Abstract
Inflammatory bowel disease, including Crohn's disease (CD) and ulcerative colitis (UC), and type 1 diabetes (T1D) are autoimmune diseases that may share common susceptibility pathways. We examined known susceptibility loci for these diseases in a cohort of 1689 CD cases, 777 UC cases, 989 T1D cases and 6197 shared control subjects of European ancestry, who were genotyped by the Illumina HumanHap550 SNP arrays. We identified multiple previously unreported or unconfirmed disease associations, including known CD loci (ICOSLG and TNFSF15) and T1D loci (TNFAIP3) that confer UC risk, known UC loci (HERC2 and IL26) that confer T1D risk and known UC loci (IL10 and CCNY) that confer CD risk. Additionally, we show that T1D risk alleles residing at the PTPN22, IL27, IL18RAP and IL10 loci protect against CD. Furthermore, the strongest risk alleles for T1D within the major histocompatibility complex (MHC) confer strong protection against CD and UC; however, given the multi-allelic nature of the MHC haplotypes, sequencing of the MHC locus will be required to interpret this observation. These results extend our current knowledge on genetic variants that predispose to autoimmunity, and suggest that many loci involved in autoimmunity may be under a balancing selection due to antagonistic pleiotropic effect. Our analysis implies that variants with opposite effects on different diseases may facilitate the maintenance of common susceptibility alleles in human populations, making autoimmune diseases especially amenable to genetic dissection by genome-wide association studies.
INTRODUCTION
Genome-wide association studies (GWAS) have been fruitful in identifying common variants underlying many human diseases (1,2), with notable success especially in several autoimmune diseases (3). Hundreds of distinct genomic loci have been associated with various autoimmune diseases (3,4), including celiac disease (CeD), Crohn's disease (CD), ulcerative colitis (UC), multiple sclerosis (MS), rheumatoid arthritis (RA), systemic lupus erythematosus (SLE) and type 1 diabetes (T1D). Besides individual studies, recent meta-analysis of GWAS has also enabled the identification of dozens of susceptibility loci for T1D (5,6) and CD (7), since single studies are typically underpowered. Additionally, comparisons of susceptibility loci between different autoimmune diseases have revealed important insights into their common genetic architecture. For example, interleukin 23 receptor (IL23R) has been consistently implicated in multiple related autoimmune disorders including CD, UC, ankylosing spondylitis and psoriasis, suggesting that it may be a common susceptibility factor for the major seronegative diseases (8–11). Another study compared shared genetic risk factors for T1D and CeD and reported multiple identical risk alleles (12), suggesting that common biologic mechanisms may be etiologic features of both diseases. Several similar studies that examined known CD susceptibility loci in GWAS for UC identified previously unreported susceptibility loci shared by these related disorders (13–15). Taken together, these studies suggest that examination of related autoimmune diseases can help reveal shared genetic pathways, and that evaluation of known susceptibility loci for one disease in GWAS for another disease may uncover novel disease-loci relationships.
In the present study, we interrogated GWAS data sets on CD, UC and T1D for known susceptibility loci implicated in these diseases. Our comparative analysis serves several important roles: first, the ability to identify additional susceptibility loci for one disease by testing known loci for another disease, similar to previous studies (12,13). This approach increases statistical power by limiting the number of hypotheses tested, thus lowering the significance threshold. For each disease, rather than applying a stringent P < 5 × 10−8 threshold for the susceptibility variants, we applied a Benjamini–Hochberg false discovery rate (FDR) approach (16), aiming that less than 5% of the declared associations are likely to be false positive associations. Second, since our samples were not used in the CD meta-analysis (7) or T1D meta-analysis (5,6), our results can serve as an independent benchmark to validate results from meta-analysis of GWAS. Finally, we are also interested in determining whether opposite risk alleles exist for these diseases. Since previous studies have already shown that susceptibility alleles in PTPN22 have opposite effects in different autoimmune disorders including CD and T1D (7,17), it is likely that additional variants with opposite effects on these diseases exist but have not been reported. Altogether, our study helps better understand the genetic architecture, including shared genetic pathways as well as risk factors with opposing effects, for these related diseases.
RESULTS
Overview of the study and candidate loci
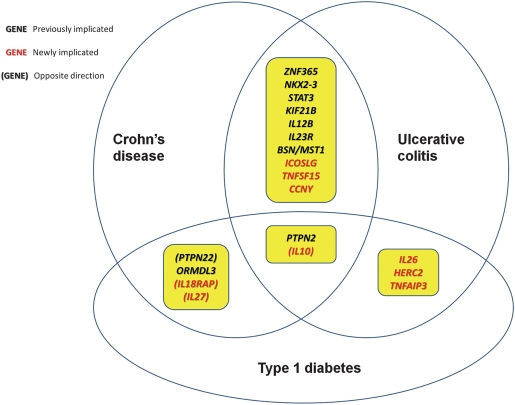
In the current study, we attempted to evaluate known susceptibility loci for CD, UC and T1D in a GWAS data set, including 1689 CD cases, 777 UC cases, 989 T1D cases and 6197 shared control subjects, all of whom were of European ancestry and were genotyped on the Illumina HumanHap550 array with ∼550 000 SNP markers (see Materials and Methods). For each disease, we tested association with multiple markers in the major histocompatibility complex (MHC) region, as well as with markers residing at 30 known CD loci, 18 known UC loci and 45 known T1D loci, for a total of 81 unique non-MHC loci (Supplementary Material, Table S1). For CD, this list is compiled from the Barrett et al. meta-analysis on CD (7), as those loci with P < 5 × 10−8, with a total of 30 loci (Supplementary Material, Table S2). For UC, since no meta-analysis has ever been reported, we instead relied on five separate individual studies (13,15,18–20) with a total of 18 loci (Supplementary Material, Table S3). For T1D, we relied on the Tables from recent Barrett et al. meta-analysis (6), with a total of 45 loci (Supplementary Material, Table S4). We did not test the susceptibility loci (such as 20q13 and 21q22) that were discovered in a subset of the data used in the current study (21). To ensure that we examine the exactly same risk SNPs reported in previously studies, we performed genotype imputation by the MACH software, and we used allelic dosage association to take into account of imputation uncertainty. A subset of shared susceptibility loci were illustrated in Figure 1 and Table 1, while the association results for all 81 loci for three diseases are summarized in Supplementary Material, Table S1 and described below.
