Abstract
Mutations in spastin are the most frequent cause of the neurodegenerative disease autosomal dominant-hereditary spastic paraplegia (AD-HSP). Drosophila melanogaster lacking spastin exhibit striking behavioral similarities to human patients suffering from AD-HSP, suggesting conservation of Spastin function between the species. Consistent with this, we show that exogenous expression of wild-type Drosophila or human spastin rescues behavioral and cellular defects in spastin null flies equivalently. This enabled us to generate genetically representative models of AD-HSP, which arises from dominant mutations in spastin rather than a complete loss of the gene. Flies co-expressing one copy of wild-type human spastin and one encoding the K388R catalytic domain mutation in the fly spastin null background, exhibit aberrant distal synapse morphology and microtubule distribution, similar to but less severe than spastin nulls. R388 or a separate nonsense mutation act dominantly and are furthermore sufficient to confer partial rescue, supporting in vitro evidence for additional, non-catalytic Spastin functions. Using this model, we tested the observation from human pedigrees that S44L and P45Q are trans-acting modifiers of mutations affecting the Spastin catalytic domain. As in humans, both L44 and Q45 are largely silent when heterozygous, but exacerbate mutant phenotypes when expressed in trans with R388. These transgenic ‘AD-HSP’ flies therefore provide a powerful and tractable model to enhance our understanding of the cellular and behavioral consequences of human spastin mutations and test hypotheses directly relevant to the human disease.
INTRODUCTION
Autosomal dominant-hereditary spastic paraplegia (AD-HSP) is a dominantly inherited, progressive neurodegenerative disease that selectively affects the terminals of the longest axons in the central nervous system (CNS) (1). Predominant symptoms are caused by aberrant motor control of the lower limbs, exhibiting as leg weakness and spasticity. These symptoms vary highly in penetrance and onset age, ranging from patients afflicted from infancy and confined to wheelchairs, to those who remain asymptomatic through life. The discovery that mutations in spastin account for approximately half of all cases of AD-HSP (2) has led to considerable interest in the neurobiological function of this protein (3,4).
Spastin monomers assemble into hexameric, ring-shaped ATPases that sever microtubules along their lengths (5–8). These severing events are distinct from the well-known disassembly mechanism of dynamic instability, which is spontaneous and occurs only at microtubule ends (9). Each Spastin monomer contains a carboxyl-terminal ‘AAA’ ATPase catalytic domain that is highly conserved between species, as well as a distinct microtubule-interacting and trafficking (MIT) domain. These two domains effect binding of the protein to microtubule polymers, promoting its hexamerization and subsequent ATP hydrolysis-dependent breakage of the microtubule substrate (8). The critical nature of the AAA domain is evidenced by the discovery that of the over 140 spastin mutations identified in AD-HSP patient cohorts to date, all but a few (see below) compromise Spastin's ATPase activity, whether through splicing, deletion, insertion, nonsense or missense errors in the gene (3,10).
Best studied of the AD-HSP disease alleles is the K388R missense mutation. Structural analysis of the AAA ATPase region reveals that the analogous lysine residue of Drosophila Spastin is required for nucleotide binding, such that mutation to arginine causes complete loss of ATPase and microtubule severing activity in vitro (5,8). Although overexpressed wild-type Spastin localizes to cytoplasmic or perinuclear aggregates in cells and causes loss of the microtubule cytoskeleton, R388 mutant Spastin associates with bundled microtubules in a filamentous pattern and fails to sever them (5,6,11–13). The disease-inducing effects of R388 have therefore been attributed to a dominant-negative mechanism (11,12,14), either through the formation of mutant and wild-type heteromers that are less effective at catalysis, or by competition between the mutant and wild-type proteins for binding sites on microtubules (12). However, strong evidence, most notably the absence of truncated protein variants in patients bearing early termination codon mutations, also supports the idea that spastin mutant alleles cause AD-HSP through haploinsufficiency (10,15–18).
Genetic analyses of AD-HSP patients have also identified a unique class of adjacent mutations outside of the catalytic domain. These are the amino-terminal S44L and P45Q missense alleles, which several studies suggest are silent polymorphisms in the human population when in trans with wild-type spastin (19–23). When in trans with a mutation affecting the Spastin catalytic domain, however, the presence of either L44 or Q45 correlates strongly with earlier onset age and exacerbated disease severity. This remains the only correlation that has been made between a specific mutation in spastin and the otherwise variable penetrance of AD-HSP.
Defects in Spastin impair the proper formation or maintenance of the microtubule cytoskeleton, compromising neuronal function. Endogenous expression studies localizing the protein to discrete punctae along microtubules within axons and dendrites of cultured neurons (24), as well as in growth cones and at branch points—regions where microtubules are highly dynamic (13,25)—support a role for Spastin in microtubule assembly during neurite outgrowth. Consistent with this idea loss of Spastin function in vivo leads to morphological, cytoskeletal and electrophysiological aberrations at presynaptic terminals (14,25,26). Endogenous spastin localization in HeLa cells to endosomal compartments and the midbody during abscission further suggests that these neuronal defects may be tied to a role in membrane trafficking (27).
Animal models bearing spastin mutations have been generated in the worm and the fly, as well as zebrafish and mouse. Although providing valuable insights into Spastin function, they have not yet led to a clear understanding of the cell biological mechanisms underlying AD-HSP pathology, due to an absence of behavioral phenotypes that correlate with representative disease genotypes. In Caenorhabditis elegans, genomic deletion of the sequence encoding the Spastin AAA region causes oogenesis and multivulva defects, but the animals move and survive normally (28). Treatment of zebrafish embryos with spastin morpholinos inhibits motor axon outgrowth and impairs motility, suggesting a role in early axon development as well as synapse formation (29). Two independent transgenic mouse models support the idea that Spastin mutations impair axon trafficking. Mice homozygous for deletions of exons 5–7 (thereby generating a premature stop codon) exhibit spinal cord axonal swellings enriched in organelles (30). Similar results are observed in mice bearing a splice site mutation that mimics an identified pathogenic human mutation (31). Cultured cortical neurons from animals heterozygous or homozygous for this deletion (expressing reduced or undetectable levels, respectively, of spastin protein) also exhibit axonal swellings correlated with reduced rates of anterograde mitochondrial transport. However, in both mouse models only mild behavioral deficits are seen despite these cellular defects, even in homozygous mutants devoid of the protein.
Homozygous deletion of spastin in flies is predominantly lethal, and ‘escaper’ flies that do survive to adulthood are severely compromised in motor function. Spastin null flies walk, climb and stand poorly, and appear to have weak legs that frequently slip from under them (25). Similarly, flies overexpressing catalytically mutant Spastin or spastin RNAi in neurons climb more slowly than wild-type animals, and decline more quickly in this ability with time (14). The behavioral similarities observed between spastin mutant flies and humans are striking, but also in accordance with the well-established utility of Drosophila melanogaster as a model system for human diseases, particularly of the nervous system (32–34). Combined with the extensive genetic tools that are available, these characteristics make Drosophila a powerful model for addressing the molecular and cellular underpinnings of AD-HSP.
Important insights into Spastin function, particularly in regards to the pathogenesis of AD-HSP, can therefore be gained from modified models that more accurately recapitulate the AD-HSP disease genotype. Orso et al. (14) generated a Drosophila model of AD-HSP in which the fly equivalent of the human K388R allele was overexpressed in neurons in a wild-type background, and found similar behavioral and cellular phenotypes in these flies as are seen in loss of function mutants. Here, we sought to generate a modular model that can accommodate the variety of allelic combinations inherent to this disease, without requiring a dominant negative mechanism of action. Using flies deleted for their endogenous spastin gene, we expressed variants of the human gene in allelic combinations that mimic human genotypes, and found, by quantifying subcellular, cellular and behavioral parameters, that the phenotypic severity of the observed genotypes conforms to that predicted by human data. Compared with animals expressing no Spastin, these flies should more accurately recapitulate the molecular and developmental series of events leading to AD-HSP, as well as the consequences of specific mutations and potential therapeutic compounds.
RESULTS
Generation and expression of Drosophila and human spastin in vivo
Drosophila Spastin shares ∼70% amino acid identity in the catalytic AAA region with its orthologous human protein. The extended amino terminus is less well conserved, but shares similar key regions, including a MIT domain involved in binding microtubules as well as other proteins related to its function (5,8,27). Alternative start and splice sites indicate that multiple transcripts are expressed in both flies and vertebrates. Only a single protein is predicted in the fly, however, and in mice and rats the longest, 616 amino acid isoform is enriched in the CNS relative to other tissues, implicating a key role for the full-length protein in AD-HSP pathogenesis (35,36).
