Abstract
Huntington's disease (HD) is an inherited autosomal dominant neurodegenerative disorder caused by an expansion of a CAG trinucleotide repeat in the huntingtin (HTT) gene [Huntington's Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell, 72, 971–983]. Despite identification of the gene in 1993, the underlying life-long disease process and effective treatments to prevent or delay it remain elusive. In an effort to fast-track treatment strategies for HD into clinical trials, we have developed a new large-animal HD transgenic ovine model. Sheep, Ovis aries L., were selected because the developmental pattern of the ovine basal ganglia and cortex (the regions primarily affected in HD) is similar to the analogous regions of the human brain. Microinjection of a full-length human HTT cDNA containing 73 polyglutamine repeats under the control of the human promotor resulted in six transgenic founders varying in copy number of the transgene. Analysis of offspring (at 1 and 7 months of age) from one of the founders showed robust expression of the full-length human HTT protein in both CNS and non-CNS tissue. Further, preliminary immunohistochemical analysis demonstrated the organization of the caudate nucleus and putamen and revealed decreased expression of medium size spiny neuron marker DARPP-32 at 7 months of age. It is anticipated that this novel transgenic animal will represent a practical model for drug/clinical trials and surgical interventions especially aimed at delaying or preventing HD initiation. New sequence accession number for ovine HTT mRNA: FJ457100.
INTRODUCTION
Huntington's disease (HD) is an inherited autosomal dominant neurological disorder of variable clinical onset but characterized by motor dysfunction, cognitive decline and in some cases severe emotional disturbance. The mutation underlying HD occurs in the huntingtin (HTT) gene and is an expanded CAG repeat in exon 1, resulting in a lengthened polyglutamine tract in the HTT protein (1). The elongated glutamine tract instigates a life-long disease process that results in neuronal degeneration of the striatum and cortex, two pathological hallmarks of the disease. At present there is no treatment or therapy to slow, let alone arrest the course of the disease. Several animal models have been generated to better understand HD and to test potential therapeutics, with the transgenic HTT exon 1 fragment R6/2 mouse line being the most widely studied model (2). However, these rodent models have a restricted life-span (<36 months) and their brains are not anatomically similar to the human brain, both of which are experimental and therapeutic barriers to studying the natural history and evaluating pre-symptomatic therapies for a chronic disease process such as HD. Therefore, our group sought to develop a large HD mammalian model.
The sheep, Ovis aries L., is an ideal alternate mammalian model for studying HD. Sheep are relatively easy to care for as they are docile and can be kept in their normal grazing environment. Importantly, they live for more than a decade, allowing for the study of the chronic effects of a full-length HTT expressing transgene. Moreover, sheep are expected to be a good model for pharmacological testing because their brain has a prosencephalon that is comparable to the human brain, and for this reason they constitute a good surgical model for testing possible cell replacement approaches, or the delivery of gene therapy agents, making feasible investigations that cannot be carried out in humans. Although not in common use, there is a considerable body of knowledge that supports the use of sheep as a model system. Currently, sheep are used for studying fetal and neonatal development, heart pathology, bone repair and to evaluate surgical procedures including repair of aneurysms, heart valves and lymphatic cannulation [reviewed in Scheerlinck et al. (3)]. There are also sheep with neurological disorders (e.g. rye grass stagger caused by neurotoxins in feed), as well as naturally arising defects in sheep with human correlates such as spina bifida lambs (4) and sheep with ceroid-lipofuscinosis (5), the latter being a well studied model for the human neurodegenerative disorder, Batten's disease. Transgenic technologies have been refined in the sheep and transgenic sheep are routinely used for the production of recombinant therapeutic proteins via their milk, and for testing wool growth and other characteristics (6,7).
Another advantage presented by a large-animal model is that in vivo brain-imaging techniques such as PET or MRI can be used for determining anatomo-pathological changes in relation to symptoms, thus giving valuable insight into the mechanisms underlying the progression of pathological processes. Furthermore, agricultural genetics is well developed in New Zealand and Australia, with easily accessible genetically defined animals. The advent of low cost sequencing has led to the largely complete sequencing of farm animal genomes and has enabled the availability of genomic tools such as microarrays. This brings into reach the more wide-spread use of large-animal models of human disease and will greatly facilitate the analysis of our ovine HD model.
RESULTS
The ovine HTT homologue was isolated using degenerate RT–PCR, enabling differentiation of the human transgene from the ovine wild-type cDNA. The full-length ovine transcript contained an open reading frame of 9381 nucleotides, corresponding to a predicted 3127 amino acid protein (GenBank accession number FJ457100). Alignment of the predicted amino acid sequence with the human sequence revealed 88% amino acid identity. The CAG/glutamine coding repeat tract in exon one of the ovine HTT gene was polymorphic within and between ovine breeds, ranging from 10 to 17 CAG repeats, overlapping with the normal CAG repeat (6–35) ranges found in humans (1).
HD transgenic sheep were generated using an 11.6 kb NruI–NotI transgene fragment containing the full-length human HTT cDNA ligated to 1.1 kb of the human HTT genomic DNA immediately upstream of exon 1, containing promotor elements. The 3′ end of the human cDNA transgene was flanked by a portion of the bovine growth hormone (BGH) UTR containing exon 4, intron 4 and exon 5. The introduction of an intron/exon structure into the transgene was an attempt to enhance transgene expression (8,9). Given that disease severity is inversely correlated with CAG repeat length (1), a pure CAG repeat tract of about 69 units, which with penultimate CAACAGCAACAG codons would encode a polyglutamine tract of 73 residues, was chosen and inserted into the appropriate position of the full-length human HTT cDNA. The ∼69 CAG repeat tract is found in human HD patients and may be of sufficient length to yield evident disease phenotypes within the ovine life-span, as a HTT polyglutamine tract of 73 units would be associated with juvenile onset of overt disease symptoms in humans.