Figure 1.
Illustration of previously known and newly identified susceptibility loci that are shared by Crohn's disease, ulcerative colitis and type 1 diabetes.
Table 1.
Newly identified disease associations at non-MHC susceptibility loci for CD, UC and T1D
| Indexa | Locus | Reported SNPb | Closest gene | Allelec | MAF | OR (CD)d | 95% CI (CD) | P (CD)e | OR (UC)d | 95% CI | P (UC)e | OR (T1D)d | 95% CI | P (T1D)e | New associationf |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1D-4/UC-3 | 1q32.1 | rs3024505 | IL10 | A/G | 0.15 | 1.24 | 1.12–1.37 | 2.10E−05 | 1.26 | 1.09–1.45 | 1.90E−03 | 0.76 | 0.66–0.88 | 1.50E−04 | (CD) |
| T1D-6 | 2q12.1 | rs917997 | IL18RAP | T/C | 0.23 | 1.23 | 1.13–1.34 | 2.20E−06 | 1.08 | 0.95–1.23 | 0.25 | 0.87 | 0.77–0.97 | 0.016 | (CD) |
| T1D-14 | 6q22.32 | rs9388489 | C6orf173 | G/A | 0.48 | 0.89 | 0.82–0.96 | 2.00E−03 | 0.99 | 0.89–1.11 | 0.87 | 1.22 | 1.11–1.35 | 3.10E−05 | (CD) |
| T1D-15 | 6q23.3 | rs2327832 | OLIG3/TNFAIP3 | G/A | 0.20 | 1.08 | 0.99–1.19 | 0.099 | 1.26 | 1.10–1.43 | 6.20E−04 | 1.1 | 0.98–1.23 | 0.11 | UC |
| T1D-32 | 16p12.3 | rs12444268 | UMOD | A/T | 0.32 | 0.9 | 0.83–0.98 | 0.011 | 1.02 | 0.90–1.14 | 0.78 | 1.09 | 0.98–1.20 | 0.1 | (CD) |
| T1D-33 | 16p11.2 | rs4788084 | NUPR1/IL27 | T/C | 0.39 | 1.23 | 1.14–1.33 | 1.40E−07 | 1.08 | 0.96–1.21 | 0.19 | 0.88 | 0.79–0.97 | 9.70E−03 | (CD) |
| T1D-44 | 22q12.2 | rs5753037 | HORMAD2 | T/C | 0.39 | 0.88 | 0.81–0.95 | 1.40E−03 | 0.87 | 0.77–0.97 | 0.015 | 1.09 | 0.99–1.20 | 0.089 | (CD) |
| CD-17 | 21q22 | rs762421 | ICOSLG | G/A | 0.37 | 1.2 | 1.11–1.30 | 4.30E−06 | 1.23 | 1.10–1.38 | 2.40E−04 | 0.95 | 0.86–1.05 | 0.3 | UC |
| CD-30 | 9q32 | rs4263839 | TNFSF15 | A/G | 0.32 | 0.79 | 0.73–0.86 | 8.60E−08 | 0.81 | 0.72–0.92 | 8.60E−04 | 1.02 | 0.92–1.13 | 0.67 | UC |
| UC-6 | 12q15 | rs1558744 | IFNG/IL26 | A/G | 0.39 | 1.09 | 1.01–1.18 | 0.024 | 1.25 | 1.12–1.40 | 8.70E−05 | 1.16 | 1.05–1.28 | 2.80E−03 | T1D |
| UC-7 | 10p11.21 | rs3936503 | CCNY | A/G | 0.33 | 1.17 | 1.08–1.26 | 1.60E−04 | 1.11 | 0.98–1.24 | 0.092 | 0.96 | 0.87–1.07 | 0.47 | CD |
| UC-9 | 15q13.1 | rs916977 | HERC2 | T/C | 0.21 | 0.97 | 0.88–1.07 | 0.51 | 1.08 | 0.95–1.24 | 0.25 | 1.21 | 1.08–1.36 | 8.10E−04 | T1D |
The complete list of association results for all 81 candidate loci can be found in Supplementary Materials.
aThe disease index for susceptibility loci was annotated in Supplementary Material, Tables S2–S4.
bReported SNP in previous publications was annotated in Supplementary Material, Tables S2–S4.
cThe minor and major alleles (based on control subjects) in forward strand in NCBI 36 human genome assembly.
dOdds ratios (OR) and 95% confidence interval (CI) were calculated with respect to minor allele.
eP-values passing FDR < 5% threshold for each disease were marked in bold fonts.
fPreviously unreported association to the diseases, where those with opposite direction were enclosed within parenthesis.
Association with non-MHC loci
Among loci examined are the 30 loci previously implicated in a meta-analysis for CD (7). We detected positive association for 24 of them in our cohort for CD (P ≤ 0.011, FDR < 5%), indicating that results from meta-analysis are highly reliable. With this threshold, assuming complete LD (D′ = 1) between marker allele and causal allele with MAF = 30% or higher, we expect a minimum power of 97, 80 and 40% for SNPs with odds ratio of 1.2, 1.15 and 1.1, respectively (Supplementary Material, Table S5). Among these 30 CD susceptibility loci, several are already known to confer UC susceptibility and were associated with UC with the same direction in our study (IL23R on 1p31, MST1/BSN on 3p21, NKX2-3 on 10q24 and IL12B on 5q33). Several CD risk loci with previously unconfirmed risk for UC were identified in our study, including ICOSLG on 21q22 (rs762421, P = 2.4 × 10−4, OR = 1.23) and TNFSF15 on 9q32 (rs4263839, P = 8.6 × 10−4, OR = 0.81), both with allelic effects in the same direction. We note that ICOSLG had been previously tested for association with UC (14); however, the evidence for association of ICOSLG with UC was weak (P = 0.016), and did not pass multiple testing threshold. Additionally, TNFSF15 was shown to be associated with CD and Inflammatory bowel disease (IBD) but did not reach significant association for UC alone in a study reported by Franke et al. (13). We next tested the CD risk loci for evidence of association with T1D. Three genes (ORMDL3, PTPN2 and PTPN22) have been previously proposed to be susceptibility genes for both T1D and CD. ORMDL3 is only marginally associated with T1D in our data (P = 0.051, OR = 1.1). PTPN2 was associated with CD in our study, but we were unable to replicate its effect on T1D (P = 0.2, OR = 1.09). We examined the T1D website (http://www.T1Dbase.org) for further evidence of association: although ORMDL3 and PTPN2 show strong association in the Barrett et al. meta-analysis (P = 3.0 × 10−7 and P = 3.6 × 10−15, respectively) (6), neither show association (P > 0.05) in the Cooper et al. meta-analysis (5). Similar to previously described (17), the risk SNP for CD within PTPN22 on 1p13 (rs2476601, P = 6.6 × 10−6, OR = 0.72 for the minor allele) is associated with T1D but with the opposite direction of association (P = 4.9 × 10−25, OR = 2.0).
