Abstract
Genome-wide association studies (GWAS) have identified numerous loci associated with various complex traits for which the underlying susceptibility gene(s) remain unknown. In a GWAS for high-density lipoprotein-cholesterol (HDL-C) level, one strongly associated locus contains at least two biologically compelling candidates, methylmalonic aciduria cblB type (MMAB) and mevalonate kinase (MVK). To detect evidence of cis-acting regulation at this locus, we measured relative allelic expression of transcribed SNPs in five genes using human hepatocyte samples heterozygous for the transcribed SNP. If an HDL-C-associated SNP allele differentially regulates mRNA level in cis, samples heterozygous both for a transcribed SNP and an HDL-C-associated SNP should display allelic expression imbalance (AEI) of the transcribed SNP. We designed statistical tests to detect AEI in a comprehensive set of linkage disequilibrium (LD) scenarios between the transcribed SNP and an HDL-C-associated SNP (rs7298565) in phase unknown samples. We observed significant AEI of 22% in MMAB (P = 1.4 × 10−13, transcribed SNP rs11067231), and the allele associated with lower HDL-C level was associated with greater MMAB transcript level. The same rs7298565 allele was also associated with higher MMAB mRNA level (P = 0.0081) and higher MMAB protein level (P = 0.0020). In contrast, MVK, UBE3B, KCTD10 and ACACB did not show significant AEI (P ≥ 0.05). These data suggest MMAB is the most likely gene influencing HDL-C levels at this locus and demonstrate that measuring AEI at loci containing more than one candidate gene can prioritize genes for functional studies.
INTRODUCTION
Recent genome-wide association studies (GWAS) have identified novel variants at ≥14 loci associated with high-density lipoprotein-cholesterol (HDL-C) levels (1–3). One HDL-C-associated locus on chromosome 12 is strongly significant (P = 1.2 × 10−10, rs2338104, n = 19 793), independently replicates across multiple studies, and is associated with a 0.48 mg/dl per allele change in HDL-C levels (2,3). This locus contains many strongly associated common SNPs [minor allele frequency (MAF) > 0.05] that exhibit high pairwise linkage disequilibrium (LD) with each other, span a 175 kb region and encompass four genes. At least two genes demonstrate biological functions relevant to cholesterol biology. MethylMalonic aciduria cblB type (MMAB) encodes the mitochondrial enzyme cob(I)alamin adenosyltransferase that catalyzes the formation of adenosylcobalamin, a critical cofactor in the catabolism of amino acids, lipids and cholesterol (4). Mevalonate kinase (MVK) encodes the essential early enzyme MVK necessary for the synthesis of isoprenoids, key intermediates in cholesterol biosynthesis (5). Two other genes at the locus not obviously relevant to cholesterol biology are KCDT10, a member of the polymerase delta-interacting protein 1 gene family (6), and UBE3B, a novel member of the ubiquitin 3 ligase family (7). A further plausible candidate located >150 kb from these genes is ACACB, which regulates fatty acid oxidation and modulates lipid metabolism (8). Given the presence of more than one biological candidate, a primary aim of this study was to identity the gene most likely to influence HDL-C level.
Genetic variation at the transcript level contributes to the phenotypic variation between individuals (9). Cis-acting variants are estimated to be responsible for at least 25–35% of inter-individual differences in gene expression (10) and functional cis-acting variants can affect transcription, mRNA-processing, splicing, stability and translation (10).
To identify evidence of cis-acting regulation associated with HDL-C level, we quantified relative allelic expression in human hepatocytes heterozygous for transcribed SNPs in MMAB, MVK, UBE3B, KCTD10 and ACACB. Hepatocytes were chosen for this study because they are a major site of HDL biosynthesis and lipidation (11), and HDL-C is transported to the liver and taken up by hepatocytes following release from peripheral cells (12). For allelic expression imbalance (AEI) analysis, measurement of allele-specific mRNA levels is made within individual samples. Therefore, in the absence of interactions, relative transcript levels are unaffected by sources of variation such as environmental and trans-acting genetic factors. Here, we use real-time PCR to detect AEI at a locus on chromosome 12 associated with HDL-C level. We designed simple-to-implement statistical tests for AEI that are tailored to the observed LD between trait-associated SNPs and transcribed tested SNPs. Our results provide evidence suggesting that MMAB is a likely susceptibility gene at this locus.
