Abstract
Plasma homocysteine (Hcy) level is associated with cardiovascular disease and may play an etiologic role in vascular damage, a precursor for atherosclerosis. We performed a genome-wide association study for Hcy in 1786 unrelated Filipino women from the Cebu Longitudinal Health and Nutrition Survey (CLHNS). The most strongly associated single-nucleotide polymorphism (SNP) (rs7422339, P = 4.7 × 10−13) encodes Thr1405Asn in the gene CPS1 and explained 3.0% of variation in the Hcy level. The widely studied MTHFR C677T SNP (rs1801133) was also highly significant (P = 8.7 × 10−10) and explained 1.6% of the trait variation. We also genotyped these two SNPs in 1679 CLHNS young adult offspring. The MTHFR C677T SNP was strongly associated with Hcy (P = 1.9 × 10−26) and explained ∼5.1% of the variation in the offspring. In contrast, the CPS1 variant was significant only in females (P = 0.11 in all; P = 0.0087 in females). Combined analysis of all samples confirmed that the MTHFR variant was more strongly associated with Hcy in the offspring (interaction P = 1.2 × 10−5). Furthermore, although there was evidence for a positive synergistic effect between the CPS1 and MTHFR SNPs in the offspring (interaction P = 0.0046), there was no significant evidence for an interaction in the mothers (P = 0.55). These data confirm a recent finding that CPS1 is a locus influencing Hcy levels in women and suggest that genetic effects on Hcy may differ across developmental stages.
INTRODUCTION
Homocysteine (Hcy) is a sulfur-containing amino acid produced in the metabolism of the essential amino acid methionine, which occurs through folic acid and vitamin B6- and vitamin B12-dependent pathways (1). Increased blood plasma Hcy levels have been shown to be a risk factor for cardiovascular disease (CVD) and may play an etiologic role in vascular damage by promoting oxidative stress, systemic inflammation and endothelial dysfunction (2). Numerous epidemiologic studies have reported that elevated Hcy is associated with both subclinical and clinical CVD, and evidence supports that the increased risk is independent of other atherosclerosis risk factors (3–8).
Genetic factors play a substantial role in Hcy levels, but whether the same variants influence Hcy across populations is unknown. Heritability estimates range from 47–70% across several genetic and environmental backgrounds (9–11). The most well-studied gene containing variants associated with Hcy levels is methylene tetrahydrofolate reductase (MTHFR), which encodes a key enzyme in folic acid metabolism and was first identified as the causal gene for three rare autosomal homocysteinurias (12). A meta-analysis of the MTHFR C677T variant (rs1801133) in 33 studies representing 35 populations showed a mean difference in serum Hcy between the TT and CC genotypes of 2.7 mmol/L (2.1–3.4 mmol/L) and between the CT and CC genotypes of 0.29 mmol/L (0.20–0.39 mmol/L) (8). More recently, genome-wide association (GWA) studies for Hcy levels in populations of European descent identified novel loci (13,14).
To further clarify the genetic contributions to normal variation in Hcy in a population of primarily non-European descent, we performed a GWA study of the Hcy level in mothers from the Cebu Longitudinal Health and Nutrition Survey (CLHNS), using single-nucleotide polymorphism (SNP) data on 1786 women from Cebu, the Philippines. We also genotyped selected SNPs and tested them for association with the Hcy level in their young adult offspring to further evaluate genetic effects in males and across a wider age range.
RESULTS
GWA results in CLHNS mothers
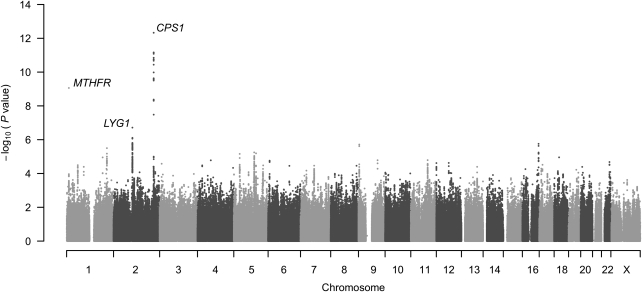
In 1786 CLHNS mothers with complete genotype and phenotype data (Supplementary Material, Table S1), we observed 22 SNPs significantly associated with natural log-transformed Hcy (log-Hcy) levels (P < 5 × 10−8), all located within or near the carbamoyl phosphate synthetase 1 (CPS1) gene on chromosome 2 (Fig. 1). The most strongly associated SNP (rs7422339, P = 4.7 × 10−13) was imputed, and encodes a Thr1405Asn substitution. Tests of association for the other CPS1 SNPs conditional on rs7422339 suggested a single CPS1 signal. The most significant SNP outside the CPS1 region was rs6722617 (P = 1.9 × 10−7), an imputed SNP located on chromosome 2q11.2 near lysozyme G-like 1 (LYG1) and thioredoxin domain containing 9 (TXNDC9), neither of which has been previously implicated as having effects on Hcy or other CVD-related traits. Several MTHFR SNPs showed modest evidence of association (best P = 2.7 × 10−4 for rs13306561). However, the widely studied MTHFR C677T variant (rs1801133) was not directly tested as part of the genome-wide panel, and the imputation quality score did not meet the minimum requirement for inclusion in analysis.
Figure 1.
GWA results for log-Hcy levels in 1786 CLHNS mothers. Result for directly genotyped SNP rs1801133 (MTHFR) is included. Loci described in the text are annotated with gene symbols.