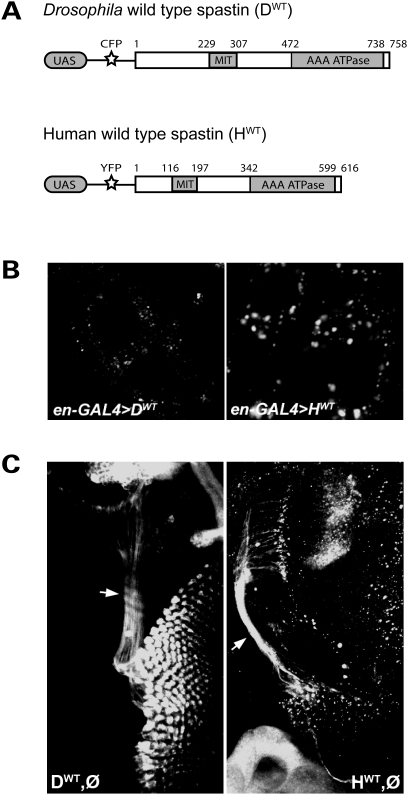
To determine the degree of functional conservation between human and fly spastin, we used the Drosophila Gateway system (T.D. Murphy, Carnegie Institute) to generate constructs containing either the full-length fly wild-type genomic or human wild-type cDNA sequence [Fig. 1A; (24)]. These insertions were preceded by yeast GAL4 binding sites (Upstream Activating Sequence or UAS), followed by either cyan- or ‘Venus’ yellow-fluorescent protein [CFP or YFP, respectively; (37)] tags at the 5′ end of the gene. Each construct was injected into wild-type Drosophila embryos to generate multiple transgenic lines, each with a single independent insertion. We confirmed transgene expression for each line by crossing them to a ‘driver’ line of flies bearing an engrailed-GAL4 transgene. Progeny from this cross, denoted as en-GAL4>UAS-spastin, expressed the yeast GAL4 protein in the tissue-specific and developmental pattern dictated by the engrailed promoter, inducing transcription of the reporter-tagged spastin. Using an anti-GFP antibody, we visualized the subcellular distribution of the transgenic Spastin protein in larval epidermal cells, which are large and flat in morphology. Drosophila Spastin was excluded from the nucleus, and localized diffusely in the cytoplasm with some aggregate formation (Fig. 1B, left). Overexpressing human spastin in the same cell type also revealed diffuse cytoplasmic protein distribution accompanied by aggregates, although the latter formed with somewhat greater frequency (Fig. 1B, right). Similar expression patterns are observed with human spastin cDNA expressed in Cos7 or HeLa cells, as well as for overexpressed Drosophila spastin cDNA in cultured S2 cells (5).
Figure 1.
Fly and human spastin transgenes have similar subcellular distributions. (A) Constructs containing GAL4 binding sites (UAS) followed by genes encoding a fluorescent tag (CFP or Venus YFP) and either the spastin full-length wild-type fly genomic (DWT) or human cDNA (HWT) sequence were generated using the Drosophila Gateway system. Transgenic flies bearing either construct could then be used to express fluorescently tagged fly or human Spastin under the spatiotemporal control of a promoter-GAL4 line. Numbers denote amino acid position. (B) en-GAL4 was used to drive expression of the constructs in larval epidermal cells to reveal their subcellular distribution. Anti-GFP immunostaining shows, in both cases, diffuse cytoplasmic signal with scattered aggregates. No expression is detected in the nucleus. (C) A single copy of wild-type Drosophila or human spastin driven by GS-elav-GAL4 in the spastin null background (genotypes DWT,Ø and HWT,Ø, respectively) induced pan-neuronal spastin transgene expression. Anti-GFP reveals cytoplasmic expression of both proteins in neuronal cell bodies as well as their processes, including the long photoreceptor axons (arrows).
To examine Spastin subcellular distribution in neurons, we induced transgene expression using the RU486 (mifepristone)-inducible pan-neuronal geneswitch elav-GAL4 driver (25,38) in a spastin null background. Although overexpression of Drosophila spastin via standard elav-GAL4 drivers is embryonic lethal (presumably due to excessive microtubule severing leading to neuronal cell death), spastin expression under the control of the inducible elav-GAL4 driver significantly rescues null defects (see below), indicating that expression occurs at physiologically relevant levels and times (25). Animals bearing the geneswitch elav-GAL4 driver, a single copy of the fly or human spastin transgene, and a homozygous deletion of endogenous Drosophila spastin (denoted as genotype ‘DWT,Ø’ for the fly transgene and ‘HWT,Ø’ for the human; Table 1), therefore expressed only transgenic Spastin in neurons, at levels induced by their ingestion of RU486. Subcellular protein distribution within neurons reflected that observed with overexpression in the epidermal cells. Both forms of Spastin distributed throughout neuronal cytoplasm, including into long, fine processes such as the photoreceptor axons of the CNS (Fig. 1C, arrows). Staining was also observed in the axons of the ventral nerve cord (VNC), although the signal was primarily within the nerve cord itself and could not be detected much more distally outside of the VNC.
Table 1.
List of fly genotypes used in this study and their abbreviations
| Abbreviation | Genotype |
|---|---|
| WCS (Control) | Canton S, w− |
| Ø,Ø (Null) | w; +/CyOKrGFP; elav-GS-Gal4, spastin5.75/spastin5.75 |
| DWT,Ø | w; UAS-CFP-Dspastin/+; elav-GS-Gal4, spastin5.75/spastin5.75 |
| HWT,Ø | w; UAS-Venus-HspastinWT/+; elav-GS-Gal4, spastin5.75/spastin5.75 |
| HL44,Ø | w; UAS-CFP-HspastinL44/+; elav-GS-Gal4, spastin5.75/spastin5.75 |
| HR388,Ø | w; UAS-Venus-HspastinR388/+; elav-GS-Gal4, spastin5.75/spastin5.75 |
| HWT,HWT (Wild-Type) | w; UAS-Venus-HspastinWT, UAS-Venus-HspastinWT/+; elav-GS-Gal4, spastin5.75/spastin5.75 |
| HWT,HL44 (S44L Heterozygote) | w; UAS-Venus-HspastinWT, UAS-CFP-HspastinL44/+; elav-GS-Gal4, spastin5.75/spastin5.75 |
| HWT,HR388 (K388R Heterozygote) | w; UAS-Venus-HspastinWT, UAS-Venus-HspastinR388/+; elav-GS-Gal4, spastin5.75/spastin5.75 |
| HL44,HR388 (S44L, K388R Compound Het) | w; UAS-CFP-HspastinL44, UAS-Venus-HspastinR388/+; elav-GS-Gal4,spastin5.75/spastin5.75 |
Canton S, w−, the control genotype, is a wild-type Drosophila strain that has been backcrossed to white− ten times (39); spastin5.75 is the Drosophila spastin null allele (25). Additional genotypes were analyzed in which HQ45 was substituted for HL44, and HR431STOP for HR388. D denotes Drosophila transgenes; H, human transgenes.
Human and Drosophila Spastin are functionally conserved
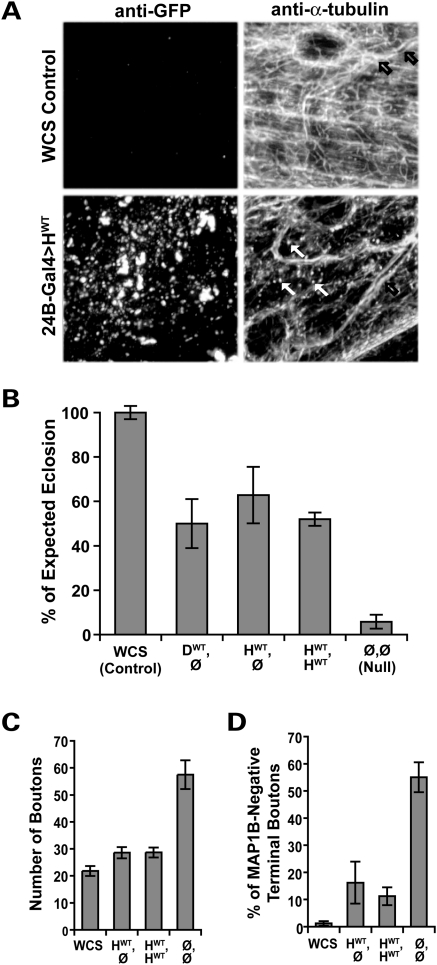
To determine whether human Spastin is functional in the context of the fly, we overexpressed the human transgene in larval muscles using the 24B-GAL4 driver line. Our earlier work demonstrated that overexpression of Drosophila spastin in these cells leads to severe disruption of the microtubule network (25). Similar to those results, overexpressed human Spastin causes a mixed pattern of filamentous and punctate microtubule staining that is noticeably reduced in overall intensity compared with controls [WCS, or Canton S, w1118; Table 1 and (39); Fig. 2A]. Punctate α-tubulin signal, consistent with fragmentation of the microtubule array, is prevalent in muscles where Spastin is overexpressed (Fig. 2A, arrows, bottom panel), and rarely observed in control muscles (top panel) or the overlying tracheal walls (open arrows, right panels). Human Spastin overexpression therefore disrupts the Drosophila microtubule network.
Figure 2.
Human and Drosophila Spastin are functionally equivalent. (A) Microtubules in the body wall muscles of wild-type third-instar Drosophila larvae were visualized using an antibody against α-tubulin, revealing a dense filamentous mesh throughout the tissue (top right). Microtubules in the walls of the trachea overlying the muscle are also seen (black arrows). Muscle-specific overexpression of human wild-type (HWT) Spastin revealed by anti-GFP (bottom left) causes an overall reduction in the intensity of α-tubulin staining (bottom right), as well as a profusion of punctae (white arrows). The punctate microtubule signal is distinct from the pattern of GFP expression, and likely reflects fragments resulting from the severing activity of the overexpressed human Spastin. (B) Pan-neuronal expression of fly or human spastin in the fly spastin null background rescues eclosion rates equivalently. Flies deleted for spastin (Ø,Ø) eclose successfully only ∼6% of the time, compared with WCS control flies, which nearly always eclose. GS-elav-GAL4-driven expression of a single copy of the fly or human spastin transgene (genotypes DWT,Ø and HWT,Ø, respectively) restores eclosion to over 50%, as does two copies of the human transgene (HWT,HWT; P < 5 × 10−6 for all groups compared with nulls, P > 0.4 between all transgenic lines). Similar results were observed for multiple independent insertions of the HWT transgene (Supplementary Material, Table S1). (C) At the cellular level, increased synaptic bouton number caused by loss of spastin is almost fully rescued by neuronal expression of the wild-type human transgene. Bouton number is 2.5-fold greater in spastin nulls (Ø,Ø) compared with WCS controls, and doubled compared with animals expressing human spastin (see also Fig. 3E). (D) Correspondingly, expression of human spastin significantly restores the penetration of microtubules into terminal synaptic boutons, such that only 16% (HWT,Ø) and 11% (HWT,HWT) are devoid of detectable MAP1B staining compared with >50% in nulls (P < 2 × 10−4 for either rescued genotype). WCS animals have <2% of terminal boutons lacking microtubules. Averages in this and subsequent figures are mean ± s.e.; P-values were calculated by one-way ANOVA; N's are detailed in the Methods.