RESULTS
AEI at a chromosome 12 locus associated with HDL-C level
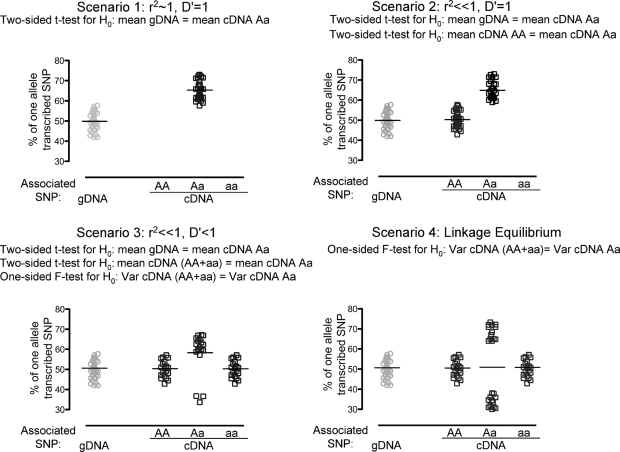
To test whether a cis-acting HDL-C-associated SNP affects transcript levels, we measured relative allelic cDNA levels of MMAB, MVK, UBE3B, KCTD10 and ACACB in human hepatocytes using quantitative real-time PCR and one to two SNPs per gene. Results for MMAB were followed up by testing for AEI using two additional SNPs. AEI can only be detected in samples heterozygous for a transcribed SNP, hence for each of 10 transcribed SNPs, 35–90 hepatocyte samples were first screened for genotype heterozygosity, and AEI was subsequently evaluated in 14–45 samples heterozygous for the transcribed SNP (Table 1). The pattern of AEI caused by an HDL-C-associated SNP will differ depending on the LD between the transcribed SNP and the HDL-C-associated SNP. We considered a comprehensive set of LD scenarios and used different statistical tests to assess AEI for different scenarios (Materials and Methods and Fig. 1).
Table 1.
Evidence of AEI for five genes at an HDL-C-associated locus (α = 0.007)
| Gene | Transcribed SNP | Number of heterozygotes tested | r2/D′ with rs7298565a,b | Statistical test to detect AEI | Power to detect AEI of 22%c | 80% Power to detect AEI (%B − %b)d | AEI (%B − %b)d | P-value |
|---|---|---|---|---|---|---|---|---|
| MMAB | rs877710 | 20 | 0.94/1.0 | t-test gDNA | 1.0 | 9.6 | 22 | 2.4 × 10−9 |
| MMAB | rs11067231 | 45 | 0.94/1.0 | t-test gDNA | 1.0 | 6.1 | 22 | 1.4 × 10−13 |
| MMAB | rs11067233 | 27 | 0.11/0.70 | t-test gDNA | 0.62 | 25.6 | 15 | 8.6 × 10−4 |
| t-test cDNA | 0.54 | 28.0 | 0.046 | |||||
| F-test cDNA | 0.10 | 0.31 | ||||||
| MMAB | rs2241201 | 22 | 0.43/1.0 | t-test gDNA | 0.83 | 21.2 | 20 | 7.1 × 10−4 |
| t-test cDNA | 0.38 | 34.2 | 6.5 × 10−3 | |||||
| MVK | rs7957619 | 16 | 0.10/1.0 | t-test gDNA | 1.0 | 10.7 | 5.7 | 0.10 |
| t-test cDNA | 1.0 | 16.8 | 0.21 | |||||
| KCTD10 | rs1477117 | 21 | 0.15/1.0 | t-test gDNA | 1.0 | 14.6 | 2.9 | 0.68 |
| t-test cDNA | 0.96 | 12.8 | 0.47 | |||||
| UBE3B | rs7298565b | 19 | 1.0/1.0 | t-test gDNA | 1.0 | 5.0 | 1.4 | 0.29 |
| UBE3B | rs2058807 | 19 | 1.0/1.0 | t-test gDNA | 1.0 | 13.2 | 0.07 | 0.73 |
| ACACB | rs3742023 | 34 | 0.23/0.75 | t-test gDNA | 0.93 | 9.7 | 3.7 | 0.051 |
| t-test cDNA | 0.89 | 10.3 | 0.44 | |||||
| F-test cDNA | 0.71 | 0.47 | ||||||
| ACACB | rs7135947 | 14 | 0.001/0.05 | F-test cDNA | 0.69 | 28 | 1.7 | 0.40 |
| t-test gDNA | 0.009 | 0.87 |
SNP, single nucleotide polymorphism; AEI, allelic expression imbalance.
ar2 and D′ values calculated based on observed hepatocyte genotypes for all samples; brs7298565 is used as a representative HDL-C-associated; cpower to detect AEI size observed for MMAB SNP rs11067231; dSNP and also as a transcribed SNP in UBE3B.
Figure 1.
Statistical tests for analysis of AEI depend on LD between transcribed tested SNPs and HDL-C-associated SNPs (cartoon). All tested samples are heterozygous for the transcribed SNP. cDNA samples are stratified by genotype of the HDL-C-associated SNP (rSNP, x-axis). Percent allele of the transcribed SNP is displayed on the y-axis. Scenario 1, r2 ∼ 1, D′ = 1; Scenario 2, r2 ≪ 1, D′ = 1; Scenario 3, r2 ≪ 1, D′ < 1; Scenario 4, linkage equilibrium. For ease of display, the mean of gDNA is assumed to be 50%, whereas in real data mean of gDNA can deviate from 50% due to differences in chemistries of fluorescent probe binding for each allele.