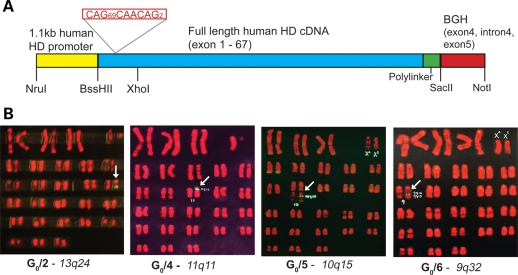
Instability of the expanded CAG polyglutamine coding repeat, cloned downstream of the HTT promotor, was encountered when propagating the transgene in Escherichia coli. Therefore, we utilized an alternative strategy, in which the promotor and full-length HTT cDNA fragments (containing the expanded repeat tract) were amplified in separate plasmids, sequence verified, digested and ligated together (Fig. 1A). The resultant 11.6 kb ligation fragment was purified and used for microinjection.
Figure 1.
HTT transgene and chromosomal location in HD founder transgenic animals (A) Schematic of the full-length human HTT cDNA ligation fragment used for transgenesis. The full-length cDNA includes exons 1–67 of the HTT gene ligated to a 5′ 1.1 kb fragment of the human HTT promotor region. The 3′-UTR is a genomic fragment of the bovine growth hormone (BGH) gene. (B) HD transgenic founder karyograms and predicted chromosome insertion sites. Location of the transgene is marked by yellow–green fluorescence. The chromosomal insertion site for G0/1 was observed to be only at 1q43 without the necessity of making a karyogram (data not shown). G0/3 is a very low-grade mosaic with no signals observed above background in any cells (data not shown). The insertion sites for the transgenes in karyograms of the other four animals are shown above.
Transgene DNA (ranging in concentration from 5 to 10 ng/µl) was microinjected into pronuclei of single-celled zygotes. Approximately 413 injected single-celled zygotes went on to cleave and form apparently healthy blastocysts and these were transferred to recipient Australian Merino ewes (∼3 embryos/ewe). There was no observable natural abortion rate during pregnancy. Of the 150 live births, 16 died shortly after birth (up to 48 h), an additional seven lambs were dead at birth, but none of these lambs were transgenic. Of the remaining 127 lambs, six lambs were transgenic as determined by PCR of tail biopsies, four of which were rams and two were ewes (data not shown). This was comparable to previously published sheep transgenesis rates (6,10). These founder (G0) animals were designated G0/1 (Ram), G0/2 (Ram), G0/3 (Ram), G0/4 (Ram), G0/5 (Ewe) and G0/6 (Ewe), with the numbers 1–6 identifying each individual familial line with respect to the original six founders.
Integration of the transgene was analysed by fluorescent in situ hybridization (FISH) on metaphase spreads (2n = 54 chromosomes) which revealed a unique single transgene integration site in five of the six animals (Fig. 1B). For G0/1, G0/2 and G0/5 the transgenes appeared to be located, respectively, in the dark R-bands 1q43 (not shown), 13q24 and 10q15. Transgene localization in animal G0/6 to a narrow dark R-band was rated more likely than localization to the adjacent faint R-band. Localization of the transgene in G0/4 was to the dark R-band 11q11 representing the procentric, non-coding satellite DNAs of Chromosome 11. The fractions of cells observed to have target-labelling were G0/1–100%, G0/2–61%, G0/4–89%, G0/5–59% and G0/6–72%, indicating that these founder-sheep were high-grade mosaics with G0/1 possibly integrating into the zygote. By comparison, it was not possible to determine the site of integration in G0/3 because the ram is a very low-grade mosaic with no target signals detected above background in any cells at metaphase on the slide.
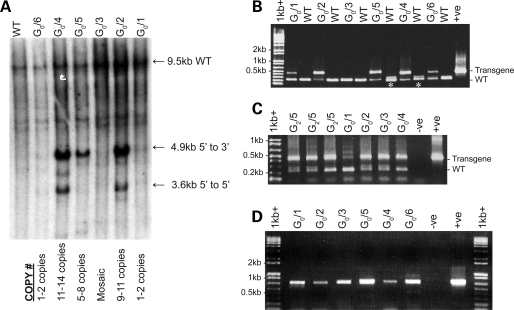
Southern blot analysis indicated the existence of multi-copy transgene loci, with the insertion of an estimated 2 (G0/1 and G0/6) through to 14 (G0/2, G0/4 and G0/5) copies of the transgene (Fig. 2A). The transgene arrays were mainly concatamers oriented in the 5′–3′ direction (head to tail as determined by Southern blot and PCR), with the higher copy number animals (G0/2, G0/4 and G0/5) also containing arrays oriented 5′–5′ and 3′–3′ (head to head and tail to tail). Copy numbers were also assessed using genomic real-time PCR assuming a single integration site of the transgene relative to the two endogenous ovine HTT alleles. The results were consistent with the Southern blot data with G0/2 containing 9–11 copies, G0/4 containing 11–14 copies, G0/5 containing 5–8 copies, G0/1, G0/6 containing 1–2 copies and G0/3 to be indeterminate, with low-grade mosaicism responsible for a very low number of transgenic cells. Although the expanded HTT CAG repeat exhibits intergenerational changes in humans (1), PCR of DNA isolated from tail biopsies in the founder animals and from the sperm of the founder (G0/2, G0/3, G0/4) and three G2/5 third generation rams did not detect large changes in CAG repeat number, suggesting limited intergenerational instability of the CAG repeat in the context of the integrated human transgenes (Fig. 2B and C). The presence of a higher molecular weight product (∼500 bp compared with the expected 413 bp product) in the sperm of founder animal G0/1 (Fig. 2C) indicated there may be some gametic mosaicism of the CAG repeat in this line. Analysis of CAG repeat length in the offspring from this founder should confirm this effect.
Figure 2.