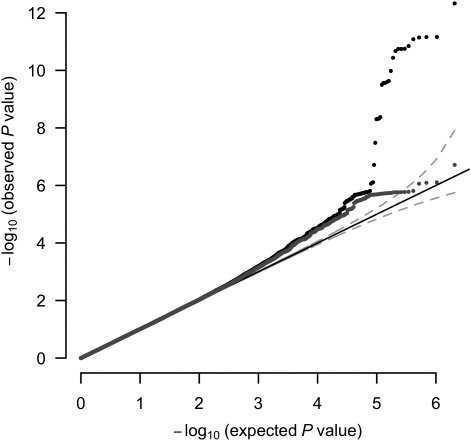
Four analyses showed little-to-no evidence for genetic heterogeneity among CLHNS subjects: principal component (PC) analysis of population substructure (Supplementary Material, Fig. S1), a minimal effect of adjusting for PCs (data not shown), the observed genomic control inflation factor (λGC) of 1.0 and a quantile–quantile plot of GWA P-values (Fig. 2).
Figure 2.
Quantile–quantile plot of SNP association with log-Hcy in CLHNS mothers. Black closed circles indicate all SNPs and dark-gray closed circles exclude all SNPs in 1 Mb windows centered on each previously reported SNP associated with log-Hcy (Table 2). A solid black line indicates the uniform expectation and the gray dashed lines indicate the 95% prediction interval.
Follow-up genotyping
The most significant variants in the CPS1 and LYG1-TXNDC9 regions were both imputed SNPs. To confirm association in the mothers and to test these variants for association with log-Hcy in the offspring (n = 1679; 54.8% male, Supplementary Material, Table S2), we directly genotyped rs7422339 and rs6722617. We also genotyped the MTHFR SNP rs1801133 in both the mothers and the offspring.
The test of association between log-Hcy and the directly genotyped CPS1 SNP rs7422339 remained significant in the CLHNS mothers (P = 5.3 × 10−12, Table 1). The estimated increase in log-Hcy for each additional A allele was 0.071 [standard error (SE) = 0.009], and the proportion of variability in the log-Hcy level explained by genotype was 3.0%. This SNP was not significantly associated with log-Hcy levels (P = 0.11) in the pooled CLHNS offspring sample (Table 1). However, evidence for association was present in the female-only subset (n = 759; P = 0.0087), with the direction of effect the same as observed in the mothers (increase of 0.040 in mean log-Hcy for each additional A allele). A formal test for a gender-by-genotype interaction in the offspring-only sample was not significant (P = 0.095), but the statistical power to detect such an interaction was only 54% for the effect size observed. The test for interaction between rs7422339 genotype and generation (mothers versus offspring) was significant (P = 0.0004). There was no significant evidence for an interaction between age and genotype in the mothers (P > 0.10).
Table 1.
Evidence of log-Hcy association with CPS1 rs7422339 and MTHFR C677T
| n | P-value for additive effecta | β-Coefficient (SE)a | R2 (%) | Genotypic least-square means for Hcy |
|||
|---|---|---|---|---|---|---|---|
| CPS1 rs7422339 | CC | CA | AA | ||||
| Mothers | 1746 | 5.3 × 10−12 | 0.071 (0.009) | 3.0 | 11.02 | 11.92 | 12.46 |
| Female offspring | 759 | 0.0087 | 0.040 (0.015) | 0.9 | 10.46 | 10.98 | 11.60 |
| Male offspring | 920 | 0.93 | 0.001 (0.017) | <0.1 | 14.36 | 14.50 | 13.78 |
| All offspring | 1679 | 0.11 | 0.021 (0.012) | 0.2 | 12.58 | 12.91 | 12.79 |
| Combined mothers + offspring | 3425 | 5.4 × 10−9 | 0.050 (0.008) | 1.1 | 11.80 | 12.43 | 12.79 |
| MTHFR C677T | CC | CT | TT | ||||
| Mothers | 1769 | 8.7×10−10 | 0.083 (0.015) | 1.6 | 11.25 | 11.94 | 22.17 |
| Female offspring | 753 | 3.6 × 10−10 | 0.136 (0.022) | 5.1 | 10.42 | 11.46 | 21.43 |
| Male offspring | 923 | 1.1 × 10−16 | 0.199 (0.023) | 7.3 | 13.81 | 16.38 | 35.11 |
| All offspring | 1676 | 1.9 × 10−26 | 0.171 (0.016) | 6.3 | 12.28 | 14.22 | 29.13 |
| Combined mothers + offspring | 3445 | 1.3 × 10−29 | 0.129 (0.012) | 3.3 | 11.80 | 12.44 | 12.86 |
Results of directly genotyped SNPs are shown.
aFor tests of association and effect estimation, natural log-Hcy was used. β-Coefficients and SEs are reported for an additive effect.
Evidence of log-Hcy association for the directly genotyped LYG1-TXNDC9 SNP rs6722617 (P = 1.2 × 10−7) was similar to the result based on its imputed genotype. The estimated increase for log-Hcy was 0.62 (SE = 0.23) for each additional C allele. No evidence for association between rs6722617 and log-Hcy was observed in the offspring-only sample (P = 0.58).
The directly genotyped MTHFR C677T SNP showed convincing evidence of association (P = 8.7 × 10−10) and explained 1.6% of the proportion of variability in log-Hcy in the CLHNS mothers (Table 1). The estimated increase in log-Hcy for each additional copy of the T allele was 0.083 (SE = 0.015). This SNP was even more strongly associated with log-Hcy levels in the gender-combined offspring sample (P = 1.9 × 10−26, Table 1). Each additional copy of the T allele was associated with an increase in log-Hcy of 0.171 (SE = 0.016), and the proportion of variability in the log-Hcy level explained by this locus was 5.1%. The gender-stratified results in the CLHNS offspring indicated that the MTHFR C677T association appeared slightly stronger in males (P = 1.1 × 10−16; β = 0.199; R2 = 7.3%) than females (P = 3.6 × 10−10; β = 0.136; R2 = 5.1%), and there was marginal evidence for a gender-by-genotype interaction (P = 0.053). The combined study of mothers and offspring for MTHFR C677T resulted in a smaller P-value (P = 1.3 × 10−29) than when analyses were restricted to the offspring alone. Evidence for an interaction between genotype and generation (P = 1.2 × 10−5) suggests that the increase in mean log-Hcy levels associated with each additional T allele is greater in the offspring cohort than in the mothers, although this result may be somewhat confounded by the marginal evidence for a gender interaction in the offspring. We did not observe evidence for an interaction between age and genotype in the mothers (P > 0.10). The MTHFR C677T genotypic means in the CLHNS mothers and offspring are most consistent with a recessive model for the T allele. The sample sizes for the 677T homozygotes (n = 7 for mothers and n = 16 for offspring), however, were too small to test this model.