We next addressed whether the human spastin gene functions closely enough to the fly gene to rescue defects observed in the nulls. Adult flies lacking Spastin rarely eclose, or emerge from their pupal cases (25). At least one reason for this may be motor weakness, as abdominal movement is required for flies to exit their pupae and the flies that fail to eclose appear fully developed. The few escapers have difficulty walking and jumping, cannot fly, and are short-lived. In addition, although the crawling behavior of larval-stage animals is grossly normal, clear morphological and cell biological phenotypes are observed at the larval neuromuscular junction (NMJ). Synaptic boutons, formed by motoneurons contacting their body wall muscle targets, are nearly doubled in number, reduced in size and exhibit a clustered rather than linear, wild-type distribution. Correlated with this defect, microtubule-associated protein 1B (MAP1B)-positive microtubules are sparse to non-existent in the most distal synaptic boutons, rather than forming the distinctive loop structures that can be seen in wild-type terminal boutons (25).
We used each of these four parameters—adult motor behavior, eclosion rate, larval NMJ bouton number and microtubule distribution within terminal boutons—to compare the ability of the fly and human spastin transgenes to rescue the null phenotypes. Pan-neuronal expression of fly wild-type spastin (DWT) in the null background (Ø), or genotype DWT,Ø, significantly rescued average eclosion rates from <6% in nulls to ∼50% of the number of expected flies recovered (Fig. 2B). This level of rescue was comparable to that attained by RU-induced neuronal expression of a Drosophila spastin cDNA construct (27). Higher levels of Spastin expression, achieved by increasing either transgene dosage by recombining two independent insertion lines (genotype ‘DWT,DWT’) or increasing RU486 concentration in the food, proved deleterious to eclosion. This suggests that the incomplete rescue of eclosion was because the pan-neuronal driver does not precisely mimic the spatiotemporal pattern of endogenous Spastin expression, rather than due to insufficient levels of expression. Nevertheless, rescued flies were noticeably improved in behavior compared with sibling spastin nulls, exhibiting the ability to fly, jump and crawl comparably to wild-type flies [(25), and data not shown].
Equally effective rescue of the eclosion phenotype was achieved when a single copy of the human wild-type spastin gene (HWT) was expressed in place of the fly transgene (genotype HWT,Ø; Fig. 2B). Although direct comparison of expression levels showed an up to 2-fold difference (presumably due to position effects, or differential transcription attributable to insertion location in the genome), four independent HWT insertion lines yielded comparable degrees of rescue that showed no relation to expression level (Supplementary Material, Fig. S1 and Table S1). Recombination of independent insertions to express two copies of the human gene (‘HWT,HWT’), mimicking the human wild-type genotype, also conferred a similar level of rescue (Fig. 2B). These animals were therefore somewhat insensitive to the absolute levels of human Spastin being overexpressed, although further increases in RU486 concentration increased lethality. A single copy of exogenous fly spastin (DWT,Ø), human spastin (HWT,Ø) or two copies of human spastin (HWT,HWT) therefore all rescued the fly null eclosion phenotype equivalently.
Detailed analysis of the cell biology of the NMJ in these animals was performed by confocal imaging of filleted larvae immunostained with antibodies against horseradish peroxidase (HRP) and the fly ortholog of the microtubule-associated protein MAP1B, Futsch (40,41). Anti-HRP delineates neuronal membranes, whereas anti-Futsch/MAP1B is uniquely advantageous for the visualization of neuronal microtubules at the NMJ since its neuronal specificity eliminates what would be a much stronger signal from the dense microtubule network in underlying muscles. In wild-type animals MAP1B staining appears as continuous bundles throughout the motor axons and into synapses, gradually tapering as the synaptic branches narrow. The signal ultimately terminates in each distal-most bouton in patterns ranging from a clear, bundled microtubule loop to diffuse or undetectable staining, thus providing a sensitive measure of microtubule distribution within synaptic boutons.
We found that the cell biological phenotypes of the NMJ synapses were, as in the eclosion assays, much closer to wild-type than nulls in flies expressing either a single copy or two copies of human spastin in the null background (HWT,Ø or HWT,HWT; Figs 2C, D and 3E). HWT expression reduced synaptic bouton numbers to near-wild-type levels (Fig. 2C) and alleviated the small and clustered bouton morphology that occurs in the absence of spastin (Fig. 3E). Quantification of the percentage of terminal boutons that lacked MAP1B signal also revealed dramatic rescue of microtubule distribution by HWT expression (Figs 2D and 3E). All but 2% of wild-type terminal boutons exhibited MAP1B immunostaining, but in spastin nulls 55% of terminal boutons were devoid of MAP1B signal, with microtubule loss most prevalent in the abnormally small terminal boutons. Neuronal expression of one or two copies of human spastin in the absence of the endogenous fly gene significantly restored microtubule distribution in terminal boutons, to 16 and 11%, respectively. Human spastin thus rescued fly null phenotypes at the cell biological level even more effectively than at the level of eclosion. Furthermore, the degree of rescue was equivalent to that achieved by neuronal expression of transgenic fly spastin (25).
Figure 3.
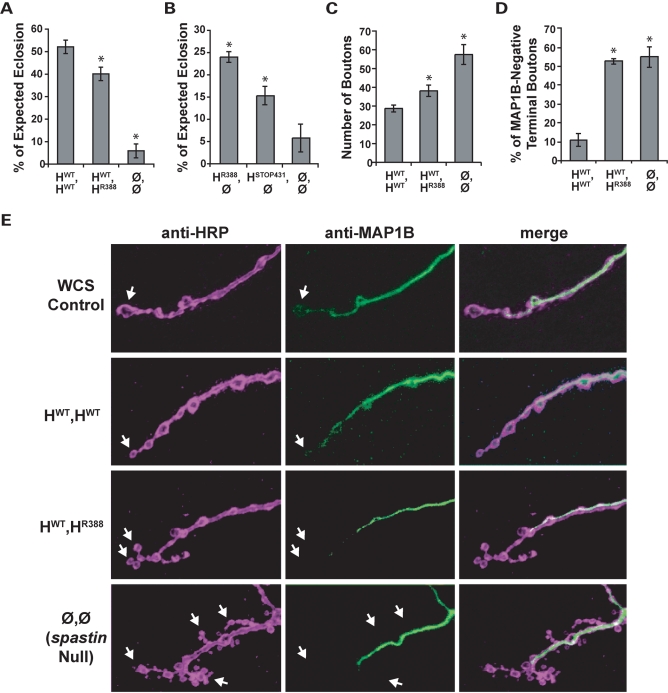
Heterozygous expression of R388 catalytic mutant Spastin causes mild loss of function phenotypes. (A) Flies mimicking the AD-HSP genotype, HWT,HR388, eclose less frequently compared with ‘wild-type’ HWT,HWT flies, but are still much healthier than nulls (Ø,Ø). *, P < 2 × 10−3 relative to wild-type. (B) Spastin retains significant function even in the likely absence of ATPase activity. Four times as many flies survive to adulthood when the R388 mutant protein is expressed alone in the spastin null background (HR388,Ø; P < 6 × 10−5 compared with nulls). STOP431, a mutant lacking over half of the catalytic domain, also confers significant rescue of eclosion to over 2-fold more than nulls (P < 8 × 10−3). (C) Larval NMJ bouton number is increased in R388 heterozygous mutants compared with wild-type controls (P < 0.02), although not as severely as in nulls (P < 2 × 10−4). (D) Microtubule distribution in terminal boutons is depleted in both HWT,HR388 heterozygotes and spastin nulls (P < 6 × 10−6 compared with HWT,HWT controls). (E) Representative synaptic bouton arbors from muscle 4 of third-instar larvae immunostained with anti-HRP (left, purple), which delineates the neuronal cell membrane, and anti-Futsch (center, green), which recognizes the Drosophila ortholog of MAP1B. WCS control boutons are linearly arrayed and MAP1B signal is typically detected throughout the arbor, including into terminal boutons where the microtubules form bundled loops (arrow). Boutons in animals deleted for spastin (‘Null’), in contrast, are highly clustered, smaller and more numerous, and in most cases lack distal MAP1B-positive microtubules. Expression of wild-type human spastin in the spastin null background (HWT,HWT) restores bouton size, linearity and microtubule penetration towards the WCS control phenotype. Expression of one copy each of wild-type and R388 mutant spastin (HWT,HR388) causes a mild loss of function phenotype, with more clustered boutons that often lack MAP1B signal in comparison to controls.
