition between G2-M by CDK1-cyclin A and B is catalyzed via dephosphorylation by a specific cysteine thiolate anion found in a shallow pocket of Cdc25B.15-17 Binding to or oxidation of this thiolate anion prevents activation of the CDK1-cyclin complex, hence triggering cell cycle arrest.1,8,19 Further cellular effects of these enzymes can be found in a recent study by Cazales et al. They reported that inhibiting Cdc25 phosphatase activity alters microtubule dynamics and impairs mitotic spindle assembly, resulting in disruption of the mitotic process.20 Furthermore, they observed with human HT29 colon cancer cells an enhancement of the antiproliferative activity of the microtubule-targeting paclitaxel when it was combined with a Cdc25B inhibitor. This emphasizes the complex and critical role of the Cdc25 family in cell cycle regulation and supports further studies on the mechanism of action of small molecule inhibitors of these protein tyrosine phosphatases.
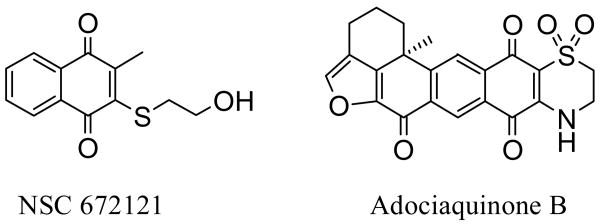
A majority of the known small molecule Cdc25B inhibitors are quinones or quinone-type compounds. Naphthoquinone derivative NSC 672121 (Figure 1, 2 μM inhibition of Cdc25B) has received considerable attention after emerging from an activity-based screening of a National Cancer Institute (NCI) Chemical Repository of 10,070 compounds.21 Since then several studies have attempted to improve this scaffold through analog synthesis.22-27 A very recent article presents the synthesis and biological evaluation of several new quinolinedione and naphthoquinone derivatives, containing carboxylic or malonic acids groups introduced to mimic the role of the phosphate moieties of Cyclin-Dependent Kinase complexes. The most efficient compounds showed inhibitory activity against Cdc25B with IC50 values in the 10 μM range, and were cytotoxic against HeLa cells.28 Furthermore, we have previously reported a number of isolates from the Indonesian sponge Xestospongia sp., and among them identified what is believed to be the most potent known inhibitor of Cdc25B, adociaquinone B (Figure 1, 80 nM).29 Herein we report the design and synthesis of simplified adociaquinone B analogs in addition to several naphthoquinone derivatives, and their subsequent ability to inhibit Cdc25B dual-specificity phosphatase. Our basic approach of evaluating why adociaquinone B might exert such potent Cdc25B activity was to systematically modify both the west and the east hemispheres of the molecule. To assess the necessity of the fused tricyclic benzofuranone moiety, chemical synthesis led to analog 9 containing a simplified west hemisphere, while analogs 10-20 were either synthesized or purchased to evaluate the importance of a basic naphthoquinone moiety in addition to an adjacent heterocyclic ring system.
Figure 1.
Structures of potent Cdc25B inhibitors.
In addition to Cdc25, several other protein tyrosine phosphatases have been identified as potential cancer chemotherapeutic targets. As examples, the mitogen-activated protein kinase phosphatases-130, 31 and -332 (MKP-1 and MKP-3), which are distal effectors for many extracellular growth factors, stress detectors, and drug sensors, have been suggested as possible targets. MKP-1 expression is elevated in prostate, breast, gastric, and renal cancer33,34 and is correlated with decreased progression-free survival.3,5 Moreover, reduction in MKP-1 expression by antisense restricts tumorigenicity.34 The activities of the target compounds against these phosphatases were determined as a measure of their selectivity for Cdc25B.
2. Results and Discussion
2.1. Synthesis
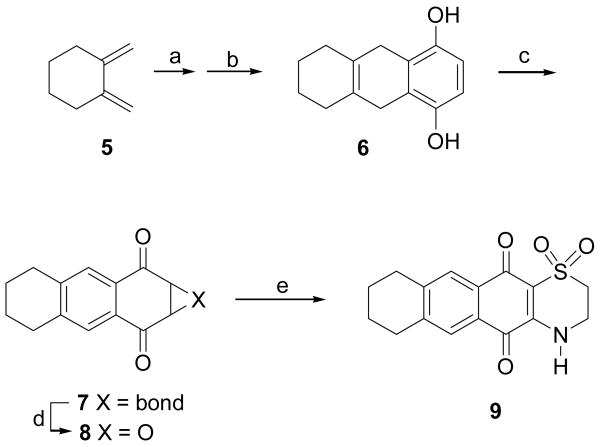
The compounds evaluated were prepared by literature methods. The synthesis of compound 9 has not previously been described, although it has been claimed in a patent,3,6 and so it is described briefly here (Scheme 1). Conjugated diene 5 was obtained from the commercially available hexahydroisobenzofuran-1,3-dione (1) by conversion to diethyl cyclohexane-1,2-dicarboxylate (2), reduction to 1,2-cyclohexanedimethanol (3), tosylation to the bis-tosylate 4, and treatment with base to give 1,2-dimethylenecyclohexane (5) as previously described.3,7,38 1,2-dimethylenecyclohexane (5) was then added to benzoquinone in a Diels-Alder reaction, followed by treatment with potassium carbonate to give the hydroquinol 6.3,9 Air oxidation of 6 gave naphthoquinone 7, as well as a small amount of epoxide 8 as an oxidation by-product. Compound 7 was coupled with hypotaurine to yield 9.40
Scheme 1.