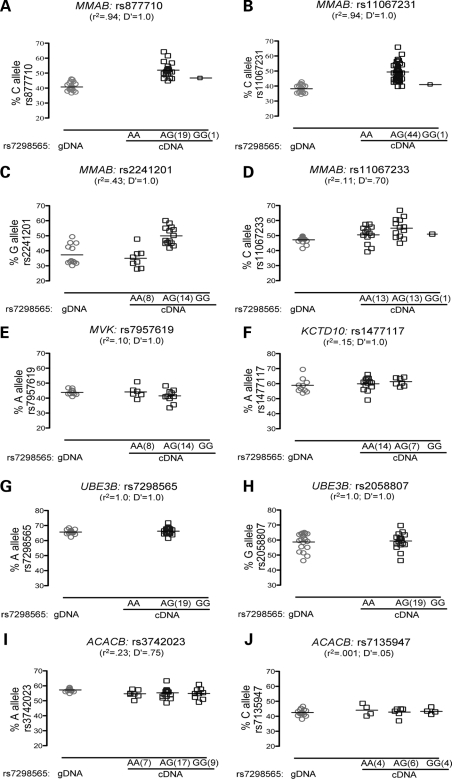
For each of 10 transcribed SNPs, Figure 2 shows the percentage of one allele present in genomic DNA (gDNA) and in cDNA stratified by genotype of representative HDL-C-associated SNP rs7298565 (G allele associated with lower HDL-C level). For an MMAB SNP in strong LD (r2 = 0.94, D′ = 1.0) with rs7298565, we observed significant AEI (rs877710; PgDNA = 2.4 × 10−9, ncDNA het = 20, Fig. 2A). The C allele of rs877710 showed 22% higher expression than the G allele. A second MMAB SNP tested in additional samples also showed strongly significant AEI (rs11067231; PgDNA = 1.4 × 10−13, ncDNA het = 45, Fig. 2B); the C allele of rs11067231 demonstrated a similar 22% higher expression than the G allele. The C alleles of both rs877710 and rs11067231 are on the same haplotype as the G allele of rs7298565 in 44 of 45 samples, thus the G allele of rs7298565 is associated with higher MMAB expression.
Figure 2.
Only SNPs in MMAB display significant AEI. AEI was measured using real-time PCR. Significant (P < 0.05) AEI was observed for transcribed SNPs in MMAB (A–D), but not for transcribed SNPs in other genes (E–J, see Table 1 for P-values). Shown on the x-axis are alleles of the rSNP rs7298565 where allele A corresponds to rSNP allele A and allele G corresponds to rSNP allele a as defined in the statistical analysis section of the methods. Data is shown as collected; all P-values were calculated based on a normalized scale (Materials and Methods).
AEI in MMAB was also tested using two SNPs in lower LD with rs7298565 and consequently with lower power to detect AEI (Table 1): rs2241201 (r2 = 0.43, D′ = 1.0; Fig. 2C) and rs11067233 (r2 = 0.11, D′ = 0.70; Fig. 2D). We observed AEI in the same direction as for rs877710 and rs11067231. The G allele of rs2241201 demonstrated 20% higher expression than the C allele (PgDNA = 7.1 × 10−4), and presence of AEI was confirmed using the cDNA t-test (PcDNA = 6.5 × 10−3), which compares the cDNA samples homozygous for the rSNP to the cDNA samples heterozygous for the rSNP and reduces the risk of bias for any technical difference present when comparing gDNA to cDNA. At rs11067233 the C allele demonstrated 15% higher expression than the G allele (PgDNA = 8.6 × 10−4), although the evidence of AEI using the cDNA t-test (PcDNA = 0.046) would not withstand correction for multiple SNPs tested (Materials and Methods).
In MVK, only one transcribed SNP with MAF > 0.01 was available for study, rs7957619. No evidence of significant AEI was observed (PgDNA = 0.10; PcDNA = 0.21; Fig. 2E). Additionally, no AEI or only marginally significant AEI was observed for five SNPs in KCTD10 (Fig. 2F), UBE3B (Fig. 2G and H) and ACACB (Fig. 2I and J). Taken together, these results argue that a cis-acting SNP is acting on MMAB leading to altered quantities of mRNA.
Power to detect AEI
We estimated the power to detect AEI in each transcribed SNP based on the 22% allelic expression difference between transcribed SNP alleles B and b observed for MMAB SNP rs11067231 (Table 1 and Supplementary Material, Tables S1, S2 and S3). For MVK, the power to detect AEI was 100%, and for KCTD10, UBE3B and ACACB power ranged from 93 to 100% for the most powerful test (Table 1). Thus, we had good power to detect AEI if it existed in these other genes with the strength observed in MMAB.
For each transcribed SNP, we also estimated the AEI that could be detected with 80% power in the samples tested (Table 1). For the four SNPs in MMAB, we estimate that we had 80% power to detect AEI of 6.1–34.2%. For the remaining genes the most powerful test had 80% power to detect AEI of 5.0–16.8%. In addition, AEI and power to detect AEI was estimated at significance level 0.05 (Supplementary Material, Table S1) and exclusively in samples of Caucasian ancestry (Supplementary Material, Tables S2 and S3). Similar results indicate that population stratification was not a factor affecting this study (Supplementary Material, Tables S2 and S3).