We next examined the 18 loci implicated in UC susceptibility in five previous studies (excluding MHC region) (13,15,18–20), and detected positive association with UC for seven of them in our cohort (P ≤ 5.6 × 10−3, FDR < 5%). With this threshold, assuming complete LD (D′ = 1) between marker allele and causal allele with MAF = 30% or higher, we expect a minimum power of 67, 37 and 13% for SNPs with odds ratio of 1.2, 1.15 and 1.1, respectively (Supplementary Material, Table S5). Four UC loci were known to confer CD susceptibility as described earlier. In addition, a variant within CCNY on 10p11.21 (rs3936503, P = 1.6 × 10−4, OR = 1.17) was associated with CD. This variant is in moderate LD (r2 = 0.66) with a variant reported in the CD meta-analysis (rs17582416); as CCNY is only weakly associated with CD in the previous study (13), our analysis rendered support that CCNY is also a shared susceptibility gene for CD. Furthermore, a variant within IL10 on 1q32.1 (rs3024505, P = 2.1 × 10−5, OR = 1.24) is associated with CD in our study, and we note that the same variant was previously investigated in CD and observed to have borderline significance albeit with similar effect size (P = 0.013, OR = 1.2 in (18)). Examination of UC susceptibility loci in T1D identified a variant within IL26 as being associated with T1D with the same direction of effect (rs1558744, P = 2.8 × 10−3, OR = 1.16). A variant within HERC2 was also associated with T1D with the same direction (rs916977, P = 8.1 × 10−4, OR = 1.21), though it did not show association with UC in our data. Furthermore, we confirmed that a variant within IL10 was associated with T1D (rs3024505, P = 1.5 × 10−4, OR = 0.76), but with opposite direction of effects to that of either CD or UC. Additionally, we interrogated the T1D website (www.T1Dbase.org) for further evidence in support of association: the SNPs for IL26 and HERC2 were not included here, but the SNP for IL10 (rs3024505) was indeed annotated in this database and showed a significant association (P = 2.2 × 10−6) in the T1D meta-analysis (6).
We next examined the 45 known T1D susceptibility loci (excluding MHC region), and detected positive association with T1D for 16 of them in our cohort (P ≤ 0.011, FDR < 5%). With this threshold, assuming complete LD (D′ = 1) between marker allele and causal allele with MAF = 30% or higher, we expect a minimum power of 85, 57 and 24% for SNPs with odds ratio of 1.2, 1.15 and 1.1, respectively (Supplementary Material, Table S5). In our study, TNFAIP3 was found for the first time to confer UC risk (rs2327832, P = 6.2 × 10−4, OR = 1.26). Additionally, we found that seven T1D loci actually confer protection against CD, including PTPN22 as described earlier, IL27 (P = 1.4 × 10−7), IL18RAP (P = 2.2 × 10−6), IL10 (P = 2.1 × 10−5), 22q12.2 (P = 1.4 × 10−3), 6q22.32 (P = 2.0 × 10−3) and 16p12.3 (P = 0.011). Genome-wide significance for IL27 (rs8049439, P = 2.41 × 10−9) was also reported in a recent GWAS with overlapping samples (22). Since the CD meta-analysis P-values from a previous study were made publicly available (7), we examined these SNPs and found further support for PTPN22 (P = 1.8 × 10−5), IL27 (P = 0.003), IL18RAP (P = 2.2 × 10−5), IL10 (P = 0.016) but less evidence for 22q12.2 (P = 0.068), 6q22.32 (P = 0.54) and 16p12.3 (P = 0.29). In addition, the loci on 22q12.2 and 16p12.3 (but not 6q22.32) did not pass the FDR threshold for T1D association in our data. Therefore, we regarded the first four loci (PTPN22, IL27, IL18RAP and IL10) as highly confident loci showing opposite directions of association between CD and T1D, all of which were outside of the MHC region.
Association of MHC loci
Given the well-known involvement of the MHC region in conferring genetic susceptibility to CD, UC and T1D (23), we next investigated whether effects of common SNPs tagging HLA alleles may differ between these diseases. For each disease, we used the ‘clumping’ function in the PLINK software (24) to identify a set of index SNPs that are highly independent of each other (r2 < 0.1) with a 5 Mb sliding window, since it is well known that long-range LD is prevalent in the MHC region. A total of 8, 12 and 80 index SNPs with P < 1 × 10−4 were found for the CD, UC and T1D data sets, respectively. For each of the three diseases, we listed the five most significant SNP variants in the MHC region and then examined the association signals for the other two diseases (Table 2). Some strikingly different signals were unveiled by this analysis. For example, the strongest T1D risk alleles are located between HLA-DQB1 and HLA-DQA2 (rs9275383, P = 2.1 × 10−138, OR = 3.77), whereas this locus confers strong protection against both CD (P = 3.9 × 10−6, OR = 0.73) and UC (P = 1.9 × 10−9, OR = 0.53). Similarly, the strongest UC protective allele is located between HLA-DRB1 and HLA-DQA1 (rs477515, P = 6.7 × 10−19, OR = 0.56), yet it confers strong risk for T1D (P = 5.6 × 10−18, OR = 1.52). Interestingly, two of the five most significant CD protective alleles (rs2187668 and rs9275383) represent the two most significant T1D risk alleles but with opposite direction of effects.
Table 2.
Association results for the most significant MHC SNPs for CD, UC and T1D
| Index SNPa | Positionb | No. of SNP in LD with index | Minor allele frequency | Minor/major allele | P (CD)c | OR (CD)d | P (UC)c | OR (UC)d | P (T1D)c | OR (T1D)d |
|---|---|---|---|---|---|---|---|---|---|---|
| Most significant MHC SNPs for CD | ||||||||||
| rs2844480 | 31672800 | 20 | 0.20 | A/G | 3.36E−07 | 1.27 | 2.94E−02 | 1.15 | 7.31E−01 | 0.98 |
| rs3129871 | 32514320 | 61 | 0.33 | A/C | 5.76E−07 | 1.22 | 3.69E−01 | 0.95 | 5.84E−87 | 0.25 |
| rs2187668 | 32713862 | 187 | 0.13 | A/G | 1.14E−06 | 0.73 | 1.45E−01 | 0.88 | 1.05E−116 | 3.40 |
| rs9275383 | 32776824 | 42 | 0.11 | T/G | 3.85E−06 | 0.73 | 1.92E−09 | 0.53 | 2.06E−138 | 3.77 |
| rs1063355 | 32735692 | 20 | 0.38 | A/C | 3.95E−06 | 1.20 | 1.61E−06 | 1.30 | 4.02E−57 | 0.39 |
| Most significant MHC SNPs for UC | ||||||||||
| rs477515 | 32677669 | 63 | 0.32 | T/C | 9.02E−03 | 0.89 | 6.67E−19 | 0.56 | 5.60E−18 | 1.52 |
| rs3104404 | 32790152 | 19 | 0.23 | A/C | 2.16E−03 | 1.15 | 5.09E−08 | 1.40 | 3.65E−46 | 0.31 |
| rs411326 | 32319295 | 38 | 0.26 | A/G | 3.40E−03 | 0.87 | 1.10E−07 | 0.70 | 1.26E−05 | 0.77 |
| rs3749946 | 31556841 | 3 | 0.09 | A/C | 6.04E−03 | 1.19 | 5.45E−07 | 1.50 | 1.91E−03 | 0.75 |
| rs2621332 | 32888295 | 43 | 0.33 | C/T | 1.59E−01 | 0.94 | 1.61E−06 | 0.75 | 1.67E−02 | 1.13 |
| Most significant MHC SNPs for T1D | ||||||||||
| rs9275383 | 32776824 | 62 | 0.11 | T/G | 3.85E−06 | 0.73 | 1.92E−09 | 0.53 | 2.06E−138 | 3.77 |
| rs2187668 | 32713862 | 197 | 0.13 | A/G | 1.14E−06 | 0.73 | 1.45E−01 | 0.88 | 1.05E−116 | 3.40 |
| rs3129871 | 32514320 | 34 | 0.33 | A/C | 5.76E−07 | 1.22 | 3.69E−01 | 0.95 | 5.84E−87 | 0.25 |
| rs2647050 | 32777745 | 28 | 0.39 | C/T | 1.71E−01 | 1.06 | 7.81E−02 | 1.10 | 3.32E−82 | 0.30 |
| rs6903608 | 32536263 | 3 | 0.32 | C/T | 5.82E−02 | 1.08 | 3.97E−08 | 1.36 | 5.96E−35 | 0.47 |
aFor each disease, the five most significant yet independent (r2 < 0.1) index SNPs are shown.
bPosition was in NCBI build 36 coordinates in chromosome 6.
cAll P < 1 × 10−4 were marked in bold fonts.
dOdds ratio (OR) was calculated for the minor allele as cases versus controls.
Given the multi-allelic nature of the MHC region, we caution that these results may not necessarily suggest that MHC risk alleles for T1D protect against CD or UC. In this regard, one could imagine that three haplotypes exist in the MHC: A, B, C and each with frequency one-third in a control population. If A is a risk haplotype for T1D and the case frequencies are half, one-fourth, one-fourth, and B is a risk haplotype for UC and cases have frequencies one-fourth, half, one-fourth, then B will appear to be ‘protective’ against T1D and A is ‘protective’ against UC, simply because the strong primary effect depletes all non-risk haplotypes in cases. This could be the case in our data as well, since all three diseases have shown strong association with HLA-DR but with different alleles (for example, DRB1*0103 for UC versus the DR3-DQ2 and DR4-DQ8 haplotypes for T1D). We note that similar discussions have been made in previous large-scale analysis of MHC risk alleles, where the investigators applied conditional regression and concluded that autoimmune diseases arise from complex, multilocus effects that span the entire region (25). For these reasons, and due to the complex structural variation and the hierarchical linkage disequilibrium patterns of MHC, we caution that additional studies are warranted, notably including sequencing of the entire MHC region, to independently correlate the SNP risk alleles and HLA alleles with respect to their effects on different diseases.
DISCUSSION
Based on comparative genetic analysis of three autoimmune disorders genotyped by whole-genome SNP arrays, we identified multiple previously unreported or unconfirmed disease-loci associations, including multiple variants with opposite effects on different diseases. Our study has significant implications for genetic studies of autoimmune disorders. A large number of autoimmune disorders are known to share etiological factors involving common genetic pathways. To identify a more comprehensive ensemble of risk factors, besides traditional single-marker analysis for single-disease, several additional analytical techniques can be used, including but not limited to: (i) meta-analysis of multiple studies with similar phenotypes, such as those performed on CD (7) and T1D (5,6); (ii) combined analysis of tightly related disorders, such as combining CD and UC into a single IBD group for association (10,21), or combining CD, UC and T1D into a single autoimmune disease group for association or some other combinations thereof (26); (iii) pathway-based approaches that try to identify groups of consistently yet probably moderately associated genes with disease (27,28) or (iv) evaluation of known risk alleles for one disease in cohort for another disease (12,13), but without assuming the same direction or same magnitude of effects. Our study is an example of the successful application of the last approach above, and has markedly enhanced our current understanding of these three disorders, and collectively resulted in the discovery of multiple novel disease-associated loci excluding MHC. Although these loci passed a FDR threshold of <5%, we caution that they still need to be examined in additional follow-up studies for further confirmation.
Furthermore, the opposite direction of association at multiple loci between CD and T1D, as well as the opposite direction of association within multiple independent MHC loci (between UC/CD and T1D), suggests that predisposition to related diseases may be controlled or regulated by both an ‘overdose’ or ‘underdose’ of genes and genomic elements in relevant biological pathways. This is not surprising, as the mechanisms for effector function in host defense and regulatory function in self-tolerance (that is, negative selection in the thymus, generation of regulatory lymphocytes and activation-induced apoptosis) rely on closely related molecular events which, in both cases, depend on antigen-induced immune response. This makes the effect of small functional changes tip the balance in one or the other direction, in a context-dependent fashion. Therefore, if a variant is associated with multiple autoimmune diseases but with different directions, it is much more likely to function in pathways related to immunological functions (as opposed to insulin production or autophagy, etc.). For example, the protein tyrosine phosphatase non-receptor type 22 (PTPN22) gene is a member of the PTPs that negatively regulate T-cell activation, and a missense SNP (R620W, rs2476601) was associated with several autoimmune diseases but with opposite directions. Recent biochemical studies show that the mutation results in gain of PTPN22 phosphatase activity in T cells, which is predicted to increase the threshold for TCR signaling (17,29), suggesting that TCR signaling augments RA, Graves disease, hashimoto thyroiditis, SLE and juvenile arthritis, but not for other diseases where TCR signaling plays a less important role. Additionally, it is likely that a MHC allele may be a good antigen presenter for particular epitopes for a virus that links to the development of T1D (perhaps due to similar epitopes as those on pancreatic beta cells), but it may not present epitopes for certain bacteria efficiently and such bacteria are opportunistic pathogens for IBDs. In any case, since some of the susceptibility loci may not have clear functional candidate or the hypothetical gene is not well characterized for function yet, focusing on immunological pathways may help reveal the causal genes and characterize their functional roles. For example, the most significant SNP (rs4788084) on 16p11.2 is much closer to NUPR1 than to IL27, but given the known immunological function for interleukins, it is quite confident to assume that IL27 is the true causal gene at the locus.
Relatively few studies have investigated the potential existence or prevalence of balancing selection in genome evolution, compared with other types of selective pressures (30). In humans, besides sporadic studies on interleukins (31,32) and various forms of heterozygosity advantage on sickle cell anemia (33,34) and cystic fibrosis (35–37), most other studies on balancing selection have focused on the MHC region alone. Based on segregation analysis of isolated populations, multiple studies have reported strong evidence for balancing selection at the MHC loci at the population level (38,39). Additionally, examination of particular groups of patients showed that maximum MHC heterozygosity of class I loci delayed acquired immunodeficiency syndrome (AIDS) onset among patients infected with HIV-1, whereas homozygotes rapidly progress to AIDS and death (40). Many more studies on MHC have also been performed in animal models. For example, in mice, MHC heterozygosity was shown to confer advantage by enhancing resistance to multiple strains of Salmonella and one of Listeria; heterozygotes were more resistant than the average of parental homozygotes, but they were not more resistant than both (dominance but not over-dominance) (38). Similarly, the San Nicolas Island fox is genetically the most monomorphic sexually reproducing animal population, with virtually no variation in most hypervariable genetic markers; however, remarkably high variation is present only in the MHC regions, suggesting the importance of balancing selection as a mechanism to maintain variation in non-human populations (41).
In our study, an interesting corollary of the sharing of opposite alleles relates to the predicted allelic structure of autoimmune diseases. Unlike susceptibility loci for most complex disorders that may be under a purifying or negative selection in human populations, the susceptibility loci for autoimmune diseases (both inside and outside of MHC) are potentially under a balancing selection that are dependent on heterogeneity in environmental factors. We stress here that the balancing selection does not act on the phenotype (autoimmune diseases) per se (which requires tens of thousands of years), but impacts immune responses towards different infectious agents, predisposing to different diseases (which act during recent times). In fact, several non-MHC genes, including IL10 (31) (shown in our study to have opposite directions of association) and five additional interleukins (32), have already been suggested to be under balancing selection. This does not necessarily suggest heterozygosity advantage, but simply reflects a compromise of battle between multiple distinct immunological pathways for distinct pathogens; in fact, at least for MHC, multiple theoretical studies already showed that heterozygosity advantage does not explain MHC variation (42), in the absence of host-pathogen co-evolution (43,44). This hypothesis does not contradict the ‘hygiene hypothesis’, which specifies that lack of exposure to bacteria/viruses in modern populations results in augmented susceptibility to the development of autoimmunity; instead, it simply suggests that host response to different pathogens might have utilized different defense mechanism or machinery (such as humoral response versus cellular immunity). Finally, this hypothesis predicts that the allelic architecture of autoimmune diseases are more likely to harbor multiple common susceptibility alleles (possibly with relatively large effects), as opposed to a collection of rare alleles (45). This hypothesis is in keeping with the notion that autoimmune diseases, compared with other complex and common diseases, are more readily interrogated by GWAS, with over a hundred loci implicated at this time.
The sample collections for the three diseases (CD, UC and T1D) used in our study are all ascertained from early-onset patients (before the age 19). Although T1D is itself a pediatric-onset disease, CD and UC are often diagnosed in early adulthood, so this raised a question how different sample ascertainment schemes may affect results. Our previous studies (21) demonstrate that pediatric-onset samples can help identify disease genes, and that these genes replicate in adult-onset cohorts such as WTCCC, albeit with smaller effect sizes. Additionally, CD-affected children are more likely to have colonic CD than adults (46,47), whereas UC-affected children are more likely to have more extensive colitis than adults. In addition, a young age of IBD onset is typically associated with a stronger family history of IBD. Furthermore, pediatric-onset cases may be less amenable to environmental risk factors. Therefore, as recently discussed (48), pediatric-onset samples may have unique characteristics that enable more powerful replication of disease genes detected from adult-onset samples as well as identification of novel disease genes.
In conclusion, our study represents a successful application of cross-disease comparative analysis to extract additional biological insights from existing data sets generated from GWAS. We identified multiple previously unreported or unconfirmed disease-loci associations, but many of them have opposite direction of association for T1D and CD, suggesting the potential presence of a ‘genetic switch’ for progression to either one of these two diseases, in addition to shared genetic risk factors. Our results suggest an interesting hypothesis that susceptibility loci involved in the pathogenesis of autoimmune diseases may have antagonistic pleiotropic effects, where risk alleles for one disease may confer selective advantage for another disease or infection resistance, suggesting that more in-depth study on gene/environmental interactions are necessary to better understand the genetic etiology of autoimmune disorders.
MATERIALS AND METHODS
Sample collection
Early-onset inflammatory bowel disease (IBD)
The cases were recruited from multiple centers from four geographically discrete countries (Italy, Scotland, Canada, and the USA). We have previously collected 647 pediatric-onset CD cases and 317 pediatric-onset UC cases from the Children's Hospital of Wisconsin and Medical College of Wisconsin, Children's Hospital of Philadelphia and Cincinnati Children's Hopsital Medical Center (21), and they were all included in the current analysis. The cases used in the current study were also included in a recent large-scale IBD analysis (22). All patients were diagnosed prior to their 19th birthday and fulfilled standard IBD diagnostic criteria, and phenotypic characterization was based on a modification of the Montreal classification (49). The multi-dimensional scaling analysis on genotype data was used to identify subjects of genetically inferred European ancestry. Since our study focused on CD and UC, we removed 53 cases whom were diagnosed as ‘IBD-unclassified’ from the case group. We additionally removed 11 CD cases and 3 UC cases, since they show evidence of cryptic relatedness, due to identity-by-descent estimate higher than 0.2 (see details below). A total of 1689 CD cases and 777 UC cases were included in the final analysis.
Type 1 diabetes (T1D)
The cases were identified through pediatric diabetes clinics at the Children's Hospital of Montreal and at Children's Hospital of Philadelphia (CHOP). We have previously collected ∼563 patients in a GWAS (50) and additional cases from a follow-up study (51), and they were included in the current analysis. The multi-dimensional scaling analysis on genotype data was used to identify subjects of genetically inferred European ancestry. We removed five subjects from related pairs and focused on subjects genotyped on the HumanHap550 platform only, resulting in the total number of 989 independent cases.
Disease-free controls
The control group was recruited by CHOP clinicians and nursing staff within the CHOP Health Care Network, which includes primary care clinics and outpatient practices. The control subjects did not have any autoimmune diseases based on self-reported intake questionnaire, clinician-based assessment or electronic health care records. The specific subsets of control subjects were selected using a matching algorithm implemented in MATLAB as previously described (22). This algorithm determines a distance for each case–control pair after mapping each sample to coordinates on the basis of the top k Eigen value-scaled principal components. The control subjects were originally selected by matching to the CD cases, resulting in a total of 6197 control subjects. The same control subjects were also compared with UC cases and T1D cases. As previously discussed (1,26), the use of shared control subjects may result in potential genome-wide biases; although we are only looking at specific loci, we also examined genome-wide inflations to address this concern. All the 81 loci interrogated in the current study were previously identified using different and independent control subjects, further reducing the concern on potential biases. The Research Ethics Board of CHOP and other participating centers approved the study, and written informed consent was obtained from all subjects or their parents.
Genotyping and association tests
All the case and control samples were genotyped on the Illumina Infinium™ HumanHap550 array (Illumina, San Diego) with ∼550 000 SNP markers, at the Center for Applied Genomics, the Children's Hospital of Philadelphia. Following genotyping, we excluded samples with greater than 2% missing genotypes. We used the PLINK software version 1.06 (24) for multi-dimensional scaling (MDS) on markers not in LD to identify the ancestry origin (via the—mds-plot—cluster argument), and only samples of genetically inferred European ancestry were used in the association analysis (Supplementary Material, Fig. S1). The genomic control inflation factors (λ) (52) were 1.07, 1.14 and 1.05 for CD, UC and T1D, respectively (Supplementary Material, Fig. S2). Furthermore, based on the whole-genome identity-by-descent estimate reported by PLINK (24) version 1.06 (via the –genome argument), we calculated the pairwise identity-by-descent measure as P(identity-by-descent =2) + 0.5*P(identity-by-descent = 1); next, we eliminated pairs of samples showing cryptic relatedness (identity-by-descent score > 0.2), similar to the previously described procedure (53). The PLINK software (24) version 1.06 was also used for data quality control, association analysis and for the SNP ‘clumping’ at the MHC locus by association test statistics.
To ensure that we examine the exactly identical SNPs that were reported in previous studies, we used genotype imputation on the SNP genotype data. The MACH software (http://www.sph.umich.edu/csg/abecasis/MaCH/) was utilized and the default two-step imputation procedure is adopted for imputation. For imputation, we used the HapMap phased haplotypes (release 22) on CEU (Utah residents with ancestry from northern and western Europe) subjects, as downloaded from the HapMap database (http://www.hapmap.org). To handle imputation uncertainty, rather than taking the best guess imputed genotypes, we used allelic dosage in association tests. The allelic dosage is the weighted sum of the genotype class probabilities: for example, if the genotype probabilities are 0.7 for AA, 0.2 Aa and 0.1 for aa in MACH imputation output, then the dosage for A allele is 2 × 0.7 + 1 × 0.2 + 0 × 0.1 = 1.6. We applied a FDR approach to assess the statistical significance of candidate variants, by controlling FDR q-value to be less than 5%. We used the QVALUE software (54) version 1.0 for the FDR calculation. We set the lambda value at zero, which is a tuning parameter used in estimating π0 (no. of true null tests/no. of total tests) (55), and effectively achieve the more stringent step-down FDR procedure originally proposed by Benjamini and Hochberg (16). We used the Genetic Power Calculator (56) for assessing the power for each disease given the FDR threshold. We used the ‘case–control for discrete traits’ module assuming multiplicative model of disease risk and assuming complete linkage disequilibrium (D′ = 1) between the marker allele and the risk allele with varying minor allele frequencies (Supplementary Material, Table S5).
SUPPLEMENTARY MATERIAL
FUNDING
This research was financially supported by the Children's Hospital of Philadelphia, Genome Canada through the Ontario Genomics Institute and the Juvenile Diabetes Research Foundation, the Primary Children's Medical Center Foundation, DK069513, M01-RR00064, M01 RR002172-26 and C06-RR11234 from the National Center for Research Resources. All genome-wide genotyping by the Illumina arrays was funded by an Institute Development Award to the Center for Applied Genomics from the Children's Hospital of Philadelphia.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully thank all the patients and their families who were enrolled in this study, as well as all the control subjects who donated blood samples to Children's Hospital of Philadelphia (CHOP) for genetic research purposes. We thank the technical staff at the Center for Applied Genomics (CAG) at CHOP for producing the genotypes used for analyses and the nursing, medical assistant and medical staff for their invaluable help with recruitment of patients and control subjects for the study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. doi:10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Altshuler D., Daly M.J., Lander E.S. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. doi:10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lettre G., Rioux J.D. Autoimmune diseases: insights from genome-wide association studies. Hum. Mol. Genet. 2008;17:R116–R121. doi: 10.1093/hmg/ddn246. doi:10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper J.D., Smyth D.J., Smiles A.M., Plagnol V., Walker N.M., Allen J.E., Downes K., Barrett J.C., Healy B.C., Mychaleckyj J.C., et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. doi:10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. doi:10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cargill M., Schrodi S.J., Chang M., Garcia V.E., Brandon R., Callis K.P., Matsunami N., Ardlie K.G., Civello D., Catanese J.J., et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 2007;80:273–290. doi: 10.1086/511051. doi:10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton P.R., Clayton D.G., Cardon L.R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D.P., McCarthy M.I., Ouwehand W.H., Samani N.J., et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. doi:10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duerr R.H., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. doi:10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tremelling M., Cummings F., Fisher S.A., Mansfield J., Gwilliam R., Keniry A., Nimmo E.R., Drummond H., Onnie C.M., Prescott N.J., et al. IL23R variation determines susceptibility but not disease phenotype in inflammatory bowel disease. Gastroenterology. 2007;132:1657–1664. doi: 10.1053/j.gastro.2007.02.051. doi:10.1053/j.gastro.2007.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smyth D.J., Plagnol V., Walker N.M., Cooper J.D., Downes K., Yang J.H., Howson J.M., Stevens H., McManus R., Wijmenga C., et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. doi:10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke A., Balschun T., Karlsen T.H., Hedderich J., May S., Lu T., Schuldt D., Nikolaus S., Rosenstiel P., Krawczak M., et al. Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat. Genet. 2008;40:713–715. doi: 10.1038/ng.148. doi:10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 14.Anderson C.A., Massey D.C., Barrett J.C., Prescott N.J., Tremelling M., Fisher S.A., Gwilliam R., Jacob J., Nimmo E.R., Drummond H., et al. Investigation of Crohn's disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology. 2009;136:523–529. doi: 10.1053/j.gastro.2008.10.032. e523 doi:10.1053/j.gastro.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher S.A., Tremelling M., Anderson C.A., Gwilliam R., Bumpstead S., Prescott N.J., Nimmo E.R., Massey D., Berzuini C., Johnson C., et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat. Genet. 2008;40:710–712. doi: 10.1038/ng.145. doi:10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 17.Vang T., Congia M., Macis M.D., Musumeci L., Orru V., Zavattari P., Nika K., Tautz L., Tasken K., Cucca F., et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat. Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. doi:10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 18.Franke A., Balschun T., Karlsen T.H., Sventoraityte J., Nikolaus S., Mayr G., Domingues F.S., Albrecht M., Nothnagel M., Ellinghaus D., et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat. Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. doi:10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 19.Silverberg M.S., Cho J.H., Rioux J.D., McGovern D.P., Wu J., Annese V., Achkar J.P., Goyette P., Scott R., Xu W., et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat. Genet. 2009;41:216–220. doi: 10.1038/ng.275. doi:10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett J.C., Lee J.C., Lees C.W., Prescott N.J., Anderson C.A., Phillips A., Wesley E., Parnell K., Zhang H., Drummond H., et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. doi:10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kugathasan S., Baldassano R.N., Bradfield J.P., Sleiman P.M., Imielinski M., Guthery S.L., Cucchiara S., Kim C.E., Frackelton E.C., Annaiah K., et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. doi:10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imielinski M., Baldassano R.N., Griffiths A., Russell R.K., Annese V., Dubinsky M., Kugathasan S., Bradfield J.P., Walters T.D., Sleiman P., et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. doi:10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernando M.M., Stevens C.R., Walsh E.C., De Jager P.L., Goyette P., Plenge R.M., Vyse T.J., Rioux J.D. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. doi:10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rioux J.D., Goyette P., Vyse T.J., Hammarstrom L., Fernando M.M., Green T., De Jager P.L., Foisy S., Wang J., de Bakker P.I., et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc. Natl Acad. Sci. USA. 2009;106:18680–18685. doi: 10.1073/pnas.0909307106. doi:10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. doi:10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Zhang H., Kugathasan S., Annese V., Bradfield J.P., Russell R.K., Sleiman P.M., Imielinski M., Glessner J., Hou C., et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am. J. Hum. Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. doi:10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baranzini S.E., Galwey N.W., Wang J., Khankhanian P., Lindberg R., Pelletier D., Wu W., Uitdehaag B.M., Kappos L., Polman C.H., et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum. Mol. Genet. 2009;18:2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregersen P.K. Gaining insight into PTPN22 and autoimmunity. Nat. Genet. 2005;37:1300–1302. doi: 10.1038/ng1205-1300. doi:10.1038/ng1205-1300. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell-Olds T., Willis J.H., Goldstein D.B. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat. Rev. Genet. 2007;8:845–856. doi: 10.1038/nrg2207. doi:10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- 31.Wilson J.N., Rockett K., Keating B., Jallow M., Pinder M., Sisay-Joof F., Newport M., Kwiatkowski D. A hallmark of balancing selection is present at the promoter region of interleukin 10. Genes Immun. 2006;7:680–683. doi: 10.1038/sj.gene.6364336. doi:10.1038/sj.gene.6364336. [DOI] [PubMed] [Google Scholar]