Generation and characterization of ‘AD-HSP’ flies
Having established that human spastin functions equally as well as fly spastin to rescue both behavioral and cellular null phenotypes, we proceeded to generate flies that genetically mimic human AD-HSP, bearing one copy of wild-type human spastin and one copy encoding the K388R catalytic domain mutation (genotype HWT,HR388). We found, in our eclosion assays, that only ∼75% as many HWT,HR388 heterozygotes emerged from their pupal cases as compared with HWT,HWT genotype flies (Fig. 3A). These transgenic lines all expressed spastin at similar levels (Supplementary Material, Fig. S1 and Table 2), indicating that the reduction in eclosion was due to the R388 mutation affecting spastin function. Adult HWT,HR388 flies were visibly compromised in movement, including walking rate, jumping and flying (Supplementary Material, Movie S1). These behavioral defects were similar to those observed in spastin nulls, although much milder in severity.
Table 2.
Summary of human spastin transgenic lines and relative expression levels
| Figure | Panel | Genotypes (line #) assayed | Relative spastin expression | Comments |
|---|---|---|---|---|
| 2 | B–D | HWT(4),Ø | 1X | Expression levels between these control genotypes (HWT,Ø versus HWT,HWT) differ, but do not cause phenotypic consequences. |
| HWT(4),HWT(6) | 1X, 1X (=2X) | |||
| 3 | A and D | HWT(4),HWT(6) | 1X, 1X | Expression levels are equivalent between genotypes. Differences are therefore due to a deleterious effect of R388. |
| HWT(4),HR388(6) | 1X, 1X | |||
| B | HR388(6),Ø | 1X | Dosage may account for difference in degree of rescue conferred between R388 and STOP431. | |
| HSTOP431(9),Ø | 0.5X | |||
| C | HWT(4),HWT(6) | 1X, 1X | One copy of HWT rescues as well as two (see Fig. 2B). HWT,HR388 defects are therefore due to the R388 mutation rather than reduced expression. | |
| HWT(4),HR388(4) | 1X, 0.5X | |||
| 4 | A | HWT(4),HWT(6) | 1X, 1X* | *No differences observed despite 1X more expression in HL44. |
| HWT(6),HL44(6) | 1X, 2X* | |||
| HL44(6),Ø | 2X | |||
| HWT(4),HR388(6) | 1X, 1X** | **See Figure 3A and D comment. | ||
| HL44(6),HR388(6) | 2X, 1X*** | ***Significantly worse than HWT,HR388 despite 1X more of HL44, which functions equivalently to HWT and should therefore be alleviatory. Deleterious effects of R388 are therefore exacerbated by L44. | ||
| B and C | HWT(4 or 6),Ø | 1X* | *No differences observed despite fold-differences in expression. | |
| HWT(4),HWT(6) | 1X, 1X* | |||
| HWT(6),HL44(6) | 1X, 2X* | |||
| HWT(4),HR388(6) | 1X, 1X** | **See Figure 3A and D comment. | ||
| HL44(6),HR388(6) | 2X, 1X*** | *** Significantly worse than HWT,HR388 despite 1X more of HL44, which functions equivalently to HWT and should therefore be alleviatory. Deleterious effects of R388 are therefore exacerbated by L44. | ||
| D | HWT(4),HWT(6) | 1X, 1X* | *Difference may be due to additional HL44 expression or to the mutation itself. | |
| HWT(6),HL44(6) | 1X, 2X* | |||
| HWT(4),HR388(4) or HWT(4),HR388(6) | 1X,0.5X or 1X,1X** | **Both R388 insertions yield significant differences versus HWT,HWT, regardless of reduced or equivalent expression levels, supporting a deleterious effect caused by the mutation. | ||
| HL44(6),HR388(1) or HL44(6),HR388(6) | 2X, N.D. or 2X,1X*** | ***Significantly worse than HWT,HR388 for all pair-wise combinations of recombinant lines, regardless of dosage (P < 0.001). Effects of R388 are therefore exacerbated by L44. | ||
| E | HWT(4),HWT(6) | 1X, 1X* | *L44 dose or mutation could underlie phenotype. | |
| HWT(6),HL44(6) | 1X, 2X* | |||
| HWT(4),HR388(6) | 1X, 1X** | **Significantly different from HWT,HWT despite equivalent expression levels. Differences are therefore due to a deleterious effect of R388. | ||
| HL44(6),HR388(6) | 2X, 1X*** | ***Different dosage as HWT,HR388, but equivalent phenotype. Same dosage but different phenotype versus HWT,HL44. Phenotypes correlate with genotype and not dosage. | ||
| 5 | A and B | HWT(4),Ø | 1X | Reduced dosage of HQ45 alone may account for differences from HWT. However, HQ45 still rescues as well or better than HWT,HR388, and exacerbates the effects of HR388, consistent with genotype, and not dosage, underlying observed phenotypes. |
| HQ45(2),Ø | 0.75X | |||
| HWT(4),HR388(6) | 1X, 1X | |||
| HQ45(2),HR388(6) | 0.75X, 1X |
A total of 11 different transgenic lines expressing human spastin were used in these studies. Specific insertion lines (number denoted in parentheses) of the recombinant strains used in each experiment are listed, together with the estimated spastin expression level for each transgene, normalized to line 4 of HWT (Supplementary Material, Fig. S1). Comparison between genotypes indicates that mutant phenotypes cannot be accounted for by differences in spastin dosage.
N.D., Not determined.
Surprisingly, even flies expressing only a single copy of the R388 mutant spastin, HR388,Ø, exhibited a significantly enhanced eclosion rate of over 4-fold greater than nulls (Fig. 3B). We tested this further using another spastin allele bearing a catalytic domain mutation, the nonsense mutant R431STOP. Although predicted to lack more than half of the AAA cassette including the functionally important Walker B motif and pore loops 2 and 3, neuronal expression of this truncated protein in the spastin null background enhanced eclosion over 2-fold (Fig. 3B). While neither the STOP431 nor R388 mutants completely deletes the AAA cassette and therefore could, hypothetically, retain very low levels of microtubule severing activity in vivo, in vitro experiments using point mutants in analogous regions of Drosophila Spastin indicate that they are devoid of catalytic activity (5,8). These results thus support the idea that Spastin has important function(s) beyond its ability to catalyze microtubule severing, which are sufficient to confer low levels of rescue.
HWT,HR388 animals also exhibited phenotypic similarities to spastin nulls at the cellular level. Synaptic bouton number in HWT,HR388 mutants showed a modest but significant increase compared with HWT,HWT animals (Fig. 3C). As in nulls, these boutons tended to be small and often exhibited a clustered distribution, rather than the linear arrangement of boutons seen in WCS and HWT,HWT controls (Fig. 3E). Furthermore, over 50% of the terminal synaptic boutons in HWT,HR388 larvae lacked detectable MAP1B staining, 5-fold more frequently than observed in HWT,HWT and indistinguishable from the null phenotype (Fig. 3D and E). The R388 catalytic domain mutation thus acted dominantly in these flies to cause subcellular (microtubule), cellular (synapse morphology) and behavioral (eclosion and motor ability) loss of function phenotypes reminiscent of spastin nulls. Furthermore, although not strictly representative of a human pathogenic mutation, parallel experiments using the R431STOP mutant yielded similar results (Supplementary Material, Fig. S2). Our observations of reduced terminal bouton microtubules, abnormal synapse morphology and compromised motor behavior in heterozygous spastin mutants is therefore consistent with the neurodegenerative consequences of heterozygous spastin catalytic domain mutations in humans, supporting the validity of these flies as a model of AD-HSP.
Use of the fly ‘AD-HSP’ model to test the S44L intragenic modifier hypothesis
The previous data illustrates the ability of our human transgene Drosophila model to recapitulate salient features of the AD-HSP disease phenotype. We next employed this model to investigate genotype–phenotype correlations uncovered in human AD-HSP families, for which there has been little or no molecular confirmation.
The variable onset age and penetrance of AD-HSP is poorly understood. No correlation has been found between disease severity and specific mutations that disrupt the catalytic domain (42). Another plausible cause of disparate penetrance is the occurrence of secondary mutations that either directly or indirectly affect Spastin function. Only one such class has been identified to date: one of two neighboring mutations in the amino terminus of Spastin itself.
Pedigree analyses have revealed that the consequences of spastin catalytic domain mutations are likely exacerbated by the presence of either the S44L or P45Q missense variant in trans (19,20,22,43,44). L44 and Q45 are themselves silent polymorphisms, as persons heterozygous for either (but otherwise wild-type for spastin) are asymptomatic. When either polymorphism is present in the compound heterozygous state with a mutation affecting the AAA ATPase domain, however, disease onset age is also dramatically decreased, and disease severity increased. Although reported for several pedigrees, a direct test of this intragenic modifier hypothesis has not been possible. The AD-HSP model flies enabled us to perform such a test, by examining an allelic series of genotypes ranging from flies expressing the amino terminal mutations alone to those in trans with the K388R catalytic mutation.
Consistent with the AD-HSP family studies, as well as expression studies showing that this region of the Spastin amino terminus is unnecessary for microtubule association and severing (5), we found that flies carrying one wild-type copy of human spastin and one encoding the L44 missense allele (genotype HWT,HL44) were unaffected in eclosion and adult behavior. HWT,HL44 flies eclosed as frequently and survived for as many days as those carrying two wild-type copies (Fig. 4A–C). Indeed, animals bearing a single copy of the L44 mutant spastin alone (genotype HL44,Ø) also eclosed comparably, suggesting that even with this amino-terminus mutation the protein retains near-normal function. Expression of only R388 mutant spastin (HR388,Ø), in contrast, rescued eclosion only half as well as L44 alone (Fig. 3B).