Higher hepatocyte MMAB mRNA level is associated with the rs7298565 allele associated with lower HDL-C level
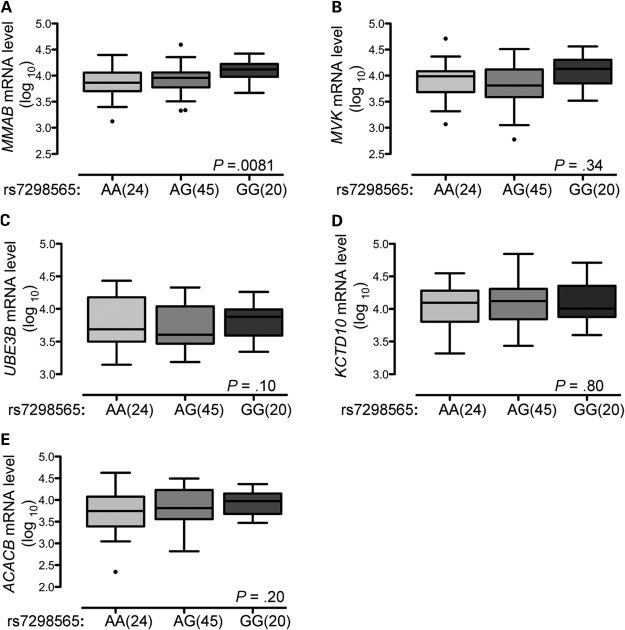
To determine whether total level of mRNA was associated with genotypes of the HDL-C-associated SNP rs7298565, mRNA level was measured in all available hepatocyte samples (n = 89). We observed a significant association between RNA batch and mRNA level for each gene (P < 0.001), and consequently included RNA batch as a covariate in the analysis. Using linear regression under an additive genetic model, we found a significant association between higher levels of MMAB mRNA and the rs7298565 G allele (P = 0.0081; Fig. 3A). No significant association was observed between rs7298565 and total mRNA level of MVK, KCTD10, UBE3B or ACACB (Fig. 3B–E).
Figure 3.
Higher hepatocyte MMAB mRNA level is associated with the G allele of rs7298565. Hepatocyte cDNA was analyzed by real-time PCR using gene-specific primers for MMAB (A), MVK (B), UBE3B (C), KCTD10 (D), ACACB (E), GUSB and B2M. mRNA level was normalized to B2M and GUSB mRNA. Boxplots extend from the first quartile to the third quartile; the median is indicated as a horizontal line; whiskers are drawn to a distance of 1.5 times the interquartile distance or to the highest or lowest point, whichever is shorter. Mean MMAB mRNA level increased with each G allele of HDL-C-associated SNP rs7298565.
Higher hepatocyte MMAB protein level is associated with the rs7298565 allele associated with lower HDL-C level
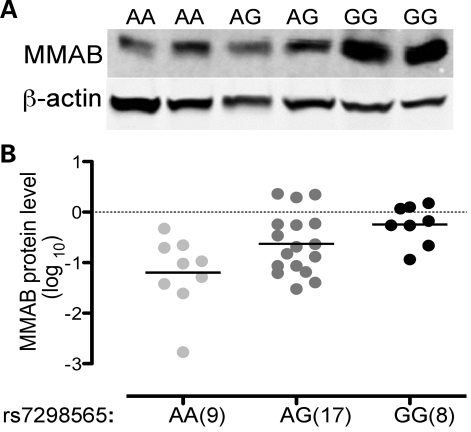
To determine whether protein level of MMAB was also associated with rs7298565 genotype, we measured protein level in 33 hepatocyte samples (Fig. 4A). Using linear regression under an additive genetic model, we found a significant association between higher levels of MMAB protein and the rs7298565 G allele (P = 0.0020; Fig. 4B). This result is consistent with existence of a functional SNP that has a cis-acting regulatory effect to increase level of MMAB and decrease HDL-C level.
Figure 4.
Higher hepatocyte MMAB protein level is associated with the G allele of rs7298565. Hepatocyte protein was analyzed by western blotting using antibodies directed against MMAB and β-actin. MMAB level was normalized to level of β-actin. (A) Representative gel showing MMAB and β-actin levels. Lanes 1–2, rs7298565 AA; lanes 3–4, rs7298565 AG; lanes 5–6, rs7298565 GG. (B) Vertical scatter plots for 9 AA, 19 AG and 8 GG independent observations. Mean MMAB protein level increased with each G allele of HDL-C-associated SNP rs7298565.
DISCUSSION
Cis-acting regulatory variants affecting gene expression are predicted to regulate expression of a large proportion of human genes (9,13,14) and are likely to account for many of the SNP-trait associations at susceptibility loci identified through GWAS. Our recent GWAS identified a region on chromosome 12 containing SNPs strongly associated with plasma HDL-C level (3). In the current study, we selected the strongly HDL-C-associated SNP rs7298565 to represent these SNPs and asked whether rs7298565 was associated with cis-acting regulatory activity in five positional candidate genes. We demonstrated that the G-allele associated with lower HDL-C level is associated both with higher levels of MMAB transcript based on AEI and gene expression and with higher MMAB protein levels.