Reagents and conditions: (a) benzoquinone, 50% DCM/DMSO, N2, RT, 24 hr; (b) K2CO3, acetone, RT, overnight; (c) air, acetone, RT; (d) 8 was a by-product the reaction of 7 with hydrogen peroxide produced during the auto-oxidation of 6 to 7; (e) 7 + hypotaurine, EtOH/MeCN/H2O (1:1:1), 40 °C, 2 hr.
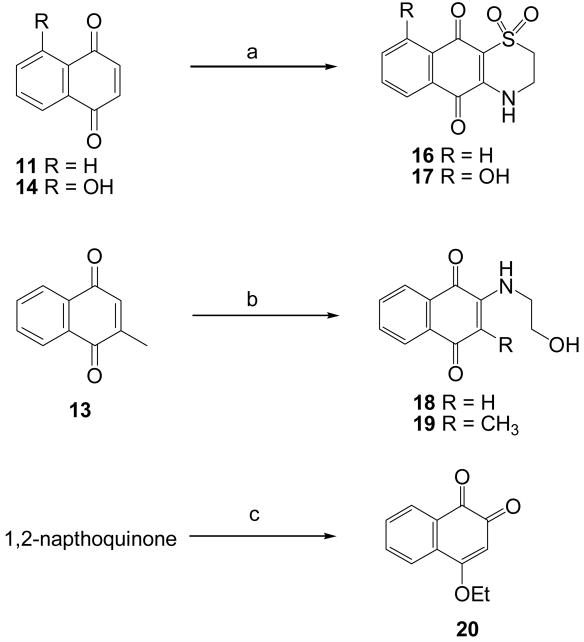
Compounds 10-15 [ 1,4-benzoquinone (10), 1,4-naphthoquinone (11), 9,10-anthraquinone (12), 2-methyl-1,4-naphthoquinone (13), 5-hydroxy-1,4-naphthoquinone (14), and 5,8-dihydroxy-1,4-naphthoquinone (15)] were purchased from commercial sources. Compounds 16-20 were prepared via coupling reactions of 1,4-naphthoquinone derivatives with hypotaurine, ethanolamine, and ethanol (Scheme 2). The synthesis of 18 from 2-methylnaphthoquinone involved the loss of the C2 methyl group by an interesting previously described mechanism involving conjugate addition of ethanolamime to the C2 methyl group followed by the loss of methyleneimino-ethanol in a reverse-Mannich type reaction.4,1,42
Scheme 2.
Reagents and conditions: (a) hypotaurine in H2O:MeCN:EtOH (1:1:1), 40 °C, 4 hr; (b) ethanolamine in MeCN, 45 °C, 2 hr; (c) 50% MeCN/EtOH, DMAP, 40 °C, 5 hr.
2.2. Bioactivities
Quinones 7 – 20, together with the lead compound adociaquinone B, were evaluated as inhibitors of Cdc25B, MKP-1, and MKP-3. The bioassay for inhibition of Cdc25B was conducted with an epitope-tagged (histidine6) catalytic domain of human recombinant Cdc25B, which contained amino acids 275–539 of the full-length protein and has been previously described.18 The results are shown in Table 1. The antiproliferative activities of the compounds against the A2780 human ovarian cancer cell line were also determined, to establish their ability to penetrate cells and demonstrate in vivo activity.
Table 1.
Biological activities of compounds 7-20 (μM)
| Compound | Cdc25Ba | MKP-1a | MKP-3a | A2780b |
|---|---|---|---|---|
| Adociaquinone B | 0.08/0.11 | 1.10 | 1.53 | 26 ± 0.06 |
| 7 | 9.53 | 21.8 | >50 | 0.75 ± 0.03 |
| 8 | 22.3 | > 50 | 49.9 | 1.4 ± 0.07 |
| 9 | 2.3 | 38.7 | > 50 | 6.9 ± 0.1 |
| 10 | > 50 | > 50 | > 50 | 7.8 ± 0.4 |
| 11 | 2.76 | 8.45 | 19.8 | 2.3 ± 0.03 |
| 12 | 9.51 | > 50 | > 50 | 0.58 ± 0.45 |
| 13 | 3.38 | 24.0 | 20.5 | 2.6 ± 0.06 |
| 14 | 1.98 | 13.0 | 12.4 | 1.4 ± 0.05 |
| 15 | 1.00 | 9.37 | 6.90 | 0.08 ± 0.001 |
| 16 | 0.94 | 17.8 | 42.6 | 4.3 ± 2.3 |
| 17 | 0.88 | 33.8 | > 50 | 2.1 ± 0.02 |
| 18 | > 50 | > 50 | > 50 | 64 ± 1.2 |
| 19 | > 50 | > 50 | > 50 | * |
| 20 | 0.27 | 0.82 | 1.35 | 0.37 ± 0.01 |
IC50 values (μM) for activity against the catalytic domain of Cdc25B. The data for adociaquinone B are for activity against the catalytic domain (0.11) and full length Cdc25B (0.08)
Antiproliferative activity (IC50 μM); growth of the A2780 human ovarian cell line according to the procedure described.29,4,3,44 with paclitaxel (IC50 23.4 nM) as the positive control. All compounds were tested in triplicate in this assay.