Figure 4.
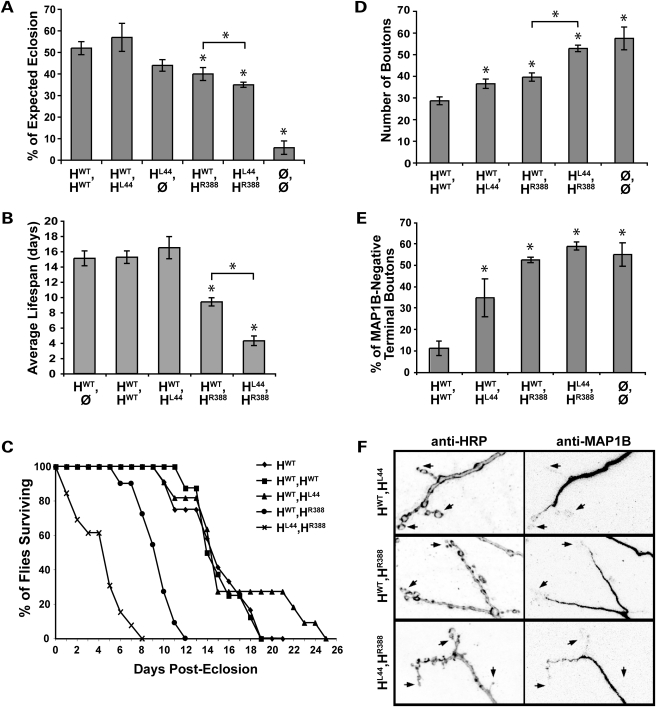
The amino-terminus S44L mutation exacerbates R388 catalytic domain mutant phenotypes but is itself only mildly deleterious. (A) L44 expressed heterozygously (HWT,HL44) or alone (HL44,Ø) rescues eclosion as effectively as one or two copies of the wild-type transgene, HWT (P > 0.1). Co-expression of R388 and L44 slightly but significantly lowers eclosion compared with the HWT,HR388 genotype, consistent with L44 being largely silent in combination with wild-type Spastin, but deleterious when in trans with a catalytic domain mutant in Spastin (P < 0.02). (B, C) Post-eclosion, survival of adult flies is dramatically affected by Spastin genotype. Compared with controls, HWT,HR388 heterozygotes survive on average only about half as long, and HL44,HR388 compound heterozygotes survive less than half as well as those (P < 9 × 10−6). No deleterious effect of the L44 allele is seen in HWT,HL44 heterozygotes compared with wild-type, however. (D, E) HWT,HL44 larvae show moderate but significant increases in bouton number and the percentage of terminal boutons lacking MAP1B compared with HWT,HWT controls, indicating that L44 is somewhat deleterious at the cellular level (P < 0.01 for both parameters). When in trans with R388, however, L44 strongly enhances the R388 loss of function phenotype, to levels equivalent to spastin nulls (P > 0.3). Bouton number is significantly greater in the compound versus single mutants (P < 6 × 10−7), although both genotypes are deficient in terminal bouton microtubules, similar to nulls. (F) Representative synaptic bouton arbors with their MAP1B-positive microtubule distribution are shown for the different ‘AD-HSP’ genotypes. Presynaptic arbors in HWT,HL44 animals resemble wild-type, with large, round terminal boutons (e.g. arrows) sometimes penetrated by microtubules. HWT,HR388 larvae exhibit smaller, more clustered boutons often devoid of microtubule staining, and HL44,HR388 synapses are similarly, but much more severely, affected.
Despite rescuing eclosion and survival equivalently as well as wild-type spastin, the L44 mutation was not silent at the cellular and subcellular levels, even when present heterozygously with wild-type spastin (Fig. 4D and E). Bouton number in HWT,HL44 larvae was slightly greater than in HWT,HWT larvae, as was the percentage of terminal boutons lacking microtubules (Fig. 4D–F). In fact, neither bouton number nor microtubule distribution differed significantly between L44 and R388 heterozygotes. Although L44 heterozygotes expressed higher levels of Spastin than R388 heterozygotes (Table 2 and Supplementary Material, Fig. S1), it is unlikely that their cellular phenotypes were due simply to excessive overexpression of functional Spastin, which causes fewer, rather than more, synaptic boutons (25).
We next tested whether the L44 mutation exacerbates the R388 mutant phenotype, by comparing compound heterozygotes of both mutations (genotype HL44,HR388) to animals bearing only the catalytic mutation (genotype HWT,HR388). HL44,HR388 eclosion was modestly, but significantly, lower than in HWT,HR388 animals (Fig. 4A). Post-eclosion, behavioral differences were dramatic. Surviving HL44,HR388 flies exhibited much weaker mobility than HWT,HR388 flies, comparable to spastin nulls (Supplementary Material, Movie S1). In addition, although average lifespan and survival rates were unaffected by the L44 heterozygous mutation, compound heterozygotes showed a >50% reduction in average lifespan compared with HWT,HR388 flies, which in turn survived only half as long as controls (HWT,HWT; Fig. 4B and C).
Examination of bouton number confirmed the increased phenotypic severity of the HL44,HR388 genotype compared with HWT,HR388 (Fig. 4D and F). Although HWT,HL44 and HWT,HR388 larvae had similar average bouton numbers, HL44,HR388 larvae had nearly 30% more boutons than either and did not differ significantly from nulls. Nearly 70% more terminal boutons were devoid of detectable microtubules in HL44,HR388 compared with HWT,HL44 larvae (Fig. 4E and F). No difference was seen between HWT,HR388 and HL44,HR388 microtubule distribution, however. Both were already indistinguishable from the null phenotype, which may represent the upper limit of this measure.
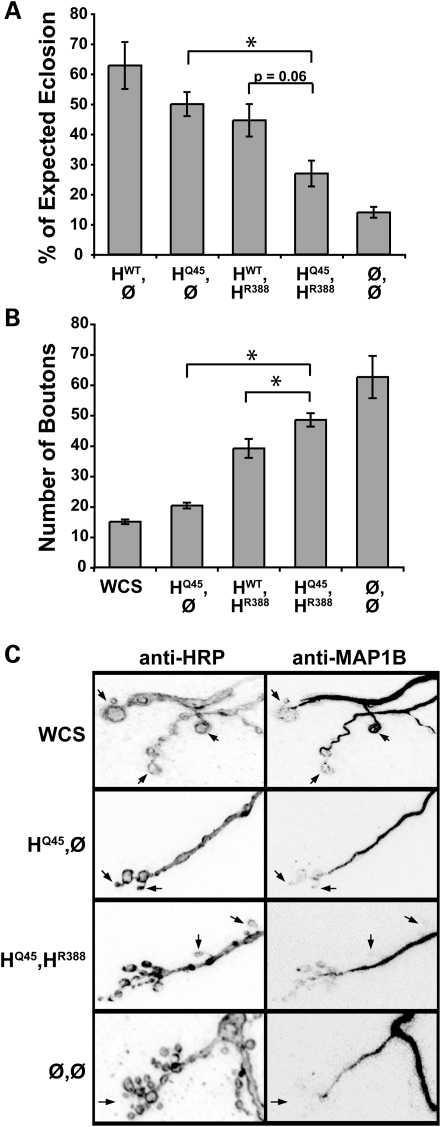
We repeated the above analysis using the neighboring polymorphism Q45 in place of L44; P45Q is also thought to intragenically modify catalytic domain mutations in Spastin, although fewer cases are reported. As for L44, expression of Q45 alone in the spastin null background conferred rescue of eclosion to levels approaching that of wild-type, and over three times the frequency of nulls (Fig. 5A). In support of Q45 acting deleteriously when in combination with R388, HQ45,HR388 compound heterozygotes eclosed only ∼60% as well as HWT,HR388 mutants, although this reduction was just short of statistical significance (P = 0.06). However, synapse morphology, bouton number and microtubule distribution also supported the predicted progression of phenotypic severity (Fig. 5B and C). HQ45,HR388 animals had small, clustered, microtubule-deficient boutons that numbered ∼25% more per muscle than HWT,HR388 animals, approaching the average bouton number and morphology observed in spastin nulls.
Figure 5.
Like L44, Q45 enhances R388 mutant effects in trans. (A) Expression of the Q45 mutation alone in spastin null flies (HQ45,Ø) rescues eclosion to the same level as HWT (P > 0.2). In contrast, ∼40% fewer HQ45,HR388 compound heterozygotes eclose compared with HWT,HR388 single mutants, although for the numbers tested this effect did not achieve a P-value of <0.05 (P = 0.06). Q45 may therefore be a weaker intragenic modifier of catalytic mutations compared with L44. (B, C) Bouton number in HQ45,HR388 compound heterozygotes is increased, similar to nulls (P > 0.07), and significantly greater than in single heterozygotes (HWT,HR388 or HQ45,Ø; P < 0.03). Expression of the HQ45 mutation alone in the null background restores bouton number and microtubule distribution to near wild-type levels. Arrows in (C) denote examples of terminal boutons.
Taken together, these data provide a direct demonstration of intragenic modification by the S44L and P45Q amino-terminus mutations on a catalytic domain mutation in Spastin. The statistically weaker modulation by Q45 may reflect actual functional differences between S44 and P45, differences in expression level, or merely greater noise in this set of experiments. Nevertheless, we find that as in humans, these amino-terminus mutations are effectively silent at the level of motor behavior in heterozygotes. At the cellular level, however, our data suggest that there are still some consequences to the neuron caused by these non-catalytic domain mutations, particularly L44.