AEI has been used to identify genes influenced by cis-acting regulatory SNPs (rSNPs; 15–18), and our results suggest that measuring AEI has good potential to help prioritize functional gene candidates in association regions containing more than one candidate gene. However, successful AEI experiments require that each candidate gene contain at least one common transcribed SNP such that sufficient informative heterozygous samples are available. In addition, the genes in which AEI is to be tested must be expressed in an available tissue relevant to the associated trait. In this study, we investigated common transcribed SNPs (MAF > 0.01) in each gene of interest in human hepatocytes.
We describe a statistical strategy that allowed us to evaluate AEI for transcribed SNPs that exhibit any observed level of LD with an HDL-C-associated SNP (Fig. 1 and Table 1) in the absence of information regarding haplotype phase. This strategy allowed us to evaluate all genes of interest. In each test, we compared the AEI in cDNA from samples heterozygous for the HDL-C-associated SNP to a reference, either gDNA from the entire set of heterozygous samples or cDNA from samples homozygous for the HDL-C-associated SNP. Pairs of SNPs with r2 = 1.0, D′ = 1.0 have the highest power to detect association because the gDNA reference group has the largest sample size. However, this comparison may also be subject to bias if, in the absence of AEI, there is a technical difference in the percent allele detected in gDNA compared with cDNA. For example, MMAB transcribed SNP rs11067233 shows ∼47% C allele in the gDNA but ∼51% C allele in the cDNA of HDL-C SNP homozygotes, and this difference could cause an over (or under) estimation of AEI. For SNPs in which r2 < 1 between the transcribed SNP and the HDL-C associated SNP, using the cDNA reference group reduces power but also the risk of bias because the compared values are all based on cDNA. Testing transcribed SNPs with varying degrees of LD with the trait-associated SNP should allow evaluation of any possible bias in the more powerful test. For studies in which phase can be determined across markers with high accuracy, testing the phased samples for allelic expression differences could provide greater power.
AEI may have more power to detect allelic effects on mRNA level compared with stratifying mRNA level by genotype. AEI offers increased sensitivity because each sample acts as its own internal control for sample-to-sample differences in cellular environment. In contrast, total mRNA levels can be influenced by inter-individual variation that arises from differences in environmental factors, physiological states, trans-acting factors or overall gene expression between individuals of different ancestry. We observed stronger evidence for association of rs7298565 allele with transcript level using AEI (PgDNA = 1.4 × 10−13, n = 45) than by evaluating total MMAB mRNA level stratified by genotype (P = 0.0081, n = 89). The observation of increased MMAB protein with each copy of the G allele of rs7298565 is consistent with observations at the mRNA level and provides further support for a regulatory variant being responsible for the association with HDL-C level at this locus.
These results provide evidence of AEI in human hepatocytes and suggest a cis-acting SNP affects expression of MMAB. MMAB is expressed in many tissues, with high expression in the liver, which may be most relevant for processes influencing cholesterol level. Tests for the presence of AEI in other cell types important in regulating HDL-C level may produce different results. Expression quantitative trait locus (eQTL) studies also detected association of rs7298565 with MMAB expression in 400 human liver samples (P = 5.5 × 10−15), a result confirmed in 950 liver samples (P = 4 × 10−23; 2,19), but not all genome-wide eQTL analyses of primary or transformed lymphocytes reported SNP association with MMAB (9,13,14). In addition, a recent genome-wide study in lymphoblastoid cell lines detected association between SNPs in moderate LD with rs7298565 (r2 = 0.11–0.67; D′ = 1) and allelic expression in unspecified genes located in a 167 kb region that includes MMAB, MVK, UBE3B and KCTD10 (18). Further study is necessary to determine the strength of association between HDL-C associated SNPs and MMAB expression in other cell types.
The role of MMAB in HDL-C biology is unclear. MMAB is a mitochondrial enzyme that catalyzes conversion of vitamin B12 into its active form, adenosylcobalamin (4). MMAB is an essential participant in the catabolism of certain amino acids, short chain fatty acids and the side chain of cholesterol to the citric acid cycle intermediate succinyl-CoA (20). An increase in expression of MMAB might therefore be expected to lower levels of total cholesterol, including levels of HDL-C. Given the severity of methylmalonic acidemia, a disorder resulting from deleterious mutations leading to impaired MMAB activity (21), and the evidence presented here of cis-acting variation in MMAB expression, we hypothesize that a variant in high LD with the HDL-C-associated SNP rs7298565 is likely to be a rSNP that has a modest effect on synthesis or stability of mRNA.