The fluorescence reading was not indicative of the true biological activity of 19. The fluorescence reading indicated that 19 was not active, but microscopic analysis clearly showed that significant inhibition of the growth of A2780 cells occurred.
2.3. Discussion
Several conclusions can be drawn from these results. Firstly, with the exception of the amino derivatives compounds 18 and 19, the current data clearly support the notion that naphthoquinone-type molecules have the potential to be effective therapeutic agents, though their exact mechanism of Cdc25 inhibition is still a topic of discussion. A few computational studies have attempted to utilize modeling to predict various binding interactions of the Cdc25 dual-specificity phosphatases. The docking orientation of Cdc25B with its Cdk2-cyclin A protein substrate has previously been reported, and therein the possibility was recognized of targeting several potential small molecule binding pockets to achieve disruption of protein substrate recognition.12 Additionally, in an effort to postulate a mechanism of enzyme inhibition, attempts were made to dock known small molecule inhibitors of Cdc25B within its shallow active site.4,5 Using a naphthoquinone derivative, one docking program showed hydrogen bonding between the quinone carbonyl oxygens with Arg482 and Arg544 of the shallow pocket. As previously suggested,21 this implies that the quinone functionality is necessary for specific enzyme-ligand binding that propagates the observed activity. An alternate docking program bound the naphthoquinone inhibitors in such a way that the quinone moiety was in close proximity to the Cys473 thiol residue.33 Indeed, the structurally similar para-quinolinediones DA3003-1 and JUN1111 were shown to engage in redox cycling, thus irreversibly oxidizing the Cys473 thiol to its sulfonic product via production of reactive oxygen species (ROS).4,6
In regard to our study the most active compound was the ortho-quinone 20. However, its activity is relatively non-selective, suggesting that it operates by a non-selective mechanism and is thus unlikely to have development potential.
The simplification of adociaquinone B by the deletion of two rings on the west hemisphere gave compound 9, with activity against Cdc25B of 2.3 μM, at least 20-fold less than that of the lead compound (0.08 or 0.11 μM). The deletion of these rings may have diminished its ability to bind effectively with residues in the active pocket of Cdc25B, thus reducing its potency. Quinones 16 and 17, lacking three rings on the west hemisphere of adociaquinone B, nevertheless were more potent than 9 toward Cdc25B, with IC50 values of 0.94 and 0.88 μM, respectively, so the simple idea that deletion of the western rings reduces activity is clearly inadequate to explain the observed effects. It should also be noted that the antiproliferative activities of 16 and 17 differ by a factor of over 20, even though their inhibitory effects on Cdc25B are very similar. Since the compounds are structurally similar it is unlikely that this difference in activity is solely due to cell permeability factors, suggesting that these compounds may have other mechanisms of action. In context of the other protein kinase phosphatase assays, 16 and 17 are moderately selective for Cdc25B as opposed to MKP-1 and MKP-3, indicating that these compounds have attractive properties for further development as selective inhibitors of Cdc25B.
Further structure activity analysis showed that simplification of 16 and 17 to basic naphthoquinone derivatives (compounds 11, 13, 14, 15; IC50 = 2.76, 3.38, 1.98, 1.00 μM, respectively) resulted in a two to three-fold decrease in activity (with the exception of 15), suggesting that the heterocyclic ring of 16 and 17 does play a role in either binding within the active pocket, or some other aspect of the mechanism of action. This hypothesis is strengthened by the observed 10-fold loss in activity when the heterocyclic ring is substituted by a six-membered aromatic ring (12, IC50 = 9.51 μM). Interestingly, in the cases of 18 and 19 a complete loss of activity is observed, presumably due to either the substitution of the sulfoxide moiety with that of an amine, or the absence of cyclization to the naphthoquinone, or both. The complete loss of enzyme activity is also observed with compound 10, thus suggesting that the basic naphthoquinone skeleton is a primary requirement for bioactivity. The aforementioned data contribute to the quest of determining exactly how such naphthoquinone derivatives exert their specific Cdc25B enzymatic bioactivity, and may highlight the presence of mechanistic complexities of potent Cdc25B inhibitors such as adociaquinone B.
Experimental evidence from our previous study29 allowed us to hypothesize that the mechanism of action of adociaquinone B is oxidation of the catalytic cysteine of Cdc25B. If this is correct, the current bioassay data suggest that the fused tricyclic benzofuranone moiety may play a pivotal role in effectively positioning the quinone for Cys473 thiolate oxidation. Comparison of 9 with 16 and 17 suggests that the absence of three rings on the west hemisphere of adociaquinone B may grant the resulting small molecule more versatility by allowing increased access to the binding site, though further studies are needed to prove such a hypothesis.