Additional covariate adjustment for estimated folate, vitamin B6 and vitamin B12 intake levels in our linear models did not appreciably change any tests of genotype association or genetic effect size estimates for CPS1 rs7422339 or MTHFR C677T in either the CLHNS mother or offspring cohorts (data not shown). Furthermore, we found no evidence of interaction between MTHFR C677T genotype and dietary intake levels of folate (data not shown).
Replication of previously identified loci
To investigate whether this Filipino sample replicated evidence for loci reported (P < 1 × 10−6) in a GWA scan for Hcy in white American women (13), we defined positive evidence for replication as P < 0.10, with an effect size in the same direction as observed for the original study (i.e. one-sided test with α = 0.05). In addition to MTHFR and CPS1, which showed stronger estimated effects in the CLHNS, association signals were replicated with similar effect sizes in NOX4, CHMPIA-DPEP1 and CENPQ-MUT, but not AKAP13, nor a second signal near MTHFR (Table 2). For the reported CBS signal, neither the SNP nor a suitable proxy was available in the CLHNS samples.
Table 2.
Evidence of association in CLHNS mothers at previously reported GWA loci
| SNP | Nearby gene | Chr | Position | Allele | Identifying sample |
CLHNS mothers |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele frequency | βa ± SE | P-value | Allele frequency | βa ± SE | P-value | |||||
| rs7422339 | CPS1 | 2 | 211 248 752 | A | 0.31 | 0.027 ± 0.004 | 1.9 × 10−11 | 0.24 | 0.076 ± 0.011 | 4.7 × 10−13 |
| rs1801133b | MTHFR | 1 | 11 778 965 | T | 0.33 | 0.048 ± 0.004 | 8.1 × 10−35 | 0.22 | 0.083 ± 0.015 | 8.7 × 10−10 |
| rs11018628 | NOX4 | 11 | 88 846 159 | G | 0.07 | −0.050 ± 0.007 | 9.6 × 10−12 | 0.07 | −0.059 ± 0.015 | 9.8 × 10−5 |
| rs460879 | CHMP1A | 16 | 88 240 390 | A | 0.46 | 0.026 ± 0.004 | 3.8 × 10−12 | 0.53 | 0.024 ± 0.008 | 0.0039 |
| rs4267943 | CENPQ | 6 | 49 547 764 | A | 0.36 | 0.024 ± 0.004 | 2.0 × 10−9 | 0.26 | 0.018 ± 0.009 | 0.050 |
| rs10986018 | GPR51 | 9 | 98 202 891 | C | 0.22 | −0.06 ± 0.01 | 2.1 × 10−8 | <0.01 | −0.120 ± 0.092 | 0.19 |
| rs2061821 | AKAP13 | 15 | 83 923 658 | A | 0.38 | 0.019 ± 0.004 | 5.5 × 10−7 | 0.39 | 0.002 ± 0.008 | 0.79 |
| rs1999594 | MTHFR | 1 | 11 893 482 | G | 0.43 | 0.04 ± 0.01 | 6.2 × 10−10 | 0.30 | −0.002 ± 0.009 | 0.87 |
| rs6586282 | CBS | 21 | 43 351 566 | A | 0.18 | −0.030 ± 0.005 | 3.2 × 10−10 | — | — | — |
SNPs previously reported to be associated with Hcy in GWA studies are from reference (13) except rs10986018 and rs1999594, which are from reference (14). Chromosomal positions are Build 36. SNPs are arranged in ascending order according to CLHNS P-value.
aβ-Coefficient for additive effect with respect to the reference allele.
bSNP rs1801133 was poorly imputed, instead the directly genotyped results are reported here. The following SNPs were imputed (imputation quality score): rs7422339 (0.76), rs11018628 (>0.99), rs460879 (0.96), rs10986018 (0.65), rs2061821 (0.97), rs1999594 (0.83). SNP rs10986018 was not analyzed in the GWA analysis due to MAF (<0.01). CBS SNP rs6586282 was neither genotyped nor successfully imputed.
Multi-locus association results
We examined additive and interaction effects for SNPs at the CPS1 and MTHFR loci in the CLHNS mothers. The cumulative proportion of variability explained by rs7422339 and rs1801133 was 4.7% in the CLHNS mothers, and there was no evidence for an interaction between the two loci (P = 0.30). However in the offspring sample, the test of interaction between the loci was statistically significant (P = 0.0046). The cumulative proportion of variance of log-Hcy explained by both loci and their interaction in the offspring sample was 5.8%. We also estimated the cumulative proportion of log-Hcy variability explained by five loci reported previously (13) that had at least nominal evidence of significance in the CLHNS mothers (CPS1 rs7422339, MTHFR rs1801133, NOX4 rs11018628, CHMPIA-DPEP1 rs460879 and CENPQ-MUT rs4267943) to be 5.9%.
Association of CPS1 rs7422339 genotype with other CVD-related traits
To characterize the CPS1 gene with respect to associations with CVD-related phenotypes, we further evaluated other possible effects of the rs7422339 SNP and additional traits of total cholesterol, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), triglycerides, C-reactive protein (CRP), fasting glucose and body mass index (BMI) in the CLHNS mothers (Supplementary Material, Table S3). The A allele of rs7422339 was nominally associated with higher LDL-C (P = 0.0010) and higher total cholesterol (P = 0.0094). After adjustment for Hcy, the P-value for LDL-C was 0.021, suggesting that rs7422339 may have some effect on LDL-C independent of Hcy. No evidence for association was observed between rs7422339 and total cholesterol after adjustment for Hcy (P = 0.11).