Our data cannot completely exclude the biological candidate MVK, nor the other nearby genes KCTD10, UBE3B and ACACB, as functional targets of an HDL-C-associated SNP. Of the transcribed SNPs tested in our study, rs7957619 in MVK had the lowest MAF and only 16 heterozygote samples were available. We had 100% power to detect AEI in MVK based on the amount of AEI observed in MMAB. A recent study in mice reports that Mmab and Mvk share a conserved promoter region and are both regulated by binding of sterol regulatory element binding protein 2, suggesting a potential novel link between the functions of these genes (22). In humans, MVK and MMAB are also arranged in a head-to-head orientation on chromosome 12 and are predicted to share a 258 bp bidirectional promoter (L. Elnitski, Ph.D, unpublished data, 2008). Although neither our data nor existing liver eQTL data (2,21) suggest evidence of cis-acting regulation in MVK, chromosome 12 HDL-C-associated variants could regulate levels of MVK in untested tissues or through functional effects unrelated to total mRNA levels.
A primary challenge remains to identify the functional SNP(s) that affect MMAB transcript levels at this HDL-C locus. Associated SNPs include a non-synonymous SNP in MMAB, rs9593. Although both alleles of rs9593, encoding lysine and methionine, are predicted to have normal physiological activity (20), we cannot exclude that this variant may also affect HDL-C level.
In summary, these data show that increased expression of MMAB is associated with SNPs associated with decreased HDL-C level, strengthening the evidence that MMAB is a target of an HDL-C-associated SNP and providing evidence of an effect on protein levels. Further studies are necessary to identify the functional SNP(s) affecting MMAB, and to determine how variation in MMAB gene expression may affect HDL-C level.
MATERIALS AND METHODS
Samples
Fresh or cryopreserved lots of human hepatocytes, harvested from subjects with various causes of death, were purchased from ADMET Technologies, Inc. (Durham, NC, USA). The ancestry of collected samples was 76% Caucasian, 12% African American, 10% Hispanic, 1% Asian and 1% Native American. Samples were received in four batches. Batches 1 (n = 33) and 4 (n = 9) were received as cryopreserved lots of cells, batch 2 (n = 12) was received directly after isolation (fresh) and batch 3 (n = 44) was received as frozen RNA samples. Initial AEI studies for all SNPs were carried out on batch 1 samples, and AEI of six SNPs (rs11067231, rs11067233, rs2241201, rs7957619, rs1477117 and rs3742023) was further tested on batches 2 and 3. Total MMAB mRNA levels were measured in all samples. MMAB protein levels were measured in available samples from batch 1 (n = 18) and all samples from batches 2 (n = 12) and 4 (n = 9). The University of North Carolina at Chapel Hill IRB determined that this study did not require approval.
Isolation of nucleic acids and cDNA preparation
gDNA was isolated using Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) and total cytosolic RNA was isolated using Tri reagent (Applied Biosystems, Foster City, CA, USA). RNA was treated with rDNase I (DNA-free kit; Applied Biosystems), and DNA and RNA concentrations were determined using a Nanodrop 1000 (Thermo Scientific, Wilmington, DE, USA). For real-time reverse transcription (RT)–PCR, first-strand cDNA was synthesized using 0.8 µg of total RNA in a 20 µl reverse transcriptase reaction mixture (Superscript III First strand synthesis kit; Invitrogen, Carlsbad, CA, USA). cDNA was diluted to contain equivalent to 4 ng/μl input RNA.
SNP selection and genotyping
Among SNPs strongly associated with HDL-C level in the chromosome 12 region (3), we selected rs7298565 (r2 = 1 with reported SNP, rs2338104) as a tag SNP to minimize the number of SNPs to be tested. One to four transcribed SNPs were selected for each gene in which AEI was to be tested. To maximize the number of informative heterozygous samples, transcribed SNPs with minor allele frequencies >0.1 in HapMap CEU [Utah residents with ancestry from northern and Western Europe (CEPH sample)] were chosen. SNPs in strong LD (r2 > 0.8, D′ = 1.0; HapMap CEU samples) with the HDL-C-associated SNP were given preference as they were expected to have the highest power to detect AEI and the direction of the AEI could be observed. Genotyping of gDNA samples were performed with TaqMan allelic discrimination assays (Applied Biosystems) according to manufacturer's instructions.
Real-time PCR for AEI and total mRNA levels
Hepatic expression of MMAB, MVK, UBE3B, KCTD10 and ACACB was confirmed using real-time PCR (M. Fogarty, unpublished data) and published expression array data (23). All PCR reactions were performed in triplicate in a 10 µl volume using a 7900 real-time PCR system. Allele-specific PCR was carried out on hepatocyte gDNA and cDNA using TaqMan allelic discrimination assays (as above).