3. Experimental
3.1. General experimental methods
All of the reagents and solvents received from commercial sources were used as purchased, at greater than 98% purity. 1H and 13C NMR spectra were obtained on Varian Unity 400 MHz, Inova 400 MHz, and JEOL Eclipse 500 MHz spectrometers. High resolution FAB and low resolution EI mass spectra were obtained on a JEOL HX-110 instrument, and a Finnigan LTQ LC/MS, respectively. Reaction mixtures were worked up by the standard procedure of quenching the reaction, extracting the resulting aqueous mixture with EtOAc or Et2O or dichloromethane, washing the organic solution with water and brine, drying over Na2SO4, filtering, and concentrating to give crude product. Final compounds were purified to homogeneity by preparative TLC, and their purity was assessed by 1H NMR spectroscopy. All the reported compounds had purities of 95% or better as estimated from analysis of their NMR spectra.
3.2 Synthesis
3.2.1. Synthesis of compound 9
A solution of compound 5 (12 mg, 111.1 μmol)37,38 in dry dichloromethane (1 mL) was added to the stirred solution of benzoquinone (72 mg, 666.7 μmol) in dry DMSO (2 mL) under a nitrogen atmosphere.39 The mixture was stirred at RT for 24 hr, which was then dried under nitrogen stream. Complete removal of the DMSO was achieved by washing the film with water and drying under a nitrogen stream.4,7 The dried residue was dissolved in dry acetone (2 mL). A spatula tip of potassium carbonate was added, and the mixture was stirred at RT overnight under a nitrogen atmosphere.39 The reaction mixture was worked up by the standard procedure of quenching the reaction, extracting the resulting aqueous mixture with EtOAc or Et2O or dichloromethane, washing the organic solution with water and brine, drying over Na2SO4, filtering, and concentrating to give crude product. The residue was purified by silica gel TLC (EtOAc/hexanes 1:6) to give 6 (Rf 0.50, 21.6 mg, 90%, and 8 (Rf 0.30, 12.5 mg, 10%). Compound 7 (silica gel TLC, EtOAc/hexanes 1:6, Rf 0.40, 5 mg, 30%) was an automatically oxidized product of 6 (5,6,7,8,9,10-hexahydroanthracene-1,4-diol) in acetone under an air atmosphere. To a solution of 7 (4 mg, 18.9 μmol) in 1.5 mL ethanol/acetonitrile/water (1:1:1) was added hypotaurine (6.2 mg, 56.9 μmol), and the reaction mixture was stirred at 40 °C for 2 hr.40 The crude product obtained was purified by silica gel TLC (DCM/EtOAc 1:1) yielding compound 9 (Rf 0.25, 3 mg, 50%).
5,6,7,8,9,10-hexahydroanthracene-1,4-diol (6). 1H NMR (C5D5N) δ 7.02 (2H, s), 3.63 (4H, s), 2.01 (4H, m), 1.58 (4H, m); 13C NMR (C5D5N) δ 149.1, 126.0, 124.5, 112.9, 31.9, 30.2, 23.1; EIMS m/z 217.1 [ M + H] + (calcd for C14H17O2, 217.1).
7. 1H NMR (CDCl3) δ 7.73 (2H, s), 6.87 (2H, s), 2.86 (4H, m), 1.81 (4H, m); 13C NMR (CDCl3) δ 185.3, 144.3, 138.6, 129.4, 127.2, 29.8, 22.5; EIMS m/z 213.2 [ M + H] + (calcd for C14H13O2, 213.1).
8. 1H NMR (CDCl3) δ 7.68 (4H, s), 3.95 (4H, s), 2.87 (8H, m), 1.82 (8H, m); 13C NMR (CDCl3) δ 191.0, 145.5, 129.2, 128.0, 55.4, 29.9, 22.5; CIMS m/z 229.0 [ M + H] + (calcd for C14H13O3, 229.1).
9. 1H NMR (CDCl3) δ 7.71 (1H, s), 7.68 (1H, s), 3.84 (1H, t, J = 6.3 Hz), 3.35 (1H, t, J = 6.3 Hz), 2.87 (4H, m), 1.76 (4H, m); 13C NMR (CDCl3) δ 178.6, 174.8, 146.7, 146.0, 142.5, 130.0, 127.3, 127.1, 126.4, 111.0, 48.3, 40.0, 29.3, 28.7, 22.0, 22.0; EIMS m/z 318.1 [ M + H] + (calcd for C16H16NO4S, 318.1).