Discussion
We observed strong evidence for association between CPS1 variation and log-Hcy level in this GWA study of 1786 Filipino mothers from the CLHNS cohort. The most significantly associated SNP (P = 4.7 × 10−13) encodes Thr1405Asn in CPS1 and explains ∼3% of the variability in log-Hcy levels in this cohort. This finding confirms a recent report by the Women's Genome Health Study (WGHS), which observed significant evidence for an association between Hcy levels and rs7422339 (P = 1.9 × 10−11) in a sample of 13 974 Caucasian women. The protein encoded by CPS1 catalyzes the first committed step of the hepatic urea cycle, and rs7422339 has been previously reported to be associated with nitric oxide level and vascular smooth muscle activity, necrotizing enterocolitis, pulmonary hypertension and, also in the WGHS, fibrinogen level (15–18). Consistent with our evidence of association in the female but not male CLHNS offspring, the WGHS did not observe replication evidence for CPS1 in samples from the Precocious Coronary Artery Disease Study (PROCARDIS), a subset of 840 post-myocardial infarction patients from several European countries, until they performed a gender-stratified analysis (male n = 658, P > 0.05; female n = 161, P = 0.0009). Although the P-value for gender-by-genotype interaction in the CLHNS offspring was not statistically significant (P = 0.095), together these results suggest that the effects of this gene may be stronger in women than in men.
The CLHNS mothers also showed evidence for association with the widely studied MTHFR C677T SNP (rs1801133), which explained an estimated 1.6% of the variation in log-Hcy levels. Follow-up analysis of the CLHNS young adult offspring (age 20–22 years) suggests that MTHFR C677T has a much stronger effect in this younger generation, with an estimated proportion of variance explained by genotype of 6.3%, which is very large for a single SNP for a complex phenotype. We also observed marginal evidence (P = 0.053) for a gender-by-genotype interaction in the offspring, suggesting that, in contrast to CPS1, there may be a stronger effect of this SNP in males than in females. These findings are consistent with several reports that have suggested that the effect of MTHFR C677T is stronger in males than in females (10,19–21) and that the importance of MTHFR C677T genotype decreases with increasing age (22), although no evidence for gender differences was reported in the PROCARDIS replication sample (13). Hcy levels have been shown to be consistently higher in males than in females after childhood (23,24), and estrogen lowers plasma Hcy (25). In our study, CHLNS male offspring had significantly higher mean Hcy levels (P < 0.0001) than female offspring despite higher mean dietary intake of folate, vitamin B6 and vitamin B12 (Supplementary Material, Table S2), suggesting that any increase in the effect of MTHFR C677T genotype in CLHNS male offspring compared with female offspring is not due to lower folate intake.
Evidence for differences in genotype effects between the CLHNS mothers and offspring was also detected at CPS1. In contrast to MTHFR, the observed effect of CPS1 was stronger in the mothers than in the offspring (Table 1). The difference in effects for CPS1 could be due to bias based on the identification of this locus in the mothers and only following up the most significant results in the offspring, although this bias is mitigated by the report of the CPS1 association in the WGHS. The differences in genetic effect strength between CLHNS mothers and their offspring could also be attributed to age or other factors. We observed no evidence for a CPS1–age interaction in the mothers, where there is an age range of 34 years (data not shown). Nonetheless, there remains the possibility that the difference may not be a linear effect with age, but rather a consequence of different life stages or environmental exposures between younger and older adults.
Effect sizes appear substantially different between CLHNS and WGHS women at both the CPS1 and MTHFR loci. Overall mean levels of Hcy were the same (CLHNS mean = 11.4, WGHS mean = 11.4), but CLHNS mothers’ Hcy levels had less variability [CLHNS standard deviation (SD) = 3.0, WGHS SD = 4.7]. CPS1 SNP rs7422339 was estimated to explain a greater proportion of the variability in log-Hcy levels in the CLHNS mothers (R2 = 3.0%) than in the WGHS samples (R2 = 0.3%) (13) despite a lower frequency (0.24 versus 0.31) of the minor allele in CLHNS mothers. Similarly, MTHFR C677T explained a greater proportion of the variability in log-Hcy levels in CLHNS mothers than in WGHS women (CLHNS R2 = 1.6%; WGHS R2 = 0.95%) despite a lower estimated frequency of the minor allele (0.22 versus 0.33). At both loci, a significantly higher estimated increase of log-Hcy levels per each additional copy of the minor allele was observed in CLHNS mothers compared with WGHS women (see Table 2 for effect size estimates and their associated SEs for both studies). The estimated proportions of variability explained by CPS1 SNP rs7422339 (R2 = 0.9%) and MTHFR C677T (R2 = 5.1%) in CLHNS female offspring were also higher than in WGHS women, although the estimated increase in log-Hcy levels per minor allele for rs7422339 in CLHNS female offspring was less than the estimated increase observed in WGHS women. The higher estimated proportion of variation explained by rs7422339 in CLHNS female offspring can be attributed to the lower overall variability of Hcy levels in CLHNS female offspring (SD = 3.0) compared with WGHS.