To confirm that each assay could detect both alleles in a usable range, a standard curve was generated. gDNA from samples homozygous for each of the two alleles, represented by B and b, corresponding to the VIC and FAM fluorescent reporters, were mixed in seven different ratios −10:90, 20:80, 40:60, 50:50, 60:40, 80:20 and 90:10. The standard curve was generated from observed percent allele B versus input ratio of allele B:b. Percent allele B in each experimental sample was estimated from the standard curve, and percent allele b was calculated as 100% − percent allele B. Under idealized conditions the observed percent allele B in heterozygous gDNA would be 50%. Deviation from 50% allele B indicates differences due to labeling and annealing efficiencies of allele-specific probes. To measure total mRNA levels, gene-specific primers were used for MMAB, MVK, UBE3B, KCTD10 and ACACB (Supplementary Material, Table S4). Serial 10-fold dilutions of cDNA from five pooled hepatocyte samples were used as a reference for a standard curve.
Western blotting
Protein was isolated according to the manufacturer's instructions (Supplemental Methods). Proteins were resolved by SDS/PAGE and transferred to PVDF membranes (Invitrogen). Membranes were probed with a mouse polyclonal antibody raised against full-length human MMAB protein and a rabbit polyclonal antibody raised against a synthetic peptide derived from human β-actin (Novus Biologicals, Littleton, CO, USA), followed by goat anti-mouse or anti-rabbit antibodies conjugated to Alexa Fluor 680 (Invitrogen) or IRdye 800 (Rockland, Gilbertsville, PA, USA). To confirm the specificity of MMAB detection, a control experiment was performed using an MMAB blocking peptide. Proteins were detected using an Odyssey imaging system (LI-COR, Lincoln, NE, USA). MMAB (∼27 kDa) and β-actin (43 kDa) bands were quantified by densitometry (LI-COR software).
Statistical analysis
We describe three tests for AEI that differ both in their potential for providing unbiased AEI estimates and in their power to detect AEI, depending on the LD between the transcribed SNP and the HDL-C-associated potential rSNP. All samples tested are heterozygous for the transcribed SNP. To correct for testing multiple hypotheses, we set the significance level to 0.007 to account for the varying levels of LD between seven SNPs tested across five genes. Assume the transcribed SNP has alleles B and b and the rSNP has alleles A and a. In the absence of both AEI and experimental bias, the %B/%b ratio should have a mean of 1. To account for experimental bias, we normalized the observed gDNA or cDNA %B/%b ratio by dividing it by the mean %B/%b ratio of gDNA from individuals heterozygous for the transcribed SNP. The normalized %B allele [%Bn = normalized ratio/(1+ normalized ratio)) was used for all AEI tests. In each test, we compared the %Bn allele in samples expected to show AEI due to the rSNP (cDNA from samples heterozygous for the transcribed SNP) to the %Bn allele in samples not expected to be influenced by the rSNP (gDNA from samples heterozygous for the transcribed SNP or cDNA from samples homozygous for the transcribed SNP, termed reference samples). We used a two-sided t-test (gDNA t-test or cDNA t-test) to compare the reference samples to the cDNA from samples heterozygous for the transcribed SNP, or a one-sided F-test for increased %Bn allele variance in cDNA samples heterozygous for the rSNP relative to those homozygous for the rSNP (cDNA F-test).
The gDNA t-test can be applied to all transcribed SNPs and can detect AEI when there is little to no recombination (D′ ∼ 1, Scenarios 1 and 2 and some Scenario 3, Fig. 1). For the reference group, we used the gDNA from samples heterozygous for the transcribed SNP. When D′ ∼ 1 and AEI is present, the %Bn allele in the cDNA from samples heterozygous for the rSNP should, on average, deviate in one direction (higher or lower) from the average %Bn allele in the reference group. We compared the mean %Bn allele in the reference group to the group of samples heterozygous for the rSNP using a two-sided t-test. The number of cDNA samples heterozygous for the rSNP is highest when r2 = 1 (Scenario 1).
The cDNA t-test can be applied to SNPs with one or more cDNA samples homozygous for the rSNP and may detect AEI when D′ ∼ 1, r2 < 1, Scenarios 2 and 3 (Fig. 1). For the reference group, we used cDNA from samples homozygous for the rSNP. We compared the mean %Bn allele in the reference group to the group of samples heterozygous for the rSNP using a two-sided t-test.
The cDNA F-test can be applied to SNPs with cDNA samples homozygous for the rSNP and can be used when recombination has occurred between the transcribed and rSNP (Scenarios 3 and 4, Fig. 1). For the reference group, we used cDNA from all samples homozygous for the rSNP. When recombination is present, a sample can contain one of four haplotype pairs. Two pairs are homozygous for the rSNP (AB/Ab and aB/ab) and two heterozygous (AB/ab and aB/Ab). These latter two pairs cannot be distinguished based on the observed genotype data. If AEI is present within cDNA from samples heterozygous for the rSNP, the AB/ab haplotype pair will deviate from the reference group in one direction and the aB/Ab haplotype pair in the opposite direction. We used a one-sided F-test of variance to test for higher variance of the %Bn allele in the cDNA of samples heterozygous for the rSNP than in those homozygous for the rSNP.
We performed all plausible tests on each SNP: gDNA t-test (Scenarios 1–4, Figure 1), cDNA test (Scenarios 2–4) and cDNA F-test (Scenarios 3 and 4).