3.2.2. Synthesis of compound 16
A solution of hypotaurine (60 mg, 550.0 μmol) in water (10 mL) was added to a stirred solution of 1,4-naphthoquinone (11) (60 mg, 379.0 μmol) in 20 mL acetonitrile/ethanol (1:1), and the mixture was stirred at 40°C for four hr. The crude product solution was refrigerated to yield a yellow precipitate, and filtered to afford compound 16 (62.6 mg, 63%). Compound 16 was identified on the basis of one- and two-dimensional NMR data, including 1H, 13C, g-COSY, g-HSQC, and g-HMBC experiments in addition to MS analyses: 1H NMR (DMSO-d6) δ 9.16 (1H, br s), 7.99 (2H, dd, J = 4.0, 1.6 Hz), 7.89 (1H, t, J = 7.6 Hz), 7.77 (1H, t, J = 7.6 Hz), 3.85 (2H, br t, J = 5.5 Hz), 3.38 (2H, br t, J = 5.5 Hz); 13C NMR (DMSO-d6) δ 178.7, 174.6, 146.8, 135.7, 132.9, 132.5, 130.0, 126.4, 125.8, 111.1, 48.26, 39.18; EIMS m/z 264.1 [ M+H] + (calcd for C12H10O4NS, 264.03).
3.2.3. Synthesis of compound 17
A solution of hypotaurine (9.3 mg, 85.0 μmol) in water (10 mL) was added to a stirred solution of 5-hydroxy-1,4-naphthoquinone (14) (10 mg, 57.0 μmol) in 20 mL acetonitrile/ethanol (1:1), and the mixture was stirred at 40°C for four hr. The crude product was purified over a flash silica column (CHCl3/EtOAc 1:1) to afford yellow needle-like crystals, compound 17 (3.6 mg, 23%). This specific regioisomer was identified upon comparison of its 1H and 13C NMR data with those in the literature:48 1H NMR (DMSO-d6) δ 12.83 (1H, s), 9.50 (1H, br s), 7.64 (1H, t, J = 7.2 Hz), 7.55 (1H, dd, J = 7.6, 0.8 Hz), 7.34 (1H, dd, J = 7.6, 0.8 Hz), 3.87 (2H, m), 3.41 (2H, m); 13C NMR (DMSO-d6) δ 181.5, 177.9, 160.8, 147.8, 135.0, 130.2, 125.8, 119.1, 113.7, 110.0, 48.2, 39.8; EIMS m/z 280.1 [ M + H] + (calcd for C12H10O5NS, 280.02).
3.2.4. Synthesis of compound 18
Ethanolamine (80.5 μL, 1.34 mmol) was added to a stirred solution of 2-methyl-1,4-naphthoquinone (13) (230 mg, 134 μmol) in 5 mL acetonitrile, and the mixture was stirred at 45°C for two hours. Unexpectedly, loss of the C-2 methyl resonance was observed in the 1H NMR spectrum. The structure was thus elucidated by one and two-dimensional NMR methods and subsequently confirmed by X-ray crystallography. It is postulated that two molecules of ethanolamine are involved in a reverse-Mannich type reaction, ultimately resulting in loss of methyleneimino-ethanol at the C-2 position. Loss of a methyl group from quinones has been reported some time ago, and mechanistic details were described. The crude product was purified by silica gel TLC (EtOAc) to afford orange crystals, compound 18 (Rf 0.44, 9.6 mg, 3.3%): 1H NMR (DMSO-d6) δ 7.98 (1H, dd, J = 8.0, 1.2 Hz), 7.93 (1H, dd, J = 7.6, 1.2 Hz), 7.82 (1H, dt, J = 8.0, 1.2 Hz), 7.72 (1H, dt, J = 7.6, 1.2 Hz), 7.34 (1H, br t, J = 5.6 Hz), 5.72 (1H, s), 4.87 (1H, t, J = 5.6 Hz), 3.58 (2H, q, J = 5.6 Hz), 3.23 (2H, q, J = 5.6 Hz); 13C NMR (DMSO-d6) δ 181.6, 181.4, 148.7, 134.9, 133.1, 132.2, 130.3, 125.9, 125.3, 99.6, 58.4, 44.6; FABMS m/z 199.4 [ M–H2O] (calcd for C12H9O2N, 199.21).
3.2.5. Synthesis of compound 19
Ethanolamine (80.5 μL, 1.34 mmol) was added to a stirred solution of 2-methyl-1,4-naphthoquinone (13) (230 mg, 134 μmol) in 5 mL acetonitrile, and the mixture was stirred at 45°C for two hr. The crude product was purified by silica gel TLC (EtOAc) to afford compound 19 as red, needle-like crystals (Rf 0.75, 62.0 mg, 20%). Compound 19 was identified on the basis of one- and two-dimensional NMR data, including 1H, 13C, g-COSY, g-HSQC, and g-HMBC experiments in addition to MS analyses: 1H NMR (DMSO-d6) δ 7.87 (2H, t, J = 6.8 Hz), 7.73 (1H, t, J = 7.6 Hz), 7.64 (1H, t, J = 7.6 Hz), 6.47 (1H, br s), 4.92 (1H, br t, J = 5.2 Hz), 3.57 (4H, m), 2.06 (3H, s); 13C NMR (DMSO-d6) δ 182.1, 181.7, 146.8, 134.3, 132.7, 132.0, 130.2, 125.6, 125.4, 110.8, 60.5, 46.8, 10.7; FABMS m/z 232.1 [ M + H] + (calcd for C13H14O3N, 231.10).