- 32.Fumagalli M., Pozzoli U., Cagliani R., Comi G.P., Riva S., Clerici M., Bresolin N., Sironi M. Parasites represent a major selective force for interleukin genes and shape the genetic predisposition to autoimmune conditions. J. Exp. Med. 2009;206:1395–1408. doi: 10.1084/jem.20082779. doi:10.1084/jem.20082779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aidoo M., Terlouw D.J., Kolczak M.S., McElroy P.D., ter Kuile F.O., Kariuki S., Nahlen B.L., Lal A.A., Udhayakumar V. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet. 2002;359:1311–1312. doi: 10.1016/S0140-6736(02)08273-9. doi:10.1016/S0140-6736(02)08273-9. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski D.P. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 2005;77:171–192. doi: 10.1086/432519. doi:10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poolman E.M., Galvani A.P. Evaluating candidate agents of selective pressure for cystic fibrosis. J. R. Soc. Interface. 2007;4:91–98. doi: 10.1098/rsif.2006.0154. doi:10.1098/rsif.2006.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meindl R.S. Hypothesis: a selective advantage for cystic fibrosis heterozygotes. Am. J. Phys. Anthropol. 1987;74:39–45. doi: 10.1002/ajpa.1330740104. doi:10.1002/ajpa.1330740104. [DOI] [PubMed] [Google Scholar]
- 37.Jorde L.B., Lathrop G.M. A test of the heterozygote-advantage hypothesis in cystic fibrosis carriers. Am. J. Hum. Genet. 1988;42:808–815. [PMC free article] [PubMed] [Google Scholar]
- 38.Penn D.J., Damjanovich K., Potts W.K. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA. 2002;99:11260–11264. doi: 10.1073/pnas.162006499. doi:10.1073/pnas.162006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassilakos D., Natoli A., Dahlheim M., Hoelzel A.R. Balancing and directional selection at exon-2 of the MHC DQB1 locus among populations of odontocete cetaceans. Mol. Biol. Evol. 2009;26:681–689. doi: 10.1093/molbev/msn296. doi:10.1093/molbev/msn296. [DOI] [PubMed] [Google Scholar]
- 40.Carrington M., Nelson G.W., Martin M.P., Kissner T., Vlahov D., Goedert J.J., Kaslow R., Buchbinder S., Hoots K., O'Brien S.J. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. doi:10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 41.Aguilar A., Roemer G., Debenham S., Binns M., Garcelon D., Wayne R.K. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl Acad. Sci. USA. 2004;101:3490–3494. doi: 10.1073/pnas.0306582101. doi:10.1073/pnas.0306582101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Boer R.J., Borghans J.A., van Boven M., Kesmir C., Weissing F.J. Heterozygote advantage fails to explain the high degree of polymorphism of the MHC. Immunogenetics. 2004;55:725–731. doi: 10.1007/s00251-003-0629-y. doi:10.1007/s00251-003-0629-y. [DOI] [PubMed] [Google Scholar]
- 43.Borghans J.A., Beltman J.B., De Boer R.J. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55:732–739. doi: 10.1007/s00251-003-0630-5. doi:10.1007/s00251-003-0630-5. [DOI] [PubMed] [Google Scholar]
- 44.Tellier A., Brown J.K. Polymorphism in multilocus host parasite coevolutionary interactions. Genetics. 2007;177:1777–1790. doi: 10.1534/genetics.107.074393. doi:10.1534/genetics.107.074393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pritchard J.K., Cox N.J. The allelic architecture of human disease genes: common disease-common variant… or not? Hum. Mol. Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. doi:10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 46.Levine A., Kugathasan S., Annese V., Biank V., Leshinsky-Silver E., Davidovich O., Kimmel G., Shamir R., Palmieri O., Karban A., et al. Pediatric onset Crohn's colitis is characterized by genotype-dependent age-related susceptibility. Inflamm. Bowel Dis. 2007;13:1509–1515. doi: 10.1002/ibd.20244. doi:10.1002/ibd.20244. [DOI] [PubMed] [Google Scholar]
- 47.Meinzer U., Idestrom M., Alberti C., Peuchmaur M., Belarbi N., Bellaiche M., Mougenot J.F., Cezard J.P., Finkel Y., Hugot J.P. Ileal involvement is age dependent in pediatric Crohn's disease. Inflamm. Bowel Dis. 2005;11:639–644. doi: 10.1097/01.mib.0000165114.10687.bf. doi:10.1097/01.MIB.0000165114.10687.bf. [DOI] [PubMed] [Google Scholar]
- 48.Sauer C.G., Kugathasan S. Pediatric inflammatory bowel disease: highlighting pediatric differences in IBD. Med. Clin. North Am. 2010;94:35–52. doi: 10.1016/j.mcna.2009.10.002. doi:10.1016/j.mcna.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Satsangi J., Silverberg M.S., Vermeire S., Colombel J.F. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–753. doi: 10.1136/gut.2005.082909. doi:10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hakonarson H., Grant S.F., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Casalunovo T., Taback S.P., Frackelton E.C., et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. doi:10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 51.Grant S.F., Qu H.Q., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Taback S.P., Frackelton E.C., Eckert A.W., et al. Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes. 2009;58:290–295. doi: 10.2337/db08-1022. doi:10.2337/db08-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. doi:10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 53.Wang K., Zhang H., Ma D., Bucan M., Glessner J.T., Abrahams B.S., Salyakina D., Imielinski M., Bradfield J.P., Sleiman P.M., et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. doi:10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. doi:10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storey J.D. A direct approach to false discovery rates. J. R. Stat. Soc. B. 2002;64:479–498. doi:10.1111/1467-9868.00346. [Google Scholar]
- 56.Purcell S., Cherny S.S., Sham P.C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. doi:10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.