Molecular characterization of the HD founder transgenic animals. (A) Southern membrane hybridized with a radioactively labelled probe covering the promotor region (1.1 kb NruI/BssHII fragment). The 9.5 kb band is predicted to be the HindIII digested fragment corresponding to the wild-type ovine HD gene. About 4.9 and 3.6 kb bands are indicative of 5′–3′ and 5′–5′ transgene concatemerization, respectively. This is clearly illustrated in the multi-copy animals; G0/2, G0/4 and G0/5. Real-time PCR transgene copy numbers are indicated below the Southern blot. The DNA was isolated from tail biopsy tissue. (B) Polyglutamine repeat region genomic PCR demonstrating the presence of the 73 unit expanded transgene repeat allele (expected size = 430 bp) and the ovine wild-type alleles (expected size = ∼220 bp; * marks those heterozygous for the wild-type ovine alleles). The positive control is the transgene ligation fragment (human cDNA'BGH plasmid). The transgene PCR product for the highly chimeric G0/3 animal is not observed in this PCR reaction but the presence and size of the repeat region was confirmed by PCR analysis of sperm DNA (Fig. 2C). (C) PCR of the polyglutamine repeat region demonstrating stability of the CAG repeat in DNA isolated from sperm of the founder rams (G0/1–4) and the G2/5 transgenic rams. The positive control is genomic tail DNA from G0/2 and the negative control is dH2O. (D) RT–PCRs covering the BGH 3′ transcriptional tag of the founder transgenics. The preferentially amplified band is the unspliced product amplified from RNA isolated from tail biopsies. The positive control is the transgene ligation fragment (human cDNA'BGH plasmid) and the negative control is dH2O. Reverse transcriptase minus controls were included for each animal and produced no amplification product, excluding the possibility of genomic contamination (data not shown).
mRNA expression of the transgene detected in RNA isolated from tail biopsies was confirmed by RT–PCR in all six of the founder animals (Fig. 2D). Interestingly, the two highest transgene copy number animals (G0/2 and G0/4) appeared to have the lowest level of transgene expression. The localization of transgenes within genetically inactive heterochromatin is likely to be a contributing factor to the low transgene expression in the RNA from G0/4. In contrast to the high copy number animals, the lower copy number animals (G0/5 and G0/6) had relatively higher expression of the transgene. RT–PCR amplification of the BGH 3′-UTR (with a non-coding splice site) revealed the majority of the transcripts to be unspliced. Spliced versions of the BGH sequence in the transgene were only identified in G0/5 and G0/6 (data not shown), and were considerably lower in abundance relative to the unspliced product (confirmed by sequencing). It may be that the BGH insert was not functional in sheep, with the majority of transgene fragments not undergoing the splicing process or that the spliceosome failed to recognize the splice signals. Thus, despite previous descriptions of enhanced transgene efficiencies through splicing of the mature mRNA (8), it was not a fundamental requirement for transgene expression in our ovine model.
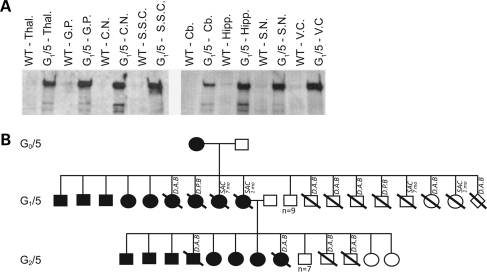
On the basis of the robust expression of the transgene in skin, and the high number of nuclei that contained the transgene, we initially bred from the transgenic ewes G0/5 and G0/6. The two lambs were stimulated at 6 weeks of age for oocyte collection, and subsequent in vitro produced embryos were transferred into cycle-synchronized adult recipient ewes (11). Nine transgenic offspring (two of which were dead within 48 h of birth) were derived from G0/5 and five from G0/6. Western blot analysis of CNS and non-CNS tissue from G0/5 offspring (G1/5) at 1 month of age and 7 months of age revealed ubiquitous expression of the human protein in the brain (Fig. 3A) and expression in representative tissues arising from each of the three primary germ layers (data not shown). It should be noted that accurate expression levels are difficult to determine by western blot due to conformational changes associated with the predominant α-helical HEAT repeat nature of the HTT protein. Interestingly, the human HTT protein was not detected in offspring G1/6, from G0/6, at 1 month of age. PCR analysis from brain and skin biopsies confirmed the transgenic status of this animal. Further work is currently being conducted to investigate possible epigenetic control of expression in this line.
Figure 3.
Expression in the G0/5 pedigree. (A) Western blot immunoprobed with the antibody MAB2166 demonstrating the expression of the human HTT transgenic protein in the first sacrificed offspring of the G0/5 line (1 month of age) as compared with the endogenous ovine HTT protein in eight different brain regions. The wild-type (WT) control animal is a true age matched sibling of the transgenic. Thal., Thalamus; G.P., Globus pallidus; C.N., Caudate nucleus; S.S.C., Somatosensory cortex; Cb., Cerebellum; Hipp., Hippocampus; S.N., Substantia nigra; V.C., Visual cortex. Gels were Coomassie stained to ensure even loading of samples (data not shown). (B) Pedigree of the G0/5 line. Shaded boxes represent those animals in the pedigree that tested positive for the transgene in DNA extracted from tail biopsy tissue. SAC 1 and 7 months = ages at which animals were sacrificed for analysis; D.A.B., dead at birth; D.P.B., dead post birth (within 48 h); open diamond = unknown sex (aborted fetus).
Following the identification of the human HTT protein in transgenic tissue from the G0/5 line, filter retardation assays were performed to establish whether insoluble SDS-resistant polyglutamine-fragment was also present. The presence of insoluble protein, particularly in the form of nuclear aggregates is a characteristic phenomenon associated with later stages of HD and the other polyglutamine diseases [reviewed in Ross (12)]. However, no insoluble protein was detected with N-675 (N-terminal HTT antibody) in the brains of 1- or 7-month-old G1/5 animals using this reported technique (data not shown). A sample of middle frontal gyrus tissue from a human HD case with a pathogenic CAG repeat length of 42 was used as a positive control.