3.2.6. Synthesis of compound 20
A solution of 1,2-naphthoquinone (30 mg, 190 μmol) in 20 mL acetonitrile/ethanol (1:1) and in the presence of a catalytic amount of DMAP, was stirred at 40°C for five hr. The crude product was purified by silica gel column chromatography (CHCl3/EtOAc 1:1) followed by an open reversed phase C18 column (MeOH/H2O 3:2) to afford compound 20 as yellow needle-like crystals (3.8 mg, 10%). Compound 20 was identified on the basis of one- and two-dimensional NMR data, including 1H, 13C, g-COSY, g-HSQC, and g-HMBC experiments in addition to MS analyses: 1H NMR (CD3CN) δ 8.00 (1H, d, J = 8.4 Hz), 7.91 (1H, d, J = 8.0 Hz), 7.75 (1H, t, J = 8.0 Hz), 7.63 (1H, t, J = 8.4 Hz), 5.94 (1H, s), 4.24 (2H, q, J = 6.8 Hz), 1.49 (3H, t, J = 6.8 Hz); 13C NMR (CD3CN) δ 180.6, 180.2, 168.5, 135.9, 133.2, 132.3, 131.6, 129.1, 125.4, 104.3, 66.9, 14.2; FABMS m/z 203.7 [ M + H] + (calcd for C12H10O3, 203.21).
3.3. Antiproliferative assay
Determination of antiproliferative activity was performed at Virginia Polytechnic Institute and State University against the A2780 ovarian cancer cell line as previously described.43 The A2780 cell line is a drug - sensitive human ovarian cancer cell line.44
3.4. In vitro phosphatase assays
3.4.1 Protein preparation
As previously noted, the bioassay for inhibitors of Cdc25B was conducted with an epitope-tagged (histidine6) catalytic domain of human recombinant Cdc25B, which contained amino acids 275–539 of the full-length protein and has been previously described.18 The histidine6-tagged catalytic domain and full-length Cdc25B were isolated and purified from E. coli with Ni-NTA resin as described previously.18 Human recombinant VHR and PTP1B phosphatases were purchased from BIOMOL (Plymouth Meeting, PA). The expression and purification of an epitope-tagged (histidine6) recombinant MKP-1 has been described previously.4,9 Briefly, His-tagged MKP-1 was expressed in BL21(DE3) competent bacterial cells transformed with the pET21a-MKP-1 expression vector and induced with IPTG. Biologically active MKP-1 was partially purified from IPTG-induced bacterial cell lysates by Cobalt-column chromatography eluted with imidazole, frozen, aliquoted and stored at − 80 °C. The relative purity of the MKP-1 enzyme preparation was visualized by separating the protein components by SDS-PAGE and staining with Coomassie Blue. The identity of the MKP-1 enzyme was confirmed by the presence of the anticipated 42 kDa band together with immuno-reactivity with antibodies against MKP-1 (Santa-Cruz, M-18, Catalog # SC-1102) and the His-Tag epitope (Cell Signaling, Catalog # 2365) on Western blots (data not shown). The expression of recombinant MKP-3 protein used a bacterial expression plasmid. A Rattus norvegicus dual specificity phosphatase6 (MKP-3)50 and pET21a-MKP3 plasmid5,1 generated a 100 μL aliquot with 0.05 μg/μL protein concentration. T7 promoter and T7 terminator primer sequencing reactions were performed, and the rat MKP-3 sequence was confirmed. BL21(DE3) pET21a-MKP3 glycerol stocks were used for protein purification.
3.4.2 Bioassay Procedures
Activities of all phosphatases were measured using the substrate O-methyl fluorescein phosphate (Sigma, St. Louis, MO) in a 96-well microtiter plate assay based on previously described methods.18,21, 52, 53 The final incubation mixtures (25 μL) were prepared with a Biomek 2000 laboratory automation workstation (Beckman Coulter, Inc., Fullerton, CA). Fluorescence emission from the product was measured after a 20 or 60 min incubation period at ambient temperature with a multiwell plate reader (PerSeptive Biosystems Cytofluor II; Framingham, MA; excitation filter, 485 nm/bandwidth 20 nm; emission filter, 530 nm/bandwidth 25 nm). For the initial bioassay-directed fractionation, samples were evaluated at one concentration (0.2–1.0 μg/mL) and subsequent fractionations were examined with a minimum of six concentrations to determine the concentration required to inhibit enzyme activity by 50% (IC50). To test for sensitivity to redox regulation, in some studies with full length Cdc25B, we adjusted the final concentration of DTT in the enzyme buffer, which contained 30 mM Tris (pH 8.5), 75 mM NaCl, 1 mM EDTA, and 0.033% bovine serum albumin, from the standard of 1 mM to a range from 0 to 100 mM. No significant difference was seen in inhibition with different DTT concentrations.
Acknowledgments
This work was supported in part by a National Cooperative Drug Discovery Group grant awarded to the University of Virginia by the National Cancer Institute (U19 CA50771), and this support is gratefully acknowledged. We thank Mr. B. Bebout for obtaining the mass spectra and Mr. T. Glass for assistance obtaining the NMR spectra.