Because environmental factors, such as dietary intake of folate, vitamin B6 and vitamin B12, and indicators of estrogen levels in women are predictive of the Hcy level, it is possible that differences across populations could play a role in how genes influence Hcy. Both CLHNS and WGHS Hcy measurements were made in environments in which folate and vitamin B12 fortification was not widely available (WGHS measurements were made prior to 1998, when fortification of refined wheat products became mainstream in the USA). Adjustment for folate, vitamin B6 and vitamin B12 did not appreciably change genetic association test results in either CLHNS sample. We also found no evidence for any interactions between either CPS1 rs7422339 or MTHFR C677T genotype and folate levels to suggest their genotypic effects are stronger in those with lower reported intake levels of folate. Other important differences to consider when comparing results from CLHNS mothers with other studies include substantially lower use of hormone replacement therapy (<1%), which was widely reported in the WGHS (44.9%), and low oral contraceptive use in CLHNS (<2%).
Three of the four other loci identified in the WGHS for which SNP genotypes were available in the CLHNS also showed evidence of association (P < 0.05, Table 2). Evidence for the NOX4 locus (rs11018628) was relatively strong in the CLHNS mothers (P = 9.8 × 10−5), and we observed modest evidence for rs460879 (P = 0.0039) at CHMP1A-DPEP1 and rs4267943 (P = 0.050) at CENPQ-MUT, but not for rs2061821 at AKAP13 (P = 0.79). The direction of estimated effects in the CLHNS mothers was consistent with those observed in the WGHS for the NOX4, CHMP1A-DPEP1 and CENPQ-MUT SNPs. We did not observe evidence for the replication of rs10986018 at GPR51 reported in a meta-analysis by Hazra et al. (14). The GPR51 C allele frequency, however, was much smaller in the CLHNS (<0.01) than in the European-ancestry based sample examined in that study.
Finally, we did not replicate an observed association for rs1999594 near MTHFR (13,14), for which a joint analysis including MTHFR SNPs rs1801133 and rs1999594 showed that both SNPs were significant (14). The frequency of the rs1999594 minor allele was lower in CLHNS mothers compared with the previous reports; however, there was no evidence of a main effect for this locus (β = −0.002) in CLHNS mothers. The different results for rs1999594 in CLHNS may be influenced by weaker linkage disequilibrium (LD) between rs1801133 and rs1999594 (HapMap CHB + JPT R2 = 0.00, D′ = 0.23; HapMap CEU R2 = 0.26, D′ = 0.91).
This study represents the first GWA results for Hcy in an Asian population. Our findings suggest that at least some of the main genetic factors influencing plasma Hcy levels are likely shared across populations of European and Asian descent, despite different genetic backgrounds and environmental and dietary exposures. These results also suggest that the effects of genotype on plasma Hcy levels may differ across gender and life stages and should motivate future studies to take these factors into consideration.
MATERIALS AND METHODS
Study population and phenotypes
Data and samples come from the CLHNS, a community-based birth cohort study that originally enrolled 3327 pregnant women in 1983–84 (3080 singleton live births), and has since followed them and their offspring to the present (26). In 2005, 1895 healthy Filipino mothers and 1775 of their offspring remained in the study and had DNA and measurement of biomarkers available.
Trained field staff conducted in-home interviews and collected anthropometric measurements and comprehensive environmental data at each visit (data available online at http://www.cpc.unc.edu/projects/cebu/). Blood samples, which were used for biomarker measurement and DNA extraction, were obtained in 2005. Plasma Hcy was measured using the automated Bayer® ADVIA Centaur chemiluminescent Hcy assay, as described (27).
Dietary intake data were collected via 24 h recall by trained study personnel during the 2005 visit. Intake was measured for 2 days, and the mean was used in the analysis. Nutrient intakes of folate, vitamin B6 and vitamin B12 were estimated using the Philippines Food Composition Tables (28,29).
Sample characteristics for the CLHNS mothers and offspring by gender are presented in Supplementary Material, Tables S1 and S2. Informed consent was obtained from all CLHNS subjects, and the study protocol was approved by the University of North Carolina Institutional Review Board for the Protection of Human Subjects.
GWA SNP genotyping
SNP genotyping was performed with the Affymetrix Genome-Wide Human SNP Array 5.0 at the Vanderbilt Microarray Shared Resource at Vanderbilt University Medical Center, Nashville, TN, USA, using the standard protocol recommended by the manufacturer. Genotype calling was performed using Birdseed (version 2). Genotyping was attempted on the 1895 unique CLHNS mothers samples, 40 CLHNS duplicates and 5 HapMap CEPH trios. Ten CLHNS samples failed genotyping, and four CLHNS samples were removed after genotyping for a call rate <97%. The final call rate across the 1881 remaining samples was 99.6%.
We applied SNP quality control checks in PLINK v1.02 on the 1881 CLHNS samples that were successfully genotyped (30). Of the initial 424 670 SNPs, we discarded 13 287 SNPs due to poor mapping, call rates <90% and/or deviation from Hardy–Weinberg equilibrium (P < 10−6). An additional 164 SNPs were dropped due to three or more discrepancies between genotypes in 40 duplicate pairs and/or Mendelian inheritance errors in five CEPH trios. Another 3113 SNPs were dropped due to three or more genotype discrepancies with HapMap genotypes.
Additional SNP genotyping
Follow-up genotyping was performed for three SNPs (rs7422339, rs1801133 and rs6722617) in the CLHNS mothers and offspring using TaqMan allelic discrimination (Applied Biosystems, Foster City, CA, USA). The SNPs rs7422339 and rs6722617 were genotyped to confirm results for the genetic regions with the strongest evidence for association with Hcy at which the most significant SNP in the region was imputed (see below), and to test for evidence of association in the CLHNS offspring. MTHFR C677T (rs1801133) was genotyped because it had a low imputation quality score (R2 < 0.3, where R is the estimated correlation between the imputed genotype calls and the true unknown genotype for the SNP) in the CLHNS mothers and was an important a priori candidate SNP. The genotype success rate for all SNPs was >98% and no discrepancies were observed among duplicate samples (discrepancy rate <0.3%).