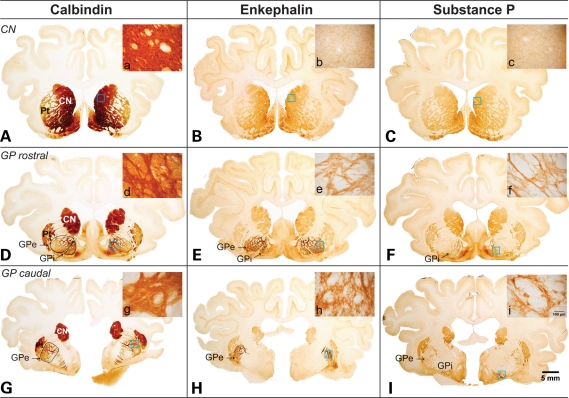
The ovine basal ganglia were characterized in wild-type sheep to enable subsequent comprehensive assessment of neuropathology in these lines. Delineation of chemospecific receptor subgroups and neurotransmitter populations comprising the nuclei of the basal ganglia provided a detailed anatomical correlate to the human basal ganglia. The biochemical distinction between the different nuclei of the wild-type ovine basal ganglia was determined primarily through immunolabelling with calbindin D28K, Substance P (SP) and enkephalin antibodies, based upon previous, well characterized studies of the cellular architecture of the human basal ganglia (13). Figure 4 displays photographs of three serial sections (rostral to caudal) of whole brain, labelled with each of these antibodies. Immunohistochemical analysis of calbindin D28K revealed a specific labelling distribution that was confined to select populations of neurons primarily located in the striatum and globus pallidus (GP; Fig. 4D). At the macroscopic level this labelling clearly delineated separate caudate nucleus, putamen and GP entities, comparable to that seen in the human brain. Enkephalin immunoreactivity was not as pronounced as that of calbindin and was primarily confined to the external segment of the globus pallidus (GPe; Fig. 4E and H), indicating that, as with the primate brain, the ovine brain is comprised of separate internal and external pallidal segments. Immunoreactive enkephalin fibres were also located in the internal segment of the globus pallidus (GPi) as shown in Figure 4E, indicating that the expression pattern of this neuropeptide in striatal output neurons was not absolute. On the whole, enkephalin appeared to be distributed in a comparable manner to that observed in the human brain and is assumed to be implicated in the ‘indirect’ pathway.
Figure 4.
Wild-type ovine brain photographs depicting the gross anatomy of the basal ganglia. These photomicrographs demonstrate the gross anatomy of the basal ganglia in the ovine brain using various markers to characterize the striatal projection neurons. Immunolabelling through the caudate nucleus and rostral and caudal ends of the GP demonstrate comparable anatomy to the human basal ganglia with separate caudate nucleus (CN), putamen (Pt) and pallidal nuclei. Furthermore (D), (E) and (F) demonstrate that the GP is sub-divided further into an enkephalin rich GPe and SP rich GPi as observed in the human brain. (A), (D) and (G) show immunoreactivity for the neurotransmitter calbindin, a marker for neuronal cell populations which use calcium dependent signalling. Microscopic insets of the neuronal specific labelling can be seen in images (a), (d) and (g). (B), (E) and (H) show immunoreactivity for the neurotransmitter enkephalin, a marker of the GABAergic medium spiny neurons involved in the ‘indirect’ pathway projecting from the striatum to the GPe. Microscopic insets of this neurotransmitter labelling pattern can be seen in (b), (e) and (h). (C), (F) and (I) show immunoreactivity for the neurotransmitter SP, a marker of the GABAergic medium spiny neurons involved in the ‘direct’ pathway projecting from the striatum to the GPi and substantia nigra (not shown). Microscopic insets of this neurotransmitter labelling pattern are shown in (c), (f) and (i).
Immunolabelling with SP confirmed the presence of this peptide in the striatal neuronal axon projection terminals of the GPi (Fig. 4F), further confirming the presence of separate internal and external segments of the GPe in the ovine brain. The lighter immunolabelling identified in the striatum represents the striatal projection neurons enriched with SP which project to the GPi. SP immunoreactivity appears to be implicated in the ‘direct’ pathway distributed in a similar fashion to that seen in the human brain. The presence of a small amount of SP immunoreactivity in the GPe (Fig. 4I) further supports the assumption that the differential expression pattern of the neuroactive peptides enkephalin and SP may not be absolute in the ovine brain. The comparable anatomy of the ovine basal ganglia to that of the human brain is of particular advantage for the neuropathological characterization and subsequent quantification of the disease progression in the HD transgenic ovine lines.
A preliminary neuropathological study has been undertaken on two offspring of the G0/5 founder. The basal ganglia of 1- and 7-month-old G1/5 transgenics and relevant age matched controls (true siblings of the transgenics) were assessed for neuropathological changes. This provisional immunohistochemical study, performed with a limited number of animals (one transgenic and control at each time point) serves as a guide for the larger characterization study which will be performed as more animals become available. Assessment of the vulnerable GABA-ergic medium spiny neurons was initially performed with calbindin, enkephalin and SP. Immunolabelling with these three antibodies did not reveal any cell loss or macro/microscopic changes in immunoreactivity of the direct and indirect basal ganglia pathways in the 1- and 7-month-old animals (data not shown). However, further investigation into receptors and cell types known to be vulnerable in HD; cannabinoid receptor type 1 (CB1) (a presynaptic receptor which is co localized on dopamine receptors) and dopamine- and cyclic AMP-regulated phosphoprotein (DARPP32) (a D1 receptor associated signalling protein found in striatal projection neurons) did reveal apparent neuropathological changes.
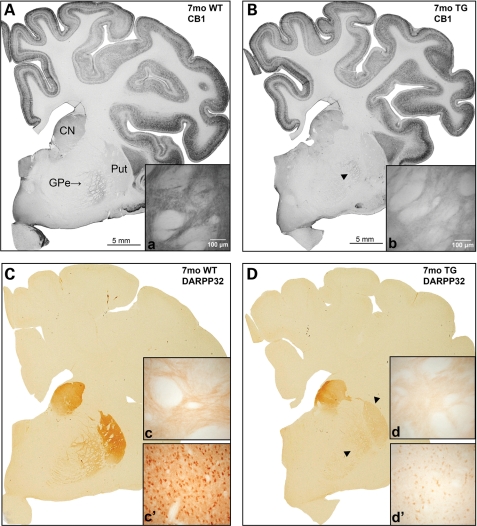
At the level of the GP, macroscopic analysis indicated subtle loss of CB1 immunoreactivity in the CN and GPe of the 1-month-old HD transgenic animal, G1/5. The loss was more marked at 7 months of age (Fig. 5B), particularly in the external pallidal region, with a decreased intensity of fine terminal labelling, as displayed microscopically (Fig. 5b). This loss was confirmed semi-qualitatively on microscopic analysis with a reduction in the intensity of CB1 immunoreactivity. Comparable immunolabelling across cortical regions in the transgenic and control animals indicated this was not a labelling anomaly. The number of axonal and dendritic processes did not appear to be reduced in the CN and GPe of the transgenic animals; however, the density of labelling along these processes was decreased indicating there may be reduced/dysfunctional CB1 expressing receptor terminals along these processes. Immunolabelling with this antibody was repeated confirming these results. Multiple fields from each section were analysed using Discovery-1 and Metamorph software revealing the decrease in CB1 immunoreactivity in the GP of the transgenic to be 95.2% of the control at 1 month of age and 93.1% of the control at 7 months of age. These decreases were statistically significant with P-values of 0.0443 and 0.0029 at 1 and 7 months of age, respectively.
Figure 5.
Loss of CB1 and DARPP32 immunoreactivity in a 7-month-old G1/5 animal as compared with control. (A) and (B) demonstrate macroscopically the loss in intensity of CB1 immunoreactivity in the GP (arrow head) of the transgenic animal (B) as compared with the control (A). In comparison, the cortex demonstrates a uniform staining intensity between the transgenic and control animals. Higher magnification illustrates the loss of immunoreactivity of fine synaptic labelling in the GP of the transgenic (b) as compared with control (a). (C) and (D) are macroscopic images of DARPP32 immunoreactivity in the control and transgenic animals, respectively. This clearly demonstrates the loss of immunolabelling intensity in the transgenic animal (arrow heads), which is observed in the absence of significant striatal cell loss. The loss of fine projection fibre labelling in the GP of the transgenic as compared with the denser labelling observed in the control is shown in (d) and (c), respectively. A photomicrograph of the putamen in the transgenic animal (d′) demonstrates loss of immunoreactivity in the absence of cell loss or dysmorphology. The corresponding control section is (c′).
The most striking neuropathological change observed was the loss of DARPP32 receptor immunoreactivity in the GP and putamen of the 7-month-old G1/5 transgenic, illustrated at the macroscopic level in the images displayed in Fig. 5D. The reduction in the intensity of DARPP32-stained cell bodies and processes was confirmed when the experiments were repeated with tissue from the same animal. These changes were not observed at 1 month of age. This loss is consistent with the pre-symptomatic striatal D1 dopamine receptor loss observed in the human disease state (14). Multiple fields from each section were analysed using Discovery-1 and Metamorph software, showing statistically significant loss of DARPP32 immunoreactivity in both the putamen (72% of control) and GP (92.4% of control) of the transgenic animal when compared with control (P = 0.001 and 0.0009, respectively). Notably, immunohistochemistry with antibodies 1C2, EM-48 and ubiquitin did not reveal any immunoreactive inclusion bodies in the tissue of transgenic animals at either age.
DISCUSSION
The longevity of sheep (∼10 years) and their analogous forebrain structure to humans (including separate GPi and GPe) make them especially suited to studying late-onset neurodegenerative disorders. We have generated six transgenic founder sheep, expressing full length human HTT with a polyglutamine region of 73 residues. We considered it important to express the full-length human HTT protein in order to permit an accurate assessment of the consequences of the HD mutation.
The six transgenic founders were generated through integration of a ligation fragment into the genome using pronuclear microinjection and all but G0/3 (indeterminate) were shown to contain single integration sites (as determined by FISH). In the cases of G0/1, G0/2, G05 and perhaps G0/6, integration was into gene-rich dark R-bands which are generally believed to contain mainly house-keeping genes. The dark R-band 11q11 into which the G0/4 transgenes are inserted is regarded as genetically inert, being composed of repetitious Satellite DNA sequences. Southern blot analysis revealed multi-copy loci in each transgenic, preferentially containing concatamers oriented 5′–3′, a common feature of stably integrated transgene arrays (15). Southern blot analysis and real-time PCR revealed G0/2 and G0/4 to have high copy numbers of the transgene (9–14 copies), but both animals had low expression of mRNA transcribed from the transgenes. As a result of random integration of the transgene into the embryonic genome, there are several possible explanations as to why RNA expression is low in the resulting transgenic animals, including DNA methylation and cis-acting silencer sequences. Elucidation of the DNA sequences adjacent to the inserted transgene may help to determine these effects. In comparison, G0/5 (having 5–8 transgene copies) and G0/1 and G0/6 (containing ∼1–2 copies) showed relatively high levels of HTT mRNA expression. At least high-grade mosaicism was found in all founder animals except G0/3, which was found by FISH analysis to be a very low-grade mosaic, indicating an integration event following many embryonic cell divisions.
Breeding from G0/5 showed stable transmission of the transgene, and mRNA and protein expression in CNS and non-CNS tissue in 1- and 7-month-old G1/5 animals. The comparable distribution of neuronal populations between the ovine and human basal ganglia is an advantage in allowing the early patterns of degeneration of the external and internal connections of the basal ganglia to be monitored. A pilot study with G1/5 animals at 1 and 7 months of age showed a significant loss in receptor density in the fine meshwork of CB1 positive fibres in the GP. No loss in enkephalin or SP (markers of the indirect and direct pathways, respectively) was observed. At 7 months of age, the G1/5 transgenic also showed a loss in DARPP32 immunoreactivity from the GP and putamen. The reduction suggests an early impairment in the regulation of the physiological state of striatal neurons via dopamine, a hallmark feature of the disease. Full immunohistochemical characterization of further brain regions, in more animals at several time points will be required to validate these preliminary results.
Interestingly, expression of the transgene was not observed as RNA or protein in the 1-month-old offspring analysed from the G0/6 founder. The transmission in this line is being investigated and this phenomenon may be explained by genetic imprinting. Breeding from the G0/5 line is currently into the third generation of animals with no gross expansion or contraction of the CAG repeat region detected. Breeding and characterization of the remaining lines is now underway and early analysis reveals that both the transgene and transgene activity are heritable in these lines. Following a detailed analysis of the transgene expression and its effects in all lines, the optimal line(s) will be selected for continued propagation.
One of our primary aims is the generation of a homozygous sire that will enable all offspring to be heterozygous transgenic. Though the transgene CAG repeat appeared stable upon transmission from ewes, it will be important to monitor repeat stability through male transmission. Moreover, additional analysis, for example by small-pool PCR, will be needed to examine the possibility of somatic repeat instability, which is another feature of the expanded human HD CAG repeat (16). PCR and sequencing analysis of sperm DNA from the founding rams (G0/1–4) and G2/5 rams revealed no gross repeat instability (Fig. 2C).