References and notes
- 1.Lyon MA, Ducret AP, Wipf P, Lazo JS. Nature Rev Drug Discov. 2002;1:961. doi: 10.1038/nrd963. [DOI] [PubMed] [Google Scholar]
- 2.Cangi MG, Cukor B, Soung P, Signoretti S, Moreira JG, Ranashinge M, Cady B, Pagano M, Loda M. J Clin Investig. 2000;106:753. doi: 10.1172/JCI9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oguri T, Singh SV, Nemoto K, Lazo JS. Cancer Res. 2003;63:771. [PubMed] [Google Scholar]
- 4.Galaktionov K, Lee AK, Eckstein J, Draetta G, Meckler J, Loda M, Beach D. Science. 1995;269:1575. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 5.Millar JBA, Blevitt J, Gerace L, Sadhu K, Featherstone C, Russell P. Proc Natl Acad Sci USA. 1991;88:10500. doi: 10.1073/pnas.88.23.10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. EMBO J. 1993;12:53. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lammer C, Wagerer S, Saffrich R, Mertens D, Ansorge W, Hoffmann I. J Cell Science. 1998;111:2445. doi: 10.1242/jcs.111.16.2445. [DOI] [PubMed] [Google Scholar]
- 8.Dunphy WG, Kumagai A. Cell. 1991;67:189. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 9.Mailand M, Podtelejnikov AV, Groth A, Mann M, Bartek J, Lukas J. EMBO J. 2002;21:5911. doi: 10.1093/emboj/cdf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutros R, Lobjois V, Ducommun B. Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 11.Lindqvist A, Kallstrom H, Lundgren A, Barsoum E, Rosenthal CK. J Cell Biol. 2005;171:35. doi: 10.1083/jcb.200503066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohn J, Parks JM, Buhrman G, Brown P, Kristjánsdóttir K, Safi A, Edelsbrunner H, Yang W, Rudolph J. Biochemistry. 2005;44:16563. doi: 10.1021/bi0516879. [DOI] [PubMed] [Google Scholar]
- 13.Rudolph J. Nat Rev Cancer. 2007;7:202. doi: 10.1038/nrc2087. [DOI] [PubMed] [Google Scholar]
- 14.Bansal P, Lazo JS. Cancer Res. 2007;67:3356. doi: 10.1158/0008-5472.CAN-06-3685. [DOI] [PubMed] [Google Scholar]
- 15.Guan KL, Dixon JE. J Biol Chem. 1991;266:17026. [PubMed] [Google Scholar]
- 16.Cho H, Krishnaraj R, Kitas E, Bannwarth W, Walsh CT, Anderson KS. J Am Chem Soc. 1992;114:7296. [Google Scholar]
- 17.Rudolph J. Biochemistry. 2002;41:14613. doi: 10.1021/bi0263513. [DOI] [PubMed] [Google Scholar]
- 18.Kar S, Lefterov IM, Wang M, Lazo JS, Scott CN, Wilcox CS, Carr BI. Biochemistry. 2003;42:10490. doi: 10.1021/bi027418p. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Dube D, Friesen RW, LeRiche TG, Bateman KP, Trimble L, Sanghara PR, Ramachandran C, Gresser MJ, Pollex R, Huang Z. Biochemistry. 2004;43:4294. doi: 10.1021/bi035986e. [DOI] [PubMed] [Google Scholar]
- 20.Cazales M, Boutros R, Brezak MC, Chaumeron S, Prevost G, Ducommun B. Mol Cancer Ther. 2007;6:318. doi: 10.1158/1535-7163.MCT-06-0299. [DOI] [PubMed] [Google Scholar]
- 21.Lazo JS, Nemoto K, Pestell KE, Cooley K, Southwick EC, Mitchell DA, Furey W, Gussio R, Zaharevitz DW, Joo B, Wipf P. Mol Pharmacol. 2002;61:720. doi: 10.1124/mol.61.4.720. [DOI] [PubMed] [Google Scholar]
- 22.Kar S, Wang M, Ham SW, Carr BI. Cancer Biol Ther. 2006;5:1340. doi: 10.4161/cbt.5.10.3223. [DOI] [PubMed] [Google Scholar]
- 23.Kar S, Wang M, Ham SW, Carr BI. Biochem Pharmacol. 2006;72:1217. doi: 10.1016/j.bcp.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 24.Tandon VK, Yadav DB, Singh RV, Vaish M, Chaturvedi AK, Shukla PK. Bioorg Med Chem. 2005;15:3463. doi: 10.1016/j.bmcl.2005.04.075. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Southwick E, Kerns J, Rosi K, Carr BI, Wilcox C, Lazo JS. Cancer Res. 2000;60:1317. [PubMed] [Google Scholar]
- 26.Brun MP, Braud E, Angotti D, Mondésert O, Quaranta M, Montes M, Miteva M, Gresh N, Ducommun B, Garbay C. Bioorg Med Chem. 2005;13:4871. doi: 10.1016/j.bmc.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Ham SW, Lee S. Bull Korean Chem Soc. 2004;25:1755. [Google Scholar]
- 28.Braud E, Goddard ML, Kolb S, Brun MP, Mondésert O, Quaranta M, Gresh N, Ducommun B, Garbay C. Bioorg Med Chem. 2008;16:9040. doi: 10.1016/j.bmc.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Cao S, Foster C, Brisson M, Lazo JS, Kingston DGI. Bioorg Med Chem. 2005;13:999. doi: 10.1016/j.bmc.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 30.Lazo JS, Nunes R, Skoko JJ, Queiroz de Oliveira PE, Vogt A, Wipf P. Bioorg Med Chem. 2006;14:5643. doi: 10.1016/j.bmc.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 31.Creus M, Zoller H. Signaling Mol Targets Cancer Ther. 2007;69 [Google Scholar]
- 32.Kikuchi K, Nakamura K, Shima H. Curr Top Biochem Res. 1999;1:75–87. [Google Scholar]
- 33.Magi-Galluzzi C, Mishra R, Fiorentino M, Montironi R, Yao H, Capodieci P, Wishnow K, Kaplan I, Stork PJ, Loda M. Lab Invest. 1997;76:37. [PubMed] [Google Scholar]
- 34.Liao Q, Guo J, Kleeff J, Zimmermann A, Büchler MW, Korc M, Friess H. Gastroenter. 2003;124:1830. doi: 10.1016/s0016-5085(03)00398-6. [DOI] [PubMed] [Google Scholar]
- 35.Denkert C, Schmitt WD, Berger S, Reles A, Pest S, Siegert A, Lichtenegger W, Dietel M, Hauptmann S. Int J Cancer. 2002;102:507. doi: 10.1002/ijc.10746. [DOI] [PubMed] [Google Scholar]
- 36.Andersen RJ, Pereira A, Huang XH, Mauk G, Vottero E, Roberge M, Balgi A. Indoleamine 2,3-Dioxygenase (Ido) Inhibitors, and Their Therapeutic Use. WO2006/005185 A1. 2006 January 19; [Google Scholar]
- 37.Baldwin JE, Chang GEC. J Org Chem. 1982;47:848. [Google Scholar]
- 38.Lin CT, Chou TC. J Org Chem. 1990;55:2252. [Google Scholar]
- 39.Thomas AD, Miller LL. J Org Chem. 1986;51:4160. [Google Scholar]
- 40.Harada N, Sugioka T, Soutome T, Hiyoshi N, Uda H, Kuriki T. Tetrahedron Lett. 1995;6:375. [Google Scholar]
- 41.Cameron DW, Scott PM. J Chem Soc. 1964:5569. [Google Scholar]
- 42.Cameron DW, Samuel EL. Aust J Chem. 1977;30:2063. [Google Scholar]
- 43.Cao S, Brodie PJ, Randrianaivo R, Ratovoson F, Callmander M, Andriantsiferana R, Rasamison VE, Kingston DGI. J Nat Prod. 2007;70:679. doi: 10.1021/np060627g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louie KG, Behrens BC, Kinsella TJ, Hamilton TC, Grotzinger KR, McKoy WM, Winker MA, Ozols RF. Cancer Res. 1985;45:2110. [PubMed] [Google Scholar]
- 45.Lavecchia A, Cosconati S, Limongelli V, Novellino E. ChemMedChem. 2006;1:540. doi: 10.1002/cmdc.200500092. [DOI] [PubMed] [Google Scholar]
- 46.Brisson M, Nguyen T, Wipf P, Joo B, Day BW, Skoko JS, Schreiber EM, Foster C, Bansal P, Lazo JS. Mol Pharmacol. 2005;68:1810. doi: 10.1124/mol.105.016360. [DOI] [PubMed] [Google Scholar]
- 47.Vié V, Van Mau N, Chaloin L, Lesniewska E, Le Grimellec C, Heitz F. Biophys J. 2000;78:846. doi: 10.1016/S0006-3495(00)76642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson RA, Pereira A, Huang XH, Mauk G, Vottero E, Roberge M, Balgi A. U. S. Patent No. WO2006005185. 2006
- 49.Vogt A, Tamewitz A, Skoko J, Sikorski RP, Giuliano KA, Lazo JS. J Biol Chem. 2005;280:19078. doi: 10.1074/jbc.M501467200. [DOI] [PubMed] [Google Scholar]
- 50.Muda M, Boschert U, Dickinson R, Martinou JC, Martinou I, Camps M, Schlegel W, Arkinstall S. J Biol Chem. 1996;271:4319. doi: 10.1074/jbc.271.8.4319. [DOI] [PubMed] [Google Scholar]
- 51.Zhou B, Wu L, Shen K, Zhang J, Lawrence DS, Zhang ZY. J Biol Chem. 2001;276:6506. doi: 10.1074/jbc.M009753200. [DOI] [PubMed] [Google Scholar]
- 52.Rice RL, Rusnak JM, Yokokawa F, Yokokawa S, Messner DJ, Boynton AL, Wipf P, Lazo JS. Biochemistry. 1997;36:15965. doi: 10.1021/bi971338h. [DOI] [PubMed] [Google Scholar]
- 53.Wipf P, Hopkins CR, Phillips EO, Lazo JS. Tetrahedron. 2002;58:6367. [Google Scholar]