Abstract
In the adult heart, regulation of fatty acid oxidation and mitochondrial genes is controlled by the PPARγ coactivator–1 (PGC-1) family of transcriptional coactivators. However, in response to pathological stressors such as hemodynamic load or ischemia, cardiac myocytes downregulate PGC-1 activity and fatty acid oxidation genes in preference for glucose metabolism pathways. Interestingly, despite the reduced PGC-1 activity, these pathological stressors are associated with mitochondrial biogenesis, at least initially. The transcription factors that regulate these changes in the setting of reduced PGC-1 are unknown, but Myc can regulate glucose metabolism and mitochondrial biogenesis during cell proliferation and tumorigenesis in cancer cells. Here we have demonstrated that Myc activation in the myocardium of adult mice increases glucose uptake and utilization, downregulates fatty acid oxidation by reducing PGC-1α levels, and induces mitochondrial biogenesis. Inactivation of Myc in the adult myocardium attenuated hypertrophic growth and decreased the expression of glycolytic and mitochondrial biogenesis genes in response to hemodynamic load. Surprisingly, the Myc-orchestrated metabolic alterations were associated with preserved cardiac function and improved recovery from ischemia. Our data suggest that Myc directly regulates glucose metabolism and mitochondrial biogenesis in cardiac myocytes and is an important regulator of energy metabolism in the heart in response to pathologic stress.
Introduction
In the fetal and neonatal heart, glucose is the primary energy substrate, and the developing heart derives most of its ATP through glycolysis (1). However, after birth, the energy substrate preference of the myocardium shifts from a dependence on glucose toward fatty acid oxidation (FAO) (2), which is coincident with a burst of mitochondrial proliferation and increase in myocardial oxygen consumption (3). This transition accounts for the large ATP-producing capacity of the adult heart. However, similar to the plasticity reported for cardiac-specific gene expression in the adult heart subjected to pathological stress (4–6), there is also a coordinated switch in energy substrate preference back toward the fetal pattern from FAO to glucose in adult myocardium in response to many stressors, including ischemia (7, 8), preconditioning (9), and pathological forms of hypertrophy (10). Investigations into the regulatory mechanisms that govern myocardial energy substrate preference have primarily focused on regulation of FAO genes by the fatty acid–activated nuclear receptor PPARα and its cofactor PPAR coactivator–1α (PGC-1α). Disrupting PPARα or PGC-1α leads to metabolic abnormalities in the heart and LV dysfunction (11, 12).
In contrast, the transcriptional regulation of genes controlling glucose metabolic pathways in the setting of cardiac growth is less well studied; however, c-Myc (Myc) has been implicated in controlling both glycolysis and mitochondrial biogenesis in malignant cells (13–15). Myc is the prototypical member of a family of sequence-specific DNA-binding proteins that are postulated to act as “third messengers” for ligand-dependent signals (16). Myc is not expressed in the adult heart under normal physiological conditions; however, it is upregulated rapidly in response to virtually all forms of pathologic stress (17, 18). Although work by our group and others has demonstrated that Myc is required for hypertrophic growth after hemodynamic stress (19–21), its role in regulating metabolism and mitochondrial biogenesis in the heart is unknown.
It has been reported that limited myocardial mitochondrial biogenesis occurs early in response to pathological growth stimuli, but ultimately the increased metabolic energy demands of the hypertrophied myocytes exceed the increase in mitochondrial number, leading to LV decompensation (22). Members of the PGC-1 family are thought to be the primary regulators of mitochondrial biogenesis from late development through normal postnatal growth (23). Thus, mitochondrial biogenesis and FAO gene expression are typically tightly coupled in the heart (24). However, PGC-1α expression in the heart is reduced after pathological stress (25), and PGC-1α–/–PGC-1β–/– mice display normal cardiac development and mitochondrial number through late gestation, suggesting that additional factors must exist that can regulate mitochondrial number and function (23). A number of other transcriptional regulators, including Myc, have been implicated in mitochondrial biogenesis, but their relative importance in mediating mitochondrial number in the heart after stress is unknown (26).