DISCUSSION
Towards understanding the mechanisms by which spastin mutations lead to neuronal dysfunction in AD-HSP and uncovering possible therapeutic approaches for the disease, we have generated a model in which flies deleted for endogenous spastin instead express the wild-type or mutated human ortholog in allelic combinations that mimic the human disease genotype. Using these animals we show that human spastin is functionally equivalent to that of the fly, rescuing the null phenotype as effectively as the fly gene when expressed at low levels in neurons. At the cellular level, rather than exhibiting small, clustered synaptic boutons containing little or no MAP1B-positive microtubules, NMJs in animals expressing human spastin in place of the fly's exhibit large, linearly arrayed boutons containing distinct microtubule loop structures, like those seen in wild-type controls or in spastin null flies expressing an exogenous wild-type Drosophila spastin transgene. Behaviorally, eclosion rates and adult mobility are also visibly and equivalently improved by the expression of human or fly spastin (Fig. 2; Supplementary Material, Movie S1).
Consistent with their functional conservation, both the fly and human spastin proteins are diffusely distributed in the cytoplasm and can permeate throughout long neuronal processes. Overexpression does reveal some differences between the two proteins, however. Human Spastin forms more aggregates than does fly Spastin; this is observed independent of the transgene insertion, and whether a genomic or cDNA fly transgene is used [Fig. 1; Supplementary Material, Fig. S1 and (25)]. Additionally, we find that two copies of the fly wild-type transgene become deleterious to rescue, although no significant differences are observed between one and two copies of the human transgene. Processing of these two proteins is therefore not identical in this overexpression context, although their ability to substitute for the loss of endogenous spastin is indistinguishable.
Although differences in transgene expression levels are one caveat of the GAL4-UAS expression system, analysis of the various insertion lines used in these experiments reveals that the mutant phenotypes we observed are relatively insensitive to expression levels, correlating instead with the different recombinant genotypes (Table 2). Pathogenic AD-HSP genotypes resemble spastin loss of function phenotypes at behavioral, cellular and subcellular levels. The ability of the human pathogenic mutations to mimic the cellular and subcellular spastin null phenotypes is even more remarkable given that this phenotype of numerous small, clustered boutons devoid of microtubules has not, to our knowledge, been observed in other mutants affecting the larval NMJ, including mutations in other closely related microtubule severing proteins (L.M.P. and N.T.S., unpublished data). This underscores the specificity of these phenotypes and their direct relation to Spastin function.
Our data support the validity of these flies as a model for the human disease, and furthermore as the only model to date demonstrating robust behavioral phenotypes in the heterozygous mutant condition that typifies most AD-HSP patients (Figs 3 and 4; Supplementary Material, Movie S1). Using this model we confirm human pedigree analyses correlating disease augmentation with trans-heterozygous expression of L44 or Q45 and mutations interfering with Spastin's catalytic domain, substantiating the significance of this amino-terminal region in normal Spastin function. Although humans heterozygous for the S44L mutation are typically asymptomatic, our studies suggest that the mutation may have some effect on protein function at the cellular and subcellular levels (Fig. 4). Consistent with this observation, abnormal electromyogram measurements have been found in L44 heterozygous individuals despite their outward lack of disease symptoms (22).
Several hypotheses have been proposed for the mechanism underlying exacerbated disease severity in the compound heterozygous condition. Modifier effects could occur at the level of the spastin protein; for instance, S44/P45 may be a phosphorylation target important in the regulation of Spastin function (19). S44 is also predicted to comprise part of a PEST sequence, such that mutations in it would interfere with degradation of full-length (but not truncated) Spastin isoforms (23). Alternatively, the base pair mutations underlying these amino acid substitutions fall within a predicted cryptic promoter in the first exon, and could affect its activity and inhibit the transcription of the shorter spastin isoform (45). Our data argue against a transcriptional mechanism, given that transgene expression in the AD-HSP model flies is under the control of an exogenous promoter. However, the precise mechanism of the exacerbating effect remains to be resolved, and can be directly tested in this model.
Surprisingly, catalytic domain mutants expressed in isolation also confer partial rescue of the null phenotype (Fig. 3). On the basis of in vitro and cell culture experiments these mutants are predicted to completely lack ATPase, and therefore microtubule severing, activity (5,8). If this is also the case in our transgenic animals, our data provide further evidence that Spastin has important functions in addition to microtubule severing that do not require its ATPase domain. Numerous in vitro experiments in which overexpressed catalytic domain mutants (including R388) associate with microtubules in a filamentous pattern suggest that Spastin bundles and/or stabilizes microtubules, in addition to severing them (4–6,11,13,24,46,47). Microtubule bundling is thought to be a key event in the growth phase of axons (48); therefore, some combination of bundling and severing may also be required to organize and enhance the establishment of the cytoskeleton in newly forming synaptic boutons.
The similarity in cellular phenotypes between spastin nulls and the ‘AD-HSP’ flies (namely, HWT,HR388, HL44,HR388 and HQ45,HR388) provides further support for the hypothesis that the cellular pathology underlying AD-HSP is a dearth of Spastin-mediated growth or transport of microtubules into the distal-most portions of the axon (25). Although this scenario is initially counterintuitive given that Spastin destroys microtubules in vitro and when highly overexpressed, increasing evidence supports the idea that microtubule severing proteins in many diverse contexts increase, rather than decrease, net microtubule mass (49,50). Loss of microtubule severing activity in such cases is therefore expected to lead to a reduction in microtubules, as we observe. Interestingly, even in vitro, addition of low concentrations of recombinant spastin protein to purified microtubules produces a high density of short microtubules, although a doubled concentration of the same protein causes microtubule destruction (46). Whether the net loss of microtubules in our disease model also causes axon transport defects, which are implicated in murine, and most recently, a bovine model of AD-HSP (51), will be determined.
A growing list of Drosophila models of HSP is contributing key insights to the function of the human spastic gait (SPG) genes: in addition to SPG4/spastin, these include SPG3A/atlastin, a novel GTPase shown in flies to be required for ER morphogenesis (52,53), SPG6/NIPA1, elucidated as a regulator of BMP signaling through studies of its Drosophila ortholog spicthyn (54) and SPG39/NTE, Drosophila Swiss-cheese, mutations in which disrupt the kinase activity and localization of PKA-C3 and cause progressive neurodegeneration (55). On the basis of our results we propose that the ‘AD-HSP’ flies presented here provide a highly tractable, relevant in vivo model that can be used to test additional hypotheses about the mechanisms underlying the disease, and for the development and screening of therapeutic approaches. Candidate therapies can be assayed in this model from the level of cell biology to the behavior of the animal, thus enabling a comprehensive understanding of the consequences of treatment vis a vis specific AD-HSP disease genotypes.
MATERIALS AND METHODS
Generation of spastin constructs
UAS-genomic fly spastin
Full-length fly genomic DNA spanning from the first ATG to the 3′-UTR was amplified by standard PCR methods using the Phusion DNA polymerase kit (New England Biolabs) and template DNA isolated from a single wild-type white, Canton S fly. The primers used were, forward, 5′-CAC CAT GGT ACG CAC TAA AAA CCA GTC-3′ (with the start ATG in bold), and reverse, 5′-GTT TCT TGA AAT CGA TTT TAT TTA GC-3′. The genomic fragment was cloned into a fluorescently tagged UAS vector using the Invitrogen Gateway cloning system modified for Drosophila (Terence McNally, Carnegie Institute). Briefly, spastin clones were inserted into the directional TOPO® pENTR™ vector (Invitrogen) and confirmed by sequencing the insertion in its entirety. LR Clonase™ Enzyme Mix (Invitrogen) was used according to the supplier's instructions to then transfer the spastin clone into the destination vector of choice. The destination vectors, PTVW or PTCW, respectively, contained an N-terminal Venus YFP or CFP tag, preceded by the UASt promoter. The resultant clones were confirmed by sequencing the UAS site, fluorescent tag and the entire spastin insert.
UAS-human spastin cDNA
The 1.87 kb long, full-length human wild-type spastin cDNA (exons 1–17) and mutated variants encoding K388R (nucleotide 1288A->G) and S44L (nt 256 C->T) were each subcloned from constructs previously generated by I. Kotowski, into Drosophila Gateway destination vectors as described above for fly spastin. Mutations were induced in the original constructs via PCR-based mutagenesis. Primers used to amplify each spastin insert were, forward: 5′-CAC CAT GAA TTC TCC GGG TGG ACG AG-3′ (exon 1 ATG marked in bold) and reverse: 5′-CAG GTT TAC AAA GGT ATT TCC-3′ (ending 23 bp after the exon 17 stop codon). Constructs encoding the human K388R and S44L mutations were generated by similar methods.
Generation and characterization of transgenic lines
All UAS constructs were injected by The Bestgene Company into yw embryos to generate transgenic flies. Four to ten independent insertion lines were recovered for each construct. Each was mapped and balanced to establish a stable stock and crossed to the en-GAL4 driver to test for expression of the transgene; no obvious differences in en-GAL4-driven expression levels were observed between any of the insertion lines for a given construct. Although expression of Venus-tagged Spastin could be observed in live animals, visualization of CFP-tagged Spastin required immunohistochemistry using an anti-GFP antibody (Invitrogen); thus, anti-GFP immunostaining was employed for all experiments. Second chromosome insertions were used to generate recombinant genotypes. Further examination of expression levels using the twist, 24B-GAL4 driver to visualize expression in body wall muscles (in contrast to en-GAL4), revealed up to 2-fold differences in expression levels between insertions of the same gene (Supplementary Material, Fig. S1). This did not affect eclosion among wild-type lines (Supplementary Material, Table S1), but may account for differences observed in eclosion levels between HR388 lines 4 and 6 (Supplementary Material, Fig. S1 and Table S2). We therefore limited our use of eclosion data to line 6 of HR388 when comparing different allelic combinations (i.e. HR388 in trans with either HWT or HL44). No differences were observed in synaptic bouton numbers between different HR388 lines, however; therefore these data were combined (Supplementary Material, Table S3). Overall, analysis of the data revealed no correlation between the expression levels of the various transgenic lines utilized and observed phenotypic differences (Table 2).
Fly genetics
The wild-type control strain used in these experiments and referred to in the text as ‘WCS controls’ was white, Canton-S, a Canton S line that has been backcrossed to white 10 times (25,39). Promoter-GAL4 lines used to drive tissue-specific expression in this study were engrailed-GAL4, 24B-GAL4, twi-GAL4 and the RU486-inducible elav-GS-GAL4 (38,56). When using the elav-GS-GAL4 driver, RU-486 was added to the food at a concentration of 2.5 µg/ml, which induced optimal levels of neuronal wild-type UAS-spastin expression to rescue the null phenotype. We therefore employed these same conditions with the mutant UAS-spastin constructs. Flies bearing either one or two copies of the spastin transgene(s) on chromosome II and heterozygous for the spastin deletion [allele spastin5.75, (25)] on chromosome III were generated by standard genetic methods. These were crossed to elav-GS-GAL4/CyOKrGFP; spastin5.75/TM6B flies. Progeny containing the GAL4 driver in trans with the UAS-spastin transgene(s) on II, and homozygous for the spastin5.75 deletion on III, were compared with sibling flies carrying either the driver or the UAS transgene(s) but homozygous for the deletion (i.e. spastin nulls), to determine the level to which transgenic spastin could compensate for loss of the endogenous gene.
Larval immunocytochemistry
Third instar larvae were live-dissected along the dorsal midline in room temperature HL3 Ringers and fixed in Bouin's fixative (Sigma) for 10 min. Primary antibodies used on larvae were rabbit anti-HRP (1:100, Cappel), rabbit anti-GFP (1:500; Invitrogen), mouse mAb DM1A (anti-α-tubulin, 1:500; Sigma) and mouse mAb 22C10 (anti-Futsch, 1:50). 22C10, developed by S. Benzer, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242, USA. Staining was visualized with Alexa-Fluor 488 conjugated goat anti-rabbit and Alexa-Fluor 568 goat anti-mouse (1:200; Molecular Probes). Fluorescently labeled samples were imaged by acquiring z-series projections with a Zeiss (Oberkochen, Germany) 510 inverted confocal microscope using 63×/1.4 n.a. or 100×/1.2 n.a. PlanApo objectives. We quantified and imaged boutons of muscle 4, selected for the visibility of synapses localized on its dorsal surface. For consistency, only larval segments A2 and A3 were analyzed; however, observed phenotypes were seen throughout the fillet. All type Ib boutons (large, glutamatergic) were scored for each NMJ in the determination of bouton number. Microtubule distribution was quantified by counting the total number of terminal (end) boutons in each Ib arbor and determining the percentage lacking MAP1B signal. Each analysis included ∼15–30 muscles per experimental genotype, collected over a minimum of two separate trials for each experiment.
Behavior analysis
The percent of expected eclosion was calculated as the ratio of flies recovered for a given genotype divided by number predicted for that genotype, based on expected genetic ratios dictated by the cross and the total number of flies recovered. Typically, more than 200 flies were counted per genotype (totaling over 1200 flies per cross); a minimum of three trials was performed for each cross. Experiments determining motor behavior of flies used age-matched flies of different genotypes (∼3 d.o.). Flies for the lifespan experiments were collected/examined for survival daily, maintained on RU486-containing food in individual vials and transferred to fresh vials every 3 days. Average lifespan was calculated by multiplying the number of flies alive on a particular day by the number of days alive, adding the values for each day tested and dividing the sum by the total number of flies tested (n > 10 for each genotype).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Spastic Paraplegia Foundation Inc. (to N.T.S.) and the National Institutes of Health (NS063896 to N.T.S.).
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the Spastic Paraplegia Foundation and their generous donors, as well as H. Willard, T. Petes, V. Bennett and R. Wharton, whose support made this work possible. We also thank the Bloomington Stock Center, Developmental Studies Hybridoma Bank, and Terence Murphy at the Carnegie Institute for their excellent reagents, A. Roll-Mecak and R. Vale for provision of constructs in the early phase of this project and helpful discussions, E. Graham and D.S. Marchuk for expert technical assistance, the invaluable resource that is Flybase, and K. Zinn, D. Tracey, A. Schmid, Z.J. Yang, D. Sherwood, and members of the N. Sherwood lab for their insights and advice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Fink J.K. Hereditary spastic paraplegia. Neurol. Clin. 2002;20:711–726. doi: 10.1016/s0733-8619(02)00007-5. [DOI] [PubMed] [Google Scholar]
- 2.Beetz C., Nygren A., Schickel J., Auer-Grumbach M., Burk K., Heide G., Kassubek J., Klimpe S., Klopstock T., Kreuz F., et al. High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology. 2006;67:1926–1930. doi: 10.1212/01.wnl.0000244413.49258.f5. [DOI] [PubMed] [Google Scholar]
- 3.Fink J.K., Rainier S. Hereditary spastic paraplegia: spastin phenotype and function. Arch. Neurol. 2004;61:830–833. doi: 10.1001/archneur.61.6.830. [DOI] [PubMed] [Google Scholar]
- 4.Salinas S., Carazo-Salas R.E., Proukakis C., Schiavo G., Warner T.T. Spastin and microtubules: Functions in health and disease. J. Neurosci. Res. 2007;85:2778–2782. doi: 10.1002/jnr.21238. [DOI] [PubMed] [Google Scholar]
- 5.Roll-Mecak A., Vale R.D. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 2005;15:650–655. doi: 10.1016/j.cub.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 6.Evans K.J., Gomes E.R., Reisenweber S.M., Gundersen G.G., Lauring B.P. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 2005;168:599–606. doi: 10.1083/jcb.200409058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White S.R., Evans K.J., Lary J., Cole J.L., Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J. Cell Biol. 2007;176:995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roll-Mecak A., Vale R.D. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchison T., Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 10.Fonknechten N., Mavel D., Byrne P., Davoine C.S., Cruaud C., Bonsch D., Samson D., Coutinho P., Hutchinson M., McMonagle P., et al. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum. Mol. Genet. 2000;9:637–644. doi: 10.1093/hmg/9.4.637. [DOI] [PubMed] [Google Scholar]
- 11.Errico A., Ballabio A., Rugarli E.I. Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum. Mol. Genet. 2002;11:153–163. doi: 10.1093/hmg/11.2.153. [DOI] [PubMed] [Google Scholar]
- 12.McDermott C.J., Grierson A.J., Wood J.D., Bingley M., Wharton S.B., Bushby K.M., Shaw P.J. Hereditary spastic paraparesis: disrupted intracellular transport associated with spastin mutation. Ann. Neurol. 2003;54:748–759. doi: 10.1002/ana.10757. [DOI] [PubMed] [Google Scholar]
- 13.Errico A., Claudiani P., D'Addio M., Rugarli E.I. Spastin interacts with the centrosomal protein NA14, and is enriched in the spindle pole, the midbody and the distal axon. Hum. Mol. Genet. 2004;13:2121–2132. doi: 10.1093/hmg/ddh223. [DOI] [PubMed] [Google Scholar]
- 14.Orso G., Martinuzzi A., Rossetto M.G., Sartori E., Feany M., Daga A. Disease-related phenotypes in a Drosophila model of hereditary spastic paraplegia are ameliorated by treatment with vinblastine. J. Clin. Invest. 2005;115:3026–3034. doi: 10.1172/JCI24694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazan J., Fonknechten N., Mavel D., Paternotte C., Samson D., Artiguenave F., Davoine C.S., Cruaud C., Durr A., Wincker P., et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 16.Charvin D., Cifuentes-Diaz C., Fonknechten N., Joshi V., Hazan J., Melki J., Betuing S. Mutations of SPG4 are responsible for a loss of function of spastin, an abundant neuronal protein localized in the nucleus. Hum. Mol. Genet. 2003;12:71–78. doi: 10.1093/hmg/ddg004. [DOI] [PubMed] [Google Scholar]