Population relatedness
We used identity-by-descent and identity-by-state estimates calculated in PLINK in combination with our prior knowledge of relationships between CLHNS participants to eliminate 81 estimated first-degree relatives. Our final sample for GWA analysis consisted of 1786 CLHNS women with available genotypes, Hcy levels and covariates from the 2005 survey.
SNP imputation
Using a hidden Markov model algorithm implemented in MACH software version 1.0 (available online at http://www.sph.umich.edu/csg/abecasis/mach/), genotype imputation was conducted using 352 264 directly genotyped SNPs that were polymorphic in both the 60 CEU founders and the 89 combined CHB + JPT HapMap samples (31). We pooled haplotypes from phased chromosomes in these two HapMap populations to better capture the LD structure in our CLHNS samples. We have shown previously that the CHB HapMap population is a reasonable proxy to the CLHNS cohort (32). For each subject, the genotype for each SNP was reported as a dosage value (a continuous number between 0 and 2) that reflects the expected number of copies of an arbitrary allele at that SNP conditional on the directly observed genotypes in both the subject and a representative sample of 200 unrelated CLHNS subjects and the phased haplotype assignments in the CEU and CHB + JPT HapMap samples. Imputation yielded genotype data for 1 878 188 new SNPs, and the 352 264 directly genotyped SNPs were also assigned imputed genotypes. We used R2 ≥ 0.3 as a quality control threshold to discard 150 177 low-quality imputed SNPs, and also excluded 30 351 SNPs with estimated minor allele frequencies (MAF) ≤ 0.01. Finally, discrete dosage values of 0, 1 or 2 were assigned for 23 750 directly genotyped SNPs non-polymorphic in either HapMap sample (and therefore not imputed) but with MAF > 0.01 in the CLHNS samples. In total, 2 073 674 SNPs were tested for association with the Hcy level.
Accounting for population substructure
We constructed PCs using the software EIGENSOFT to capture population substructure among our CHLNS subjects (33). We identified a set of 13 972 independent SNPs (estimated R2 < 0.005 between all pairs of SNPs within 1 Mb) with observed MAF > 0.05. The PCs were constructed on the basis of these 13 972 SNPs and 1571 CHLNS subjects with estimated genome-wide pair-wise identity-by-descent <0.1. The PCs for the remaining 228 subjects were subsequently calculated using the parameter coefficients obtained from the first 1571 subjects. The corresponding eigenvalues for the individual PCs were plotted (data not shown) and ‘the elbow’ of the eigenvalue plot was used to define which PCs to be included as covariates in association analyses. A plot of the first versus the second PC is shown in Supplementary Material, Figure S1. In addition, we tested for the association between each of the first 10 PCs and the Hcy level to confirm that any important ancestry explanatory PC was included. The first three PCs were included as covariates in the linear regression models. Finally, we examined the calculated genomic control value to evaluate for systematic P-value inflation due to cryptic relatedness between samples not accounted for by the analytic adjustment for PCs in the regression analyses (34).
GWA association analysis
Hcy level was natural log-transformed (log-Hcy) to satisfy the model assumption of normally distributed residuals after adjustment for the covariates of age, number of previous pregnancies (categorized as 0–4, 5–10, ≥11), menopausal status and the first three PCs from the population substructure analysis (Supplementary Material, Table S4). Array Studio software version 3.1 was used to perform the initial GWA analyses (Array Studio and all other Omicsoft products or service names are registered trademarks or trademarks of Omicsoft Corporation, Research Triangle Park, NC, USA). Multiple linear regression was used to test for the association between log-Hcy and each SNP, with adjustment for covariates. Genotype was modeled as an additive effect, with the genotype dosage values used as the primary predictor of interest. A 1 degree-of-freedom likelihood ratio test was used to assess statistical significance. A significance threshold of P < 5 × 10−8 was used to define a single result as genome-wide significant. Quantile–quantile plots of observed versus expected –log10(P-values) were constructed to help assess any cumulative inflation or deflation of statistical significance estimates compared with expectation under the null hypothesis. To compare effect sizes with one study that did not report SEs (13), SE estimates for the additive genetic effect β-coefficients were derived from the reported estimated β-coefficients and their corresponding Wald test P-values.
Follow-up association analysis on CLHNS mothers and offspring
Linear regression models were analyzed in the software package SAS version 9.3 (SAS Inc., Cary, NC, USA) to test the genotyped SNPs rs7422339, rs6722617 and rs1801133 for association with log-Hcy separately in the mother and the offspring samples. In the offspring sample, we adjusted only for gender, because the ages of all offspring were within 2 years of each other, menopause status was not relevant and the number of previous pregnancies and PCs for population stratification were not available. Genotype was modeled as an additive effect. Partial correlations were calculated for each locus to estimate the proportion of variation explained by genotype. In the offspring sample, we further performed analyses stratified by gender and tests of gender-by-genotype interaction to assess whether results were similar for males and females. We also tested for evidence of a genotype-by-genotype interaction effect between CPS1 rs7422339 and MTHFR C677T on the Hcy level.
We used general linear mixed models to evaluate genotype effects on the combined sample of mothers and offspring while accounting for the correlation of Hcy levels between mother–child pairs due to shared environmental and genetic exposures. Within this setting, we also conducted tests of interaction to assess whether the effects for each locus were similar in mothers and offspring. A significance threshold of P < 0.05 was used for tests of interaction. We repeated the mother–offspring-stratified and -combined analyses with additional adjustment for estimated dietary folate, vitamin B6 and vitamin B12 intake to assess whether adjustment for these factors appreciably changed genetic association results. Finally, we tested for interactions between genotype and folate levels to assess whether there were stronger genetic effects in subjects with lower reported folate levels.
Conditional analysis for CPS1 SNPs
To evaluate whether any of the other 21 CPS1 SNPs with P < 5 × 10−8 were independently associated with log-Hcy levels in the CLHNS mothers after accounting for the strongest associated SNP (rs7422339), we performed association analyses for the CPS1 SNPs conditioning on rs7422339 genotype. Using SAS, we examined tests of additive genotype effects for each of the 21 SNPs in individual linear regression models with the same covariates as above plus the rs7422339 genotype.
Association of CPS1 SNP rs7422339 with additional traits
We tested the CPS1 SNP rs7422339 for association with several additional biomarkers associated with CVD: total cholesterol, HDL-C, LDL-C, triglycerides, CRP, fasting glucose and BMI. Outcome traits were first natural log-transformed and adjusted for the same covariates as for the Hcy analysis. Any associations that were found to be nominally significant (P < 0.05) were further tested adding the Hcy level to the model to evaluate whether the association was independent of the phenotype's correlation with Hcy.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health grants (DK078150, TW05596, HL085144), pilot funds (RR20649, ES10126 and DK56350) and training grants (T32 GM007092 to D.C.C.-C., T32 HL69768 to A.F.M.). K.L.M. is a Pew Scholar in the Biological Sciences.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Office of Population Studies Foundation research and data collection teams and Melissa Daniels for nutrient intake estimates.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Wierzbicki A.S. Homocysteine and cardiovascular disease: a review of the evidence. Diab. Vasc. Dis. Res. 2007;4:143–150. doi: 10.3132/dvdr.2007.033. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell S.R. Coronary artery disease – free radical damage, antioxidant protection and the role of homocysteine. Basic Res. Cardiol. 2000;95(Suppl. 1):I65–I71. doi: 10.1007/s003950070012. [DOI] [PubMed] [Google Scholar]
- 3.Bots M.L., Launer L.J., Lindemans J., Hofman A., Grobbee D.E. Homocysteine, atherosclerosis and prevalent cardiovascular disease in the elderly: the Rotterdam Study. J. Intern. Med. 1997;242:339–347. doi: 10.1046/j.1365-2796.1997.00239.x. [DOI] [PubMed] [Google Scholar]
- 4.Fortin L.J., Genest J., Jr Measurement of homocyst(e)ine in the prediction of arteriosclerosis. Clin. Biochem. 1995;28:155–162. doi: 10.1016/0009-9120(94)00073-5. [DOI] [PubMed] [Google Scholar]
- 5.Nygard O., Vollset S.E., Refsum H., Stensvold I., Tverdal A., Nordrehaug J.E., Ueland M., Kvale G. Total plasma homocysteine and cardiovascular risk profile. The Hordaland Homocysteine Study. JAMA. 1995;274:1526–1533. doi: 10.1001/jama.1995.03530190040032. [DOI] [PubMed] [Google Scholar]
- 6.Perry I.J., Refsum H., Morris R.W., Ebrahim S.B., Ueland P.M., Shaper A.G. Prospective study of serum total homocysteine concentration and risk of stroke in middle-aged British men. Lancet. 1995;346:1395–1398. doi: 10.1016/s0140-6736(95)92407-8. [DOI] [PubMed] [Google Scholar]
- 7.Christen W.G., Ajani U.A., Glynn R.J., Hennekens C.H. Blood levels of homocysteine and increased risks of cardiovascular disease: causal or casual? Arch. Intern. Med. 2000;160:422–434. doi: 10.1001/archinte.160.4.422. [DOI] [PubMed] [Google Scholar]
- 8.Wald D.S., Law M., Morris J.K. The dose–response relation between serum homocysteine and cardiovascular disease: implications for treatment and screening. Eur. J. Cardiovasc. Prev. Rehabil. 2004;11:250–253. doi: 10.1097/01.hjr.0000129742.15346.ab. [DOI] [PubMed] [Google Scholar]
- 9.Siva A., De Lange M., Clayton D., Monteith S., Spector T., Brown M.J. The heritability of plasma homocysteine, and the influence of genetic variation in the homocysteine methylation pathway. Q. J. Med. 2007;100:495–499. doi: 10.1093/qjmed/hcm054. [DOI] [PubMed] [Google Scholar]
- 10.Jee S.H., Song K.S., Shim W.H., Kim H.K., Suh I., Park J.Y., Won S.Y., Beaty T.H. Major gene evidence after MTHFR-segregation analysis of serum homocysteine in families of patients undergoing coronary arteriography. Hum. Genet. 2002;111:128–135. doi: 10.1007/s00439-002-0757-8. [DOI] [PubMed] [Google Scholar]
- 11.Kullo I.J., Ding K., Boerwinkle E., Turner S.T., Mosley T.H., Jr, Kardia S.L., de Andrade M. Novel genomic loci influencing plasma homocysteine levels. Stroke. 2006;37:1703–1709. doi: 10.1161/01.STR.0000225929.96190.b3. [DOI] [PubMed] [Google Scholar]
- 12.Wilcken D.E., Wang X.L., Wilcken B. Methylenetetrahydrofolate reductase (MTHFR) mutation, homocyst(e)ine, and coronary artery disease. Circulation. 1997;96:2738–2740. [PubMed] [Google Scholar]
- 13.Paré G., Chasman D.I., Parker A.N., Robert R.Y., Zee R.R.Y., Mälarstig A., Seedorf U., Collins R., Watkins H., Hamsten A., Miletich J.P., Ridker P.M. Novel associations of CPS1, MUT, NOX4 and DPEP1 with plasma homocysteine in a healthy population: a genome-wide evaluation of 13,974 participants in the Women's Genome Health Study. Circ. Cardiovasc. Genet. 2009;2:142–150. doi: 10.1161/CIRCGENETICS.108.829804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazra A., Kraft P., Lazarus R., Chen C., Chanock S.J., Jacques P., Selhub J., Hunter D.J. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum. Mol. Genet. 2009;18:4677–4687. doi: 10.1093/hmg/ddp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summar M.L., Hall L., Christman B., Barr F., Smith H., Kallianpur A., Brown N., Yadav M., Willis A., Eeds A., et al. Environmentally determined genetic expression: clinical correlates with molecular variants of carbamyl phosphate synthetase I. Mol. Genet. Metab. 2004;81(Suppl. 1):S12–S19. doi: 10.1016/j.ymgme.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Moonen R.M., Paulussen A.D., Souren N.Y., Kessels A.G., Rubio-Gozalbo M.E., Villamor E. Carbamoyl phosphate synthetase polymorphisms as a risk factor for necrotizing enterocolitis. Pediatr. Res. 2007;62:188–190. doi: 10.1203/PDR.0b013e3180a0324e. [DOI] [PubMed] [Google Scholar]