In contrast to transgenic primates over-expressing a small fragment of the HTT gene, which display compromised longevity and reproduction (17), the ovine transgenic lines that we have generated are viable, allowing sibships to be bred for longitudinal study. At 3 years of age, the founders do not show overt abnormal phenotypes (i.e. gait or behavioural abnormalities), which is of considerable advantage in maintaining the lines and for presymptomatic drug trials. In the coming years, phenotyping using proteomic and gene expression analyses (utilizing microarrays), will allow the collection of longitudinal molecular data, to complement metabolic, behavioural and MRI assessments that will yield a detailed natural history of the disease process over a decade or more, which is not currently possible with humans. Such extensive longitudinal data will complement the data that is now beginning to emerge from studies of adult humans carrying the HTT CAG mutation who are not yet exhibiting overt disease symptoms (18–30).
Despite the discovery of the gene which causes HD over 15 years ago, there is no recognized treatment for preventing or delaying the disease. Slow disease course, low disease frequency, insidious onset and patient to patient variability makes monitoring modification of the disease process in human drug trials challenging. It may be that large animals could yield results with therapeutic relevance to human. Ultimately, it is envisaged that this ovine model (in tandem with other model systems) will facilitate the fast tracking of treatments and therapies for HD into clinical trials. This is an emerging possibility for patients with HD and this work establishes a precedent for the experimental approach in other neurodegenerative diseases, particularly chronic progressive disorders, or those which have a low prevalence, limiting the number of patients available for clinical trials.
MATERIALS AND METHODS
Construction of the transgene
The full length HTT cDNA transgene was created through manipulation of the HD1-3144Q113 construct (31). The CMV promotor was replaced with 1.1 kb of the human HTT genomic promotor using an engineered BssHII site, and a reverse transcription generated PCR fragment containing a pathogenic polyglutamine repeat of 73 units was ligated into the full length construct at a unique XhoI site in exon 4 of the HTT cDNA. The final 11.6 kb ligation fragment could be isolated with an NruI–NotI double digest. DNA sequencing was used to verify restriction enzyme ligation sites and CAG repeat length.
Transgenesis
The ligation fragment was microinjected into pronuclei of single-celled zygotes derived from South Australian Merino sheep. Genomic DNA from the resulting lambs was purified from tail biopsies via proteinase-K digestion and phenol/chloroform extraction (32) and the presence of the transgene assessed by PCR and sequencing. The transgenesis experimentation was performed at the South Australian Research and Development Institute (SARDI) in accordance with the SARDI/PIRSA Animal Ethics Committee (Approval number 19/02).
FISH analysis
The transgenes were localized using FISH. Transformed lymphocytes were labelled in the late half of S-phase with 5-bromodeoxyuridine for the final 7.5 h of a 3-day culture. The 9 kb HTT-DNA probe was labelled with biotin using nick translation, hybridized in-situ to the chromosomes and then detected using FITC-avidin with one round of signal amplification after use of biotinylated anti-avidin (Vector laboratories). R-banded chromosomes with signal were visualized under blue excitation and with clearer bands under green (Fig. 1B). This probe did not detect the endogenous ovine HTT gene.
Southern analysis
cDNA probes covering the entire 11.6 kb transgene were radioactively labelled using [α-32P]dCTP with Amersham's megaprime labelling kit and purified by ethanol precipitation prior to use. Ten microgram of digested DNA was resolved on 0.8% TAE agarose gels. Digested DNA was electrophoresed, and transferred from the gel to an N+ nylon alkali filter membrane (Amersham) using alkali Southern blotting. Filter membranes were subsequently hybridized with pre-prepared [α-32P]dCTP labelled probes, and excess probe removed by washes at high stringency (2×SSC with 0.1% SDS at 65°C for 15 min). Further washes (1×SSC with 0.1% SDS, 0.5×SSC with 0.1%SDS) were performed where necessary. Filters were exposed to film (Amersham) at −80°C and developed using standard developing chemicals (Kodak).
Polymerase chain reaction
Real-time PCR
Real-time primers were designed to the same region (exon 39) in the human and ovine HTT genes, a region which on alignment revealed significant sequence divergence between the two species, ensuring species specificity of the PCR reactions. Each real-time PCR reaction was performed in a total volume of 10 µl using SYBR green technology (Invitrogen). Each PCR reaction underwent 40 cycles of amplification (30 s at 94°C, 30 s at 55°C, 1 min at 72°C) following an initial 5 min denaturation period using the AB1 7000 machine (Applied Biosystems).
Reverse transcription PCR
Ten units of DNaseI recombinant RNase-free (Roche) was used to digest 1 µg of RNA prior to first strand synthesis. cDNA was reverse transcribed in a total reaction volume of 20 µl, using Invitrogen's SuperScript™ First-Strand Synthesis System for RT–PCR. An Oligo-dT (Invitrogen) primer and several gene specific primers located along the length of the transgene were used to prime cDNA synthesis. The resulting cDNA was used directly for amplification of the target DNA or it was stored at −20°C.
PCR primer sequences
Real-time PCR primer sequences used for amplification of the human HTT transgene and wild-type ovine HTT transcript:
Human HD RTpA 5′ GTTCTGATTTCCCAGTCAACTGAA 3′ and Human HD RTpB 5′ ACAGGAGATTAAATACGGAGAGAAGGA 3′.
Ovine HD RTpA 5′ CTGATTTCCCAGTCGACCGAG 3′ and Ovine HD RTpB 5′ CAGGAGACCAAGTTCGGAGAGA 3′.
PCR primer sequences used for amplification of the CAG repeat region in the human HTT transgene and ovine HTT transcript:
BssHII HTTpA: 5′ TTGGCGCGCACCGCCATGGCGACCCTGGAAAAGCTG 3′.
HompB3: 5′ GGTCGGTGCAGCGGCTCCTC 3′.
Primer sequences (which amplify the 3′ BGH region) used to assess expression of the HTT transgene in the mRNA:
TF1: 5′ GGGGCCGGAGCCTTTGGAAGTC 3′.
TR1: 5′ GCCATCCCCACCCGCATCAG 3′.
Sequencing
Sequencing was performed in our laboratory or at the Allan Wilson Centre Genome Sequencing Service (AWCGS), Massey using the ABI3730 genetic analyser (Applied Biosystems) with BigDye™ Terminator Version 3.1 Ready Reaction Cycle Sequencing kit chemistry and data collection software (Applied Biosystems).
Western analysis
Thirty micrograms of protein lysates from dissected tissues of transgenic and wild-type animals were resolved on 3–8% SDS–PAGE gels (Invitrogen) or 4–20% Ready Gel precast gels (BioRad) in XCell SureLock Mini-Cell (Invitrogen) and Mini-PROTEAN 3 (BioRad) electrophoresis systems, respectively. The expression profile determined using ECL-Plus (Amersham) using the anti-HTT antibody MAB2166 [Chemicon (33)] at 1:20 000 and 1F8 (34) at 1:10 000 raised against the pathogenic polyglutamine tract. MAB2166 recognizes both sheep and human isoforms and was used to determine transgene expression relative to the endogenous sheep HTT. The appropriate species-specific horse radish peroxidase-conjugated secondary antibody (Chemicon) was used at 1:5000 in Tris-buffered saline Tween-20 and the complex visualized with ECL Plus Western blotting reagents (Amersham) when exposed to ECL hyperfilm (Amersham).
Filter retardation assays
Fresh frozen transgenic and control brain samples were assessed for the presence of insoluble N-terminal HTT protein using filter retardation assays. About 100 µg of pre-prepared frontal cortex was applied to 0.2 µm cellulose acetate membranes and reacted with the N-terminal anti-HTT antibody N-675 [gift from Dr Leslie Jones (35)] at 1:1000. Following incubation with the appropriate HRP-conjugated secondary antibodies (1:5000) membranes were visualized using chemi-luminescence as previously described for Western analysis.
Immunohistochemistry
Brains were fixed by immersion (36 h) in 15% formalin in 0.1 m phosphate buffer, and cryoprotected in sucrose (graded 20–30% solutions). All sections were cut on a freezing sledge microtome at 50 µm and stored in PBS-Azide until use. Free floating sections were initially washed for 20 min in a solution of 50% methanol and 1% H2O2 followed by incubation in primary antiserum for 48 h at 4°C. Antibody concentrations were as follows: Rabbit α calbindin (gift from Piers Emson) 1:15 000; Rabbit α enkephalin (INCSTAR) 1:5000; Rabbit α SP (Watpa Enterprises) 1:10 000; Rabbit α CB1L15 (gift from Kenneth Mackie) 1:750; Rabbit α DARPP32 (Millipore) 1:1000. Following incubation in primary antiserum, biotinylated goat anti-mouse or goat anti-rabbit IgG was applied to the appropriate sections and incubated at room temperature for 24 h. The following day sections were incubated for 4 h at room temperature in avidin–biotin-horseradish peroxidase complex followed by incubation in 0.05% diaminobenzidine tetrahydrochloride with 0.01% H2O2 to visualize the tertiary complex. Sections were mounted with gelatin, dried and dehydrated in a graded ethanol series and finally cleared with three 20 min incubations in 100% xylene. Sections were mounted with DPX prior to visualization.
FUNDING
This work was supported by the Freemasons of New Zealand; the Neurological Foundation of New Zealand; the Health Research Council of New Zealand; Wellcome Trust; the Foundation of Research Science and Technology; and the National Institutes of Health NINDS grants (NS16367, NS32765).
ACKNOWLEDGEMENTS
We thank the Neurological Foundation of New Zealand Human Brain Bank for the gift of human brain tissue, Dr Kenneth Mackie for the gift of the CB1 antibody, Dr Leslie Jones for the gift of the N-675 antibody, Dr Piers Emson for the gift of the Calbindin D28K antibody and Dr Mike Dragunow for Discovery-1 and Metamorph analysis.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Mangiarini L., Sathasivam K., Seller M., Cozens B.A., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 3.Scheerlinck J.-P.Y., Snibson K.J., Bowles V.M., Sutton P. Biomedical applications of sheep models: from asthma to vaccines. Trends Biotechnol. 2008;26:259–266. doi: 10.1016/j.tibtech.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Adalsteinsson S., Basrur P.K. Inheritance of spina bifida in Icelandic lambs. J. Hered. 1984;75:378–382. doi: 10.1093/oxfordjournals.jhered.a109960. [DOI] [PubMed] [Google Scholar]
- 5.Jolly R.D., Martinus R.D., Shimada A., Fearnley I.M., Palmer D.N. Ovine ceroid-lipofuscinosis is a proteolipid proteinosis. Can. J. Comp. Med. 1990;54:15–21. [PMC free article] [PubMed] [Google Scholar]
- 6.Bawden C.S., Powell B.C., Walker S.K., Rogers G.E. Expression of a wool intermediate filament keratin transgene in sheep fibre alters structure. Transgenic Res. 1998;7:273–287. doi: 10.1023/a:1008830314386. [DOI] [PubMed] [Google Scholar]
- 7.Schnieke A.E., Kind A.J., Ritchie W.A., Mycock K., Scott A.R., Ritchie M., Wilmut I., Colman A., Campbell K.H.S. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science. 1997;278:2130–2133. doi: 10.1126/science.278.5346.2130. [DOI] [PubMed] [Google Scholar]
- 8.Brinster R.L., Allen J.M., Behringer R.R., Gelinas R.E., Palmiter R.D. Introns increase transcriptional efficiency in transgenic mice. Proc. Natl. Acad. Sci. USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang M.T.F., Gorman C.M. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bawden C.S., Sivaprasad A.V., Verma P.J., Walker S.K., Rogers G.E. Expression of bacterial cysteine biosynthesis genes in transgenic mice and sheep: toward a new in vivo amino acid biosynthesis pathway and improved wool growth. Transgenic Res. 1995;4:87–104. doi: 10.1007/BF01969411. [DOI] [PubMed] [Google Scholar]
- 11.Kelly J.M., Kleemann D.O., Walker S.K. Enhanced efficiency in the production of offspring from 4- to 8-week-old lambs. Theriogenology. 2005;63:1876–1890. doi: 10.1016/j.theriogenology.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Ross C.A. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 13.Harper P.S. Huntington's Disease. London: W.B.Saunders; 1996. [Google Scholar]