To determine the power to detect AEI of a specified amount in samples heterozygous for each transcribed SNP, we simulated data based on (i) the number of cDNA samples with each rSNP genotype, (ii) the number of genotyped gDNA samples heterozygous for the transcribed SNP, (iii) the standard deviation (SD) of the %Bn allele observed for gDNA and cDNA from specified sets of samples and (iv) the observed LD between the transcribed SNP and the rSNP. We used the tests described above to analyze the simulated data. For each transcribed SNP, we also determined the AEI necessary to obtain 80% power (Supplemental Methods).
For analysis of total mRNA levels, raw expression data were adjusted based on the average expression of two housekeeping genes, B2M and GUSB, and log10 transformed to approximate normality. We tested for association between mRNA level and the number of rs7298565 G alleles using linear regression. Age, sex, self-reported ancestry and sample batch (described above) were considered as covariates.
For analysis, levels of MMAB protein were normalized for β-actin levels and log transformed to approximate normality. We tested for association between MMAB protein level and the number of rs7298565 G alleles using linear regression. Sample batch was not included as a covariate as no significant association was observed between protein levels and sample batch.
SUPPLEMENTARY MATERIAL
FUNDING
This research was funded by the National Institutes of Health (DK072193, DK062370).
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Laura Elnitski, Ph.D for sharing unpublished data.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Manolio T.A. Cohort studies and the genetics of complex disease. Nat. Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- 2.Kathiresan S., Willer C.J., Peloso G.M., Demissie S., Musunuru K., Schadt E.E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deodato F., Boenzi S., Santorelli F.M., Dionisi-Vici C. Methylmalonic and propionic aciduria. Am. J. Med. Genet. C. Semin. Med. Genet. 2006;142C:104–112. doi: 10.1002/ajmg.c.30090. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J., Ren K., Liu X., Xiong X., Hu X., Zhang J. A novel PDIP1-related protein, KCTD10, that interacts with proliferating cell nuclear antigen and DNA polymerase delta. Biochim. Biophys. Acta. 2005;1729:200–203. doi: 10.1016/j.bbaexp.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Gong T.W., Huang L., Warner S.J., Lomax M.I. Characterization of the human UBE3B gene: structure, expression, evolution, and alternative splicing. Genomics. 2003;82:143–152. doi: 10.1016/s0888-7543(03)00111-3. [DOI] [PubMed] [Google Scholar]
- 8.Ha J., Lee J.K., Kim K.S., Witters L.A., Kim K.H. Cloning of human acetyl-CoA carboxylase-beta and its unique features. Proc. Natl Acad. Sci. USA. 1996;93:11466–11470. doi: 10.1073/pnas.93.21.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 10.Pastinen T., Hudson T.J. Cis-Acting Regulatory Variation in the Human Genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 11.Rader D.J. Molecular regulation of HDL metabolism and function: implications for novel therapies. J. Clin. Invest. 2006;116:3090–3100. doi: 10.1172/JCI30163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver D.L., Wang N., Tall A.R. Defective HDL particle uptake in ob/ob hepatocytes causes decreased recycling, degradation, and selective lipid uptake. J. Clin. Invest. 2000;105:151–159. doi: 10.1172/JCI8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 14.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastinen T., Ge B., Gurd S., Gaudin T., Dore C., Lemire M., Lepage P., Harmsen E., Hudson T.J. Mapping common regulatory variants to human haplotypes. Hum. Mol. Genet. 2005;14:3963–3971. doi: 10.1093/hmg/ddi420. [DOI] [PubMed] [Google Scholar]
- 16.Serre D., Gurd S., Ge B., Sladek R., Sinnett D., Harmsen E., Bibikova M., Chudin E., Barker D.L., Dickinson T., et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 18.Ge B., Pokholok D.K., Kwan T., Grundberg E., Morcos L., Verlaan D.J., Le J., Koka V., Lam K.C., Gagne V., et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 19.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leal N.A., Olteanu H., Banerjee R., Bobik T.A. Human ATP:Cob(I)alamin adenosyltransferase and its interaction with methionine synthase reductase. J. Biol. Chem. 2004;279:47536–47542. doi: 10.1074/jbc.M405449200. [DOI] [PubMed] [Google Scholar]
- 21.Dobson C.M., Wai T., Leclerc D., Kadir H., Narang M., Lerner-Ellis J.P., Hudson T.J., Rosenblatt D.S., Gravel R.A. Identification of the gene responsible for the cblB complementation group of vitamin B12-dependent methylmalonic aciduria. Hum. Mol. Genet. 2002;11:3361–3369. doi: 10.1093/hmg/11.26.3361. [DOI] [PubMed] [Google Scholar]
- 22.Murphy C., Murray A.M., Meaney S., Gafvels M. Regulation by SREBP-2 defines a potential link between isoprenoid and adenosylcobalamin metabolism. Biochem. Biophys. Res. Commun. 2007;355:359–364. doi: 10.1016/j.bbrc.2007.01.155. [DOI] [PubMed] [Google Scholar]
- 23.Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.