- 17.Canter J.A., Summar M.L., Smith H.B., Rice G.D., Hall L.D., Ritchie M.D., Motsinger A.A., Christian K.G., Drinkwater D.C., Jr, Scholl F.G., et al. Genetic variation in the mitochondrial enzyme carbamyl-phosphate synthetase I predisposes children to increased pulmonary artery pressure following surgical repair of congenital heart defects: a validated genetic association study. Mitochondrion. 2007;7:204–210. doi: 10.1016/j.mito.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danik J.S., Pare G., Chasman D.I., Zee R.Y.L., Kwiatkowski D.J., Parker A., Miletich J.P., Ridker P.M. Novel loci, including those related to Crohn disease, psoriasis, and inflammation, identified in a genome-wide association of fibrinogen in 17,686 women. Circ. Cardiovasc. Genet. 2009;2:134–141. doi: 10.1161/CIRCGENETICS.108.825273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chango A., Potier De Courcy G., Boisson F., Guilland J.C., Barbe F., Perrin M.O., Christides J.P., Rabhi K., Pfister M., Galan P., et al. 5,10-methylenetetrahydrofolate reductase common mutations, folate status and plasma homocysteine in healthy French adults of the Supplementation en Vitamines et Mineraux Antioxydants (SU.VI.MAX) cohort. Br. J. Nutr. 2000;84:891–896. [PubMed] [Google Scholar]
- 20.Dekou V., Whincup P., Papacosta O., Ebrahim S., Lennon L., Ueland P.M., Refsum H., Humphries S.E., Gudnason V. The effect of the C677T and A1298C polymorphisms in the methylenetetrahydrofolate reductase gene on homocysteine levels in elderly men and women from the British regional heart study. Atherosclerosis. 2001;154:659–666. doi: 10.1016/s0021-9150(00)00522-0. [DOI] [PubMed] [Google Scholar]
- 21.Papoutsakis C., Yiannakouris N., Manios Y., Papaconstantinou E., Magkos F., Schulpis K.H., Zampelas A., Matalas A.L. The effect of MTHFR(C677T) genotype on plasma homocysteine concentrations in healthy children is influenced by gender. Eur. J. Clin. Nutr. 2006;60:155–162. doi: 10.1038/sj.ejcn.1602280. [DOI] [PubMed] [Google Scholar]
- 22.Husemoen L.L., Thomsen T.F., Fenger M., Jorgensen H.L., Jorgensen T. Contribution of thermolabile methylenetetrahydrofolate reductase variant to total plasma homocysteine levels in healthy men and women. Inter99 (2) Genet. Epidemiol. 2003;24:322–330. doi: 10.1002/gepi.10239. [DOI] [PubMed] [Google Scholar]
- 23.Selhub J., Jacques P.F., Rosenberg I.H., Rogers G., Bowman B.A., Gunter E.W., Wright J.D., Johnson C.L. Serum total homocysteine concentrations in the third National Health and Nutrition Examination Survey (1991–1994): population reference ranges and contribution of vitamin status to high serum concentrations. Ann. Intern. Med. 1999;131:331–339. doi: 10.7326/0003-4819-131-5-199909070-00003. [DOI] [PubMed] [Google Scholar]
- 24.Bjorke Monsen A.L., Ueland P.M. Homocysteine and methylmalonic acid in diagnosis and risk assessment from infancy to adolescence. Am. J. Clin. Nutr. 2003;78:7–21. doi: 10.1093/ajcn/78.1.7. [DOI] [PubMed] [Google Scholar]
- 25.Morris M.S., Jacques P.F., Selhub J., Rosenberg I.H. Total homocysteine and estrogen status indicators in the Third National Health and Nutrition Examination Survey. Am. J. Epidemiol. 2000;152:140–148. doi: 10.1093/aje/152.2.140. [DOI] [PubMed] [Google Scholar]
- 26.Adair L.S., Kuzawa C.W., Borja J. Maternal energy stores and diet composition during pregnancy program adolescent blood pressure. Circulation. 2001;104:1034–1039. doi: 10.1161/hc3401.095037. [DOI] [PubMed] [Google Scholar]
- 27.Tewari P.C., Zhang B., Bluestein B.I. Analytical and clinical evaluation of the Bayer ADVIA Centaur homocysteine assay. Clin. Chim. Acta. 2004;342:171–178. doi: 10.1016/j.cccn.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Daniels M.C. Dietary Diversity as a Measure of the Micronutrient Adequacy Results from Metropolitan Cebu, Philippines Site. Washington, DC: Food and Nutrition Technical Assistance II Project, Academy for Educational Development; 2009. [Google Scholar]
- 29.Food and Nutrition Research Institute. Food Composition Table and Menu Eval. Manila, Philippines: Department of Science and Technology; 2000. [Google Scholar]
- 30.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marvelle A.F., Lange L.A., Qin L., Wang Y., Lange E.M., Adair L.S., Mohlke K.L. Comparison of ENCODE region SNPs between Cebu Filipino and Asian HapMap samples. J. Hum. Genet. 2007;52:729–737. doi: 10.1007/s10038-007-0175-9. [DOI] [PubMed] [Google Scholar]
- 33.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.