- 17.Molon A., Montagna P., Angelini C., Pegoraro E. Novel spastin mutations and their expression analysis in two Italian families. Eur. J. Hum. Genet. 2003;11:710–713. doi: 10.1038/sj.ejhg.5201027. [DOI] [PubMed] [Google Scholar]
- 18.Riano E., Martignoni M., Mancuso G., Cartelli D., Crippa F., Toldo I., Siciliano G., Di Bella D., Taroni F., Bassi M.T., et al. Pleiotropic effects of spastin on neurite growth depending on expression levels. J. Neurochem. 2009;108:1277–1288. doi: 10.1111/j.1471-4159.2009.05875.x. [DOI] [PubMed] [Google Scholar]
- 19.Svenson I.K., Kloos M.T., Gaskell P.C., Nance M.A., Garbern J.Y., Hisanaga S., Pericak-Vance M.A., Ashley-Koch A.E., Marchuk D.A. Intragenic modifiers of hereditary spastic paraplegia due to spastin gene mutations. Neurogenetics. 2004;5:157–164. doi: 10.1007/s10048-004-0186-z. [DOI] [PubMed] [Google Scholar]
- 20.Chinnery P.F., Keers S.M., Holden M.J., Ramesh V., Dalton A. Infantile hereditary spastic paraparesis due to codominant mutations in the spastin gene. Neurology. 2004;63:710–712. doi: 10.1212/01.wnl.0000135346.63675.3e. [DOI] [PubMed] [Google Scholar]
- 21.Naimi M., Tardieu S., Depienne C., Ruberg M., Brice A., Dubourg O., Leguern E. Detection of genomic rearrangements by DHPLC: a prospective study of 90 patients with inherited peripheral neuropathies associated with 17p11.2 rearrangements. Am. J. Med. Genet. A. 2005;136:136–139. doi: 10.1002/ajmg.a.30790. [DOI] [PubMed] [Google Scholar]
- 22.McDermott C.J., Burness C.E., Kirby J., Cox L.E., Rao D.G., Hewamadduma C., Sharrack B., Hadjivassiliou M., Chinnery P.F., Dalton A., et al. Clinical features of hereditary spastic paraplegia due to spastin mutation. Neurology. 2006;67:45–51. doi: 10.1212/01.wnl.0000223315.62404.00. [DOI] [PubMed] [Google Scholar]
- 23.Schickel J., Pamminger T., Ehrsam A., Munch S., Huang X., Klopstock T., Kurlemann G., Hemmerich P., Dubiel W., Deufel T., et al. Isoform-specific increase of spastin stability by N-terminal missense variants including intragenic modifiers of SPG4 hereditary spastic paraplegia. Eur. J. Neurol. 2007;14:1322–1328. doi: 10.1111/j.1468-1331.2007.01971.x. [DOI] [PubMed] [Google Scholar]
- 24.Svenson I.K., Kloos M.T., Jacon A., Gallione C., Horton A.C., Pericak-Vance M.A., Ehlers M.D., Marchuk D.A. Subcellular localization of spastin: implications for the pathogenesis of hereditary spastic paraplegia. Neurogenetics. 2005;6:135–141. doi: 10.1007/s10048-005-0219-2. [DOI] [PubMed] [Google Scholar]
- 25.Sherwood N.T., Sun Q., Xue M., Zhang B., Zinn K. Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2004;2:e429. doi: 10.1371/journal.pbio.0020429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trotta N., Orso G., Rossetto M.G., Daga A., Broadie K. The hereditary spastic paraplegia gene, spastin, regulates microtubule stability to modulate synaptic structure and function. Curr. Biol. 2004;14:1135–1147. doi: 10.1016/j.cub.2004.06.058. [DOI] [PubMed] [Google Scholar]
- 27.Connell J.W., Lindon C., Luzio J.P., Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushita-Ishiodori Y., Yamanaka K., Ogura T. The C. elegans homologue of the spastic paraplegia protein, spastin, disassembles microtubules. Biochem. Biophys. Res. Commun. 2007;359:157–162. doi: 10.1016/j.bbrc.2007.05.086. [DOI] [PubMed] [Google Scholar]
- 29.Wood J.D., Landers J.A., Bingley M., McDermott C.J., Thomas-McArthur V., Gleadall L.J., Shaw P.J., Cunliffe V.T. The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum. Mol. Genet. 2006;15:2763–2771. doi: 10.1093/hmg/ddl212. [DOI] [PubMed] [Google Scholar]
- 30.Tarrade A., Fassier C., Courageot S., Charvin D., Vitte J., Peris L., Thorel A., Mouisel E., Fonknechten N., Roblot N., et al. A mutation of spastin is responsible for swellings and impairment of transport in a region of axon characterized by changes in microtubule composition. Hum. Mol. Genet. 2006;15:3544–3558. doi: 10.1093/hmg/ddl431. [DOI] [PubMed] [Google Scholar]
- 31.Kasher P.R., De Vos K.J., Wharton S.B., Manser C., Bennett E.J., Bingley M., Wood J.D., Milner R., McDermott C.J., Miller C.C., et al. Direct evidence for axonal transport defects in a novel mouse model of mutant spastin-induced hereditary spastic paraplegia (HSP) and human HSP patients. J. Neurochem. 2009;110:34–44. doi: 10.1111/j.1471-4159.2009.06104.x. [DOI] [PubMed] [Google Scholar]
- 32.Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- 33.Bilen J., Bonini N.M. Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 34.Marsh J.L., Thompson L.M. Drosophila in the study of neurodegenerative disease. Neuron. 2006;52:169–178. doi: 10.1016/j.neuron.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 35.Claudiani P., Riano E., Errico A., Andolfi G., Rugarli E.I. Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp. Cell Res. 2005;309:358–369. doi: 10.1016/j.yexcr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Solowska J.M., Morfini G., Falnikar A., Himes B.T., Brady S.T., Huang D., Baas P.W. Quantitative and functional analyses of spastin in the nervous system: implications for hereditary spastic paraplegia. J. Neurosci. 2008;28:2147–2157. doi: 10.1523/JNEUROSCI.3159-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 38.Osterwalder T., Yoon K.S., White B.H., Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon A.F., Shih C., Mack A., Benzer S. Steroid control of longevity in Drosophila melanogaster. Science. 2003;299:1407–1410. doi: 10.1126/science.1080539. [DOI] [PubMed] [Google Scholar]
- 40.Hummel T., Krukkert K., Roos J., Davis G., Klambt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- 41.Roos J., Hummel T., Ng N., Klambt C., Davis G.W. Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron. 2000;26:371–382. doi: 10.1016/s0896-6273(00)81170-8. [DOI] [PubMed] [Google Scholar]
- 42.Yip A.G., Durr A., Marchuk D.A., Ashley-Koch A., Hentati A., Rubinsztein D.C., Reid E. Meta-analysis of age at onset in spastin-associated hereditary spastic paraplegia provides no evidence for a correlation with mutational class. J. Med. Genet. 2003;40:e106. doi: 10.1136/jmg.40.9.e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crippa F., Panzeri C., Martinuzzi A., Arnoldi A., Redaelli F., Tonelli A., Baschirotto C., Vazza G., Mostacciuolo M.L., Daga A., et al. Eight novel mutations in SPG4 in a large sample of patients with hereditary spastic paraplegia. Arch. Neurol. 2006;63:750–755. doi: 10.1001/archneur.63.5.750. [DOI] [PubMed] [Google Scholar]
- 44.Depienne C., Tallaksen C., Lephay J.Y., Bricka B., Poea-Guyon S., Fontaine B., Labauge P., Brice A., Durr A. Spastin mutations are frequent in sporadic spastic paraparesis and their spectrum is different from that observed in familial cases. J. Med. Genet. 2006;43:259–265. doi: 10.1136/jmg.2005.035311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mancuso G., Rugarli E.I. A cryptic promoter in the first exon of the SPG4 gene directs the synthesis of the 60-kDa spastin isoform. BMC Biol. 2008;6:31. doi: 10.1186/1741-7007-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salinas S., Carazo-Salas R.E., Proukakis C., Cooper J.M., Weston A.E., Schiavo G., Warner T.T. Human spastin has multiple microtubule-related functions. J. Neurochem. 2005;95:1411–1420. doi: 10.1111/j.1471-4159.2005.03472.x. [DOI] [PubMed] [Google Scholar]
- 47.Sanderson C.M., Connell J.W., Edwards T.L., Bright N.A., Duley S., Thompson A., Luzio J.P., Reid E. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum. Mol. Genet. 2006;15:307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka E.M., Kirschner M.W. Microtubule behavior in the growth cones of living neurons during axon elongation. J. Cell Biol. 1991;115:345–363. doi: 10.1083/jcb.115.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roll-Mecak A., Vale R.D. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J. Cell Biol. 2006;175:849–851. doi: 10.1083/jcb.200611149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baas P.W., Karabay A., Qiang L. Microtubules cut and run. Trends Cell Biol. 2005;15:518–524. doi: 10.1016/j.tcb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 51.Thomsen B., Nissen P.H., Agerholm J.S., Bendixen C. Congenital bovine spinal dysmyelination is caused by a missense mutation in the SPAST gene. Neurogenetics. 2009 doi: 10.1007/s10048-009-0214-0. doi:10.1007/s10048-009-0214-0; http://www.springerlink.com/content/tx85775k63772174/?p=a8569c81e3e2466381452878ffeecec1&pi=0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee M., Paik S.K., Lee M.J., Kim Y.J., Kim S., Nahm M., Oh S.J., Kim H.M., Yim J., Lee C.J., et al. Drosophila Atlastin regulates the stability of muscle microtubules and is required for synapse development. Dev. Biol. 2009;330:250–262. doi: 10.1016/j.ydbio.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 53.Orso G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., Micaroni M., Egorova A., Martinuzzi A., McNew J.A., et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 54.Wang X., Shaw W.R., Tsang H.T., Reid E., O'Kane C.J. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat. Neurosci. 2007;10:177–185. doi: 10.1038/nn1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bettencourt da Cruz A., Wentzell J., Kretzschmar D. Swiss Cheese, a protein involved in progressive neurodegeneration, acts as a noncanonical regulatory subunit for PKA-C3. J. Neurosci. 2008;28:10885–10892. doi: 10.1523/JNEUROSCI.3015-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGuire S.E., Roman G., Davis R.L. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20:384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.