- 14.Weeks R.A., Piccini P., Harding A.E., Brooks D.J. Striatal D1 and D2 dopamine receptor loss in asymptomatic mutation carriers of Huntington's disease. Ann. Neurol. 1996;40:49–54. doi: 10.1002/ana.410400110. [DOI] [PubMed] [Google Scholar]
- 15.Palmiter R.D., Brinster R.L. Germ - line transformation of mice. Annu. Rev. Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telenius H., Kremer B., Goldberg Y.P., Theilmann J., Andrew S.E., Zeisler J., Adam S., Greenberg C., Ives E.J., Clarke L.A., et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat. Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- 17.Yang S.-H., Cheng P.-H., Banta H., Piotrowska-Nitsche K., Yang J.-J., Cheng E.C.H., Snyder B., Larkin K., Liu J., Orkin J., et al. Towards a transgenic model of Huntington's disease in a non-human primate. Nature. 2008;453:921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biglan K.M., Ross C.A., Langbehn D.R., Aylward E.H., Stout J.C., Queller S., Carlozzi N.E., Duff K., Beglinger L.J., Paulsen J.S., et al. Motor abnormalities in premanifest persons with Huntington's disease: the PREDICT-HD study. Mov. Disord. 2009;24:1763–1772. doi: 10.1002/mds.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duff K., Paulsen J.S., Beglinger L.J., Langbehn D.R., Stout J.C. Psychiatric symptoms in Huntington's disease before diagnosis: the Predict-HD study. Biol. Psychiatry. 2007;62:1341–1346. doi: 10.1016/j.biopsych.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 20.Henley S.M.D., Wild E.J., Hobbs N.Z., Frost C., MacManus D.G., Barker R.A., Fox N.C., Tabrizi S.J. Whole-brain atrophy as a measure of progression in premanifest and early Huntington's disease. Mov. Disord. 2009;24:932–936. doi: 10.1002/mds.22485. [DOI] [PubMed] [Google Scholar]
- 21.Johnson S.A., Stout J.C., Solomon A.C., Langbehn D.R., Aylward E.H., Cruce C.B., Ross C.A., Nance M., Kayson E., Julian-Baros E., et al. Beyond disgust: impaired recognition of negative emotions prior to diagnosis in Huntington's disease. Brain. 2007;130:1732–1744. doi: 10.1093/brain/awm107. [DOI] [PubMed] [Google Scholar]
- 22.Kloppel S., Chu C., Tan G.C., Draganski B., Johnson H., Paulsen J.S., Kienzle W., Tabrizi S.J., Ashburner J., Frackowiak R.S.J., et al. Automatic detection of preclinical neurodegeneration: presymptomatic Huntington disease. Neurology. 2009;72:426–431. doi: 10.1212/01.wnl.0000341768.28646.b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kloppel S., Draganski B., Siebner H.R., Tabrizi S.J., Weiller C., Frackowiak R.S.J. Functional compensation of motor function in pre-symptomatic Huntington's disease. Brain. 2009;132:1624–1632. doi: 10.1093/brain/awp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulsen J.S. Biomarkers to predict and track diseases. Lancet Neurol. 2009;8:776–777. doi: 10.1016/S1474-4422(09)70203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulsen J.S., Hayden M., Stout J.C., Langbehn D.R., Aylward E., Ross C.A., Guttman M., Nance M., Kieburtz K., Oakes D., et al. Preparing for preventive clinical trials: the Predict-HD study. Arch. Neurol. 2006;63:883–890. doi: 10.1001/archneur.63.6.883. [DOI] [PubMed] [Google Scholar]
- 26.Paulsen J.S., Langbehn D.R., Stout J.C., Aylward E., Ross C.A., Nance M., Guttman M., Johnson S., MacDonald M., Beglinger L.J., et al. Detection of Huntington's disease decades before diagnosis: the Predict-HD study. J. Neurol. Neurosurg. Psychiatr. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saft C., Schüttke A., Beste C., Andrich J., Heindel W., Pfleiderer B. fMRI reveals altered auditory processing in manifest and premanifest Huntington's disease. Neuropsychologia. 2008;46:1279–1289. doi: 10.1016/j.neuropsychologia.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Schippling S., Schneider S.A., Bhatia K.P., Münchau A., Rothwell J.C., Tabrizi S.J., Orth M. Abnormal motor cortex excitability in preclinical and very early Huntington's disease. Biol. Psychiatry. 2009;65:959–965. doi: 10.1016/j.biopsych.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solomon A.C., Stout J.C., Johnson S.A., Langbehn D.R., Aylward E.H., Brandt J., Ross C.A., Beglinger L., Hayden M.R., Kieburtz K., et al. Verbal episodic memory declines prior to diagnosis in Huntington's disease. Neuropsychologia. 2007;45:1767–1776. doi: 10.1016/j.neuropsychologia.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabrizi S.J., Langbehn D.R., Leavitt B.R., Roos R.A.C., Durr A., Craufurd D., Kennard C., Hicks S.L., Fox N.C., Scahill R.I., et al. Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8:791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Persichetti F., Trettel F., Huang C.C., Fraefel C., Timmers H.T.M., Gusella J.F., MacDonald M.E. Mutant huntingtin forms in vivo complexes with distinct context-dependent conformations of the polyglutamine segment. Neurobiol. Dis. 1999;6:364–375. doi: 10.1006/nbdi.1999.0260. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Fusco F.R., Chen Q., Lamoreaux W., Figueredo-Cardenas G., Jiao Y., Coffman J.A., Surmeier D.J., Honig M.G., Carlock L.R., Reiner A. Cellular localization of huntingtin in striatal and cortical neurons in rats: lack of correlation with neuronal vulnerability in Huntington's disease. J. Neurosci. 1999;19:1189–1202. doi: 10.1523/JNEUROSCI.19-04-01189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White J.K., Auerbach W., Duyao M.P., Vonsattel J.-P., Gusella J.F., Joyner A.L., MacDonald M.E. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat. Genet. 1997;17:404–410. [Google Scholar]
- 35.Jones A.L. The localization and interactions of huntingtin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999;354:1021–1027. doi: 10.1098/rstb.1999.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]