Here, we investigate the role of Myc in regulating energy metabolism in the adult heart utilizing inducible transgenic mouse models in which Myc can be activated or inactivated specifically in adult myocardium. Induction of Myc in myocardium induced hypertrophic growth and increased glucose uptake and utilization by activating a panel of genes involved in this metabolic pathway, downregulated FAO genes by inhibiting PGC-1α expression, and induced mitochondrial biogenesis. In contrast, deletion of Myc in adult myocardium attenuated hypertrophic growth and expression of glucose metabolism and mitochondrial biogenesis genes in response to hemodynamic stimuli. Endogenous Myc also directly bound the promoters of key factors involved in glucose metabolism and mitochondrial replication in the cardiac myocytes after stress, suggesting that Myc is a direct regulator of these processes in the heart. Importantly, the Myc-orchestrated alterations in metabolism and mitochondrial biogenesis were cardioprotective and support a key role for Myc in regulating cardiac metabolism.
Results
Myc is upregulated in the heart in response to multiple pathological stressors.
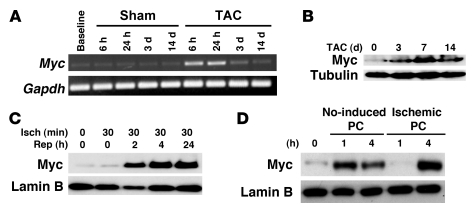
To determine the expression pattern of Myc in the heart after differing pathological stressors, we subjected wild-type mice to pressure overload, ischemia/reperfusion (I/R), and preconditioning stimuli. Myc is typically associated with hypertrophic growth in the adult heart, so we first examined Myc levels after aortic banding in vivo. Myc mRNA increased 15.3 ± 2.9–fold (n = 3/group; P < 0.05) in myocardium of mice subjected to transaortic constriction (TAC) versus sham operation (Figure 1A), with Myc protein levels remaining elevated for days after hemodynamic load (Figure 1B). To determine whether Myc is also upregulated in response to stressors not typically associated with growth, we determined Myc protein levels in ventricular lysates obtained from hearts after 30 minutes of ischemia or ischemia followed by the indicated reperfusion periods (Figure 1C). Myc protein levels increased markedly after ischemia in the reperfusion phase. To determine whether a biological stress typically associated with a cardioprotective effect also upregulated Myc, we examined Myc expression in ventricles subjected ischemic or NO-induced preconditioning. Western blot analysis revealed that both NO treatment and ischemic preconditioning upregulated Myc expression (Figure 1D). Although the exact role of Myc upregulation in response to these stressors that are not typically associated with growth is unknown, the result suggests that Myc may mediate other biological processes in addition to myocyte growth in adult myocardium.
Figure 1. Expression of Myc is upregulated in the heart in response to multiple pathological stressors.
(A) Representative time course of Myc expression in sham or transverse aortic–banded wild-type mice. Semiquantitative PCR was performed on total ventricular RNA. (B) Western blot for expression of Myc in wild-type ventricles after 0 hours, 3 days, 7 days, and 14 days of TAC together with tubulin as a loading control. (C) Temporal expression pattern of Myc protein levels in wild-type hearts as determined by immunoblot analysis at baseline, after 30 minutes of ischemia (Isch), and after 24 hours of reperfusion (Rep). Lamin B was included as loading control. (D) Immunoblotting on ventricular lysates from NO-induced and ischemic preconditioned hearts.
Altered myocardial fatty acid and glucose uptake and utilization rates in Myc-activated myocardium.
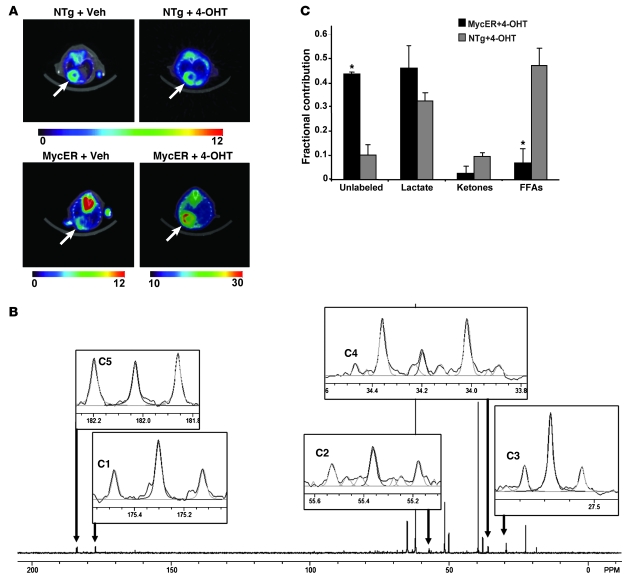
Since these pathological stressors are all associated with a switch in cardiac metabolism from FAO to glucose utilization and Myc has been implicated in regulating glycolysis in cancer cells (13, 27), we then sought to determine whether Myc activation alone could induce similar metabolic changes in the heart. Glucose uptake and utilization in Myc-induced mice was assessed by microPET using 18F-fluorodeoxyglucose (18F-FDG). Myocardial 18F-FDG uptake in 4-hydroxytamoxifen–treated (4-OHT–treated) MycER mice was increased 2-fold compared with that in MycER mice at baseline or 4-OHT–treated nontransgenic (NTg) mice (Figure 2A; P < 0.005). In order to determine the fractional contributions of metabolic substrates to mitochondrial respiration, we determined carbon labeling of glutamate from the myocardium of NTg and MycER mice treated with 4-OHT for 3 days by 13C-NMR and isotopomer analyses. Figure 2B shows representative 13C-NMR spectra obtained from NTg and MycER hearts. There was a 4.4-fold increase in the contribution of acetyl-CoA from the unlabeled fraction to the citric acid cycle (P < 0.05). Previous studies showed that the unlabeled fraction in the working mouse heart is derived predominately from exogenous glucose or glycogen stores (28), although endogenous triglycerides may also contribute. The assumption that this unlabeled fraction consists primarily of glucose is supported by the results of the microPET studies indicating increased glucose uptake after Myc induction. Isotopomer analysis also revealed a corresponding decrease in acetyl-CoA production through FAO when compared with 4-OHT–treated NTg hearts (Figure 2C). Thus, Myc activation in the adult heart alters the pattern of substrate oxidation, with a relative increase in oxidation of glucose and a decrease in fatty acids.
Figure 2. Increased glucose uptake and utilization rates in Myc-activated myocardium.
(A) MicroPET imaging on representative NTg and MycER mice with or without (Veh) 4-OHT treatment (n = 4/group). The relative amount of tracer uptake into the mouse heart 1 hour after intraperitoneal injection of 18F-FDG is indicated by the color scale (range, 0–30). The arrows indicate the cardiac field. (B) Representative NMR spectra from Myc-induced mouse heart. Labeled glutamate carbons are highlighted. Light gray lines represent peak integration with NUTS program. (C) Fractional contribution of acetyl-CoA from the substrates to the citric acid cycle after 4-OHT treatment. *P < 0.05, MycER mice versus NTg littermates; n = 3/group.
Myc induces glucose metabolism genes in adult myocardium.
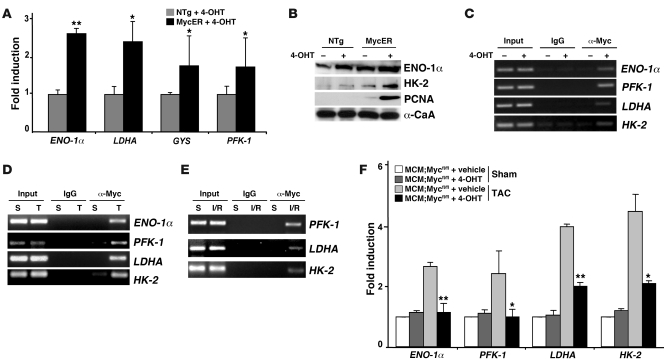
To determine the mechanisms whereby Myc might regulate the observed changes of substrate preference in the heart, we analyzed total ventricular RNA from NTg or MycER mice treated with 4-OHT for 7 days for genes involved in glucose metabolism or storage. Myc activation led to increases in mRNA levels of a panel of these genes, including enolase-1α (ENO-1α) by 2.65-fold (P < 0.001), lactate dehydrogenase A (LDHA) by 2.43-fold (P < 0.05), glycogen synthase (GYS) by 1.78-fold (P < 0.05), and phosphofructokinase 1 (PFK-1) by 1.75-fold (P < 0.05; Figure 3A). Consistent with the changes in mRNA levels, protein expression of ENO-1α and HK-2 increased (Figure 3B). To determine whether enzymes involved in glucose metabolism are direct targets of Myc transcriptional activation, we performed ChIP analysis with anti-Myc antibody and primers designed to amplify genomic sequences spanning putative Myc-binding sites within the promoter sequences of key glycolytic genes after activation of Myc. Amplification of Myc-immunoprecipitated DNA complexes from 4-OHT–treated MycER ventricles demonstrated Myc binding to the endogenous promoters of ENO-1α, PFK-1, LDHA, and HK-2 (Figure 3C). Quantitation of Myc promoter binding by ChIP followed by real-time PCR revealed that activation of Myc led to increased binding to the LDHA promoter by 7.9-fold, 2.1-fold for PFK-1, and 1.8-fold for HK-2 promoters (P < 0.05 vs. untreated MycER). To confirm that endogenous Myc also bound these promoters in the heart after stress, we subjected wild-type mice to pressure overload for 24 hours (Figure 3D) or ischemia for 30 minutes followed by 24 hours of reperfusion (Figure 3E). Association of endogenous Myc to ENO-1α, PFK-1, LDHA, and HK-2 promoters was enhanced by hypertrophic stimulus and after I/R injury (Figure 3, D and E). Thus, Myc directly regulates genes involved in glycolysis in the adult heart.
Figure 3. Myc directly regulates expression of glycolytic genes in adult myocardium.
(A) Total ventricular RNA from NTg and MycER mice with or without 4-OHT treatment for 7 days assayed by real-time PCR for glycolytic enzymes genes ENO-1α, LDHA, GYS, and PFK-1 in activated MycER versus 4-OHT–treated NTg ventricles (n = 3/group). (B) Total ventricular protein lysates from mice of the indicated genotypes were probed with antibodies against glycolysis-associated proteins (ENO-1α and HK-2) along with α-CaA as a loading control. (C) Myc binds to the glycolytic gene promoter sequences in situ. ChIP analysis performed with anti-Myc antibody and PCR primers to the selected glycolytic genes promoters. Ventricular tissues obtained from MycER mice 24 hours after 4-OHT or vehicle treatment were analyzed. Input lanes show PCR product derived from chromatin before immunoprecipitation to verify equal loading. (D and E) Endogenous Myc binds to the promoters of ENO-1α, PFK-1, LDHA, and HK-2 in response to pathologic stress. Ventricular tissue obtained from wild-type mice subjected to sham (S) operation or TAC (T; 1 day) and after I/R injury (30 minutes of ischemia and 24 hours of reperfusion) were analyzed with anti-Myc antibody and PCR primers to the glycolytic promoters of ENO-1α, PFK-1, LDHA, and HK-2. Input lanes show PCR product derived from chromatin before immunoprecipitation to verify equal loading. (F) Myc-null mice demonstrate an attenuated stress-induced increase in glycolytic genes. Total ventricular RNA from MCM;Mycfl/fl mice with or without 4-OHT treatment subjected to sham or TAC operation was assayed using quantitative real-time PCR for glycolytic genes ENO-1α, PFK-1, LDHA, and HK-2 (n = 3/group). *P < 0.05, **P < 0.001.
To evaluate whether Myc was required for the increased expression of genes involved in glucose metabolism typically seen after hypertrophic stimuli, we examined Myc-deficient mice in which Myc can be inducibly inactivated, specifically in adult myocardium with a tamoxifen-regulated Cre (21). Deletion of Myc attenuates hypertrophic growth in response to both hemodynamic and pharmacologic hypertrophic stimuli (21). Consistent with our previous observation, we found that vehicle-treated MerCreMer;Mycfl/fl (MCM;Mycfl/fl) mice with intact Myc expression demonstrated the expected hypertrophic growth after TAC compared with sham-operated animals (6.12 ± 0.24 versus 4.2 ± 0.16 mg/g, P < 0.001). In contrast, the hypertrophic response in banded Myc-deficient mice was attenuated compared with that in sham-treated animals (5.3 ± 0.29 versus 4.35 ± 0.17 mg/g, P < 0.01). The relative increase in heart weight in banded 4-OHT–treated MCM;Mycfl/fl mice was reduced 52% compared with that in vehicle-treated MCM;Mycfl/fl mice (45.7% versus 21.8%, P < 0.001). To determine whether Myc was also required for upregulation of genes involved in glucose metabolism, we analyzed the total ventricular RNA from MCM;Mycfl/fl mice with or without 4-OHT treatment. As shown in Figure 3F, TAC induced a 2.6-fold increase in ENO-1α (P < 0.001), 2.43-fold increase in PFK-1 (P < 0.05), 4-fold increase in LDHA (P < 0.001), and 4.4-fold increase in HK-2 (P < 0.05) expression in vehicle-treated MCM;Mycfl/fl mice when compared with sham-operated mice. Pretreatment of MCM;Mycfl/fl mice with 4-OHT attenuated the expression of all these genes after hemodynamic load. These data suggest that Myc is required for the expression of glucose metabolism genes in response to hemodynamic stress in adult myocardium.
Myc is required for induction of the LDHA gene in response to hypertrophic agonists.
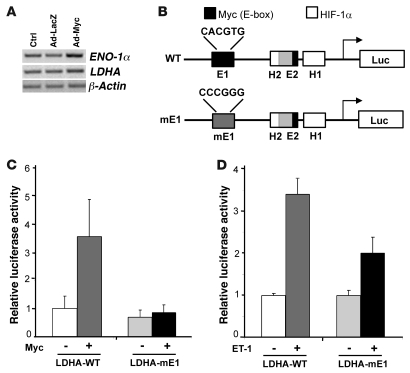
As neonatal cardiomyocytes are known to preferentially use glucose as an energy source and have a very low metabolic rate (29), their application as a model for metabolic studies is limited. However, although neonatal rat ventricular myocytes (NRVMs) express Myc and glucose metabolism genes when initially isolated, these factors are downregulated as the cells differentiate in cell culture, with a corresponding increase in FAO. To determine whether forced expression of Myc in vitro would reactivate genes involved in glucose utilization, we examined total ventricular RNA from NRVMs infected with no virus (control), AdLacZ, or AdMyc by semiquantitative PCR. Myc activation led to upregulation of a panel of genes including ENO-1α and LDHA (Figure 4A). In addition to Myc, HIF-1α has also been implicated in regulating the LDHA gene (30). Thus, to confirm that in addition to being sufficient, Myc was also necessary for LDHA induction, we utilized a luciferase-based LDHA reporter construct. The LDHA promoter contains 2 Myc-binding sites called E-boxes (31) and 2 HIF-1α–binding sites (30). Since mutation of either E-box abolishes Myc responsiveness (31), NRVMs were transfected with either the wild-type LDHA promoter driving luciferase (LDHA-WT; Figure 4B) or a mutant promoter wherein the 5ι-E-box has been mutated, preventing Myc binding but retaining full ability to bind HIF-1α (LDHA-mE1; Figure 4B) (31). Overexpression of Myc induced the LDHA-WT promoter 3.7-fold (P < 0.01) but had no effect on the LDHA-mE1 promoter (Figure 4C). Stimulation of transfected NRVMs with the hypertrophic agonist ET-1 induced LDHA-WT by 3.4-fold (Figure 4D; P < 0.001). However, mutating the Myc-binding site attenuated this ET-1–mediated induction by 38% (Figure 4D; P < 0.01), suggesting that Myc is required for LDHA upregulation in response to hypertrophic agonists and is a direct Myc target gene in cardiac myocytes.
Figure 4. Myc is required for induction of the LDHA gene in an E-box–dependent manner in response to hypertrophic agonists.
(A) Total ventricular RNA from NRVMs infected with no virus (Ctrl), AdLacZ, or AdMyc was analyzed by semiquantitative PCR for glycolytic genes (ENO-1α and LDHA) or β-actin control. (B) Schematic diagram depicting the wild-type LDHA promoter containing 2 E-boxes (E1 and E2) and 2 HIF-1α–binding (H1 and H2) sites. The E2 and H2 binding sites overlap. The mutation of the 5ι-E-box, mE1, is also shown. (C) NRVMs were transfected with either LDHA-WT or LDHA-mE1 infected with control or Myc-expressing vector. The relative luciferase activities are shown for the wild-type or mutated LDHA promoter. *P < 0.01 for LDHA-WT + Myc versus LDHA-WT without Myc or LDHA-mE1 + Myc. (D) NRVMs were transfected with either LDHA-WT or mutated promoter LDHA-mE1 and stimulated with vehicle or ET-1 for 16 hours. *P < 0.001 versus LDHA-WT without ET-1; **P < 0.01 versus LDHA-mE1 + ET-1. All data shown are representative of at least 3 independent experiments.
Myc activation downregulates FAO gene expression in the heart.
Typically, glucose oxidation and FAO are reciprocally regulated in the heart. To determine whether Myc contributed to the downregulation of FAO enzyme expression as is seen after pathological stress, RNA and protein from Myc-activated ventricles was examined for a panel of FAO genes and their transcriptional regulators. Expression levels of coactivator PGC-1α, PGC-1β, PGC-1–related coactivator (PPRC), nuclear receptor PPARα, estrogen-related receptor (ERRα), and nuclear respiratory factor 1 (NRF-1) as well as their downstream targets carnitine palmitoyltransferase 1 (CPT-1), medium chain acyl-CoA dehydrogenase (MCAD; implicated in FAO and respiratory chain enzymes), ATP synthase subunit alpha (ATP5A), ATP synthase subunit beta (ATP5B), ATP synthase subunit gamma (ATP5C), cytochrome c oxidase complex IV, subunit I (COX-I), cytochrome c oxidase complex IV, subunit IV (COX-IV), and cytochrome c (Cyt-C) were determined. Myc activation downregulated expression of CPT-1 and MCAD (Figure 5C), which corresponded to reductions in protein expression (Figure 5B). This was likely secondary to the decreased protein levels of the FAO transcriptional regulator PGC-1α. However, the finding that RNA levels of PGC-1α were unchanged suggests that the reduction in PGC-1α protein levels might be posttranslational (Figure 5A). PGC-1β, PPRC, PPARα, and ERRα transcripts were unchanged, but NRF-1 transcripts were upregulated (Figure 5A). Since no changes were observed in RNA and protein levels for respiratory chain enzymes (Figure 5, C and D), Myc activation of NRF-1 appears to be sufficient to maintain respiratory apparatus gene expression even in the setting of reduced PGC-1α–dependent transcription.
Figure 5. Expression of FAO genes is downregulated with Myc activation in the heart.
(A) Total ventricular RNA from NTg and MycER mice with or without 4-OHT treatment for 7 days assayed by semiquantitative PCR for FAO enzymes genes (PGC-1α, PGC-1β, PPRC, PPARα, ERRα, and NRF-1) together with β-actin as a loading control. (B) Total ventricular protein lysates from mice of the indicated genotypes were probed with antibodies against different FAO associated proteins (PGC-1α and MCAD) along with α-CaA as a loading control. (C) Total ventricular RNA from NTg and MycER mice with or without 4-OHT treatment for 7 days as assayed by semiquantitative PCR for target genes of PPARα and ERRα (CPT-1, MCAD, ATP5A, ATP5B, ATP5C, COX-I, COX-IV, and Cyt-C) together with β-actin as a loading control. (D) Total ventricular protein lysates from mice of the indicated genotypes were probed with antibodies against ATP5A, ATP5B, ATP5C, COX-IV, and α-CaA.
Activation of Myc in adult myocardium induces mitochondrial biogenesis.
In addition to induction of enzymes involved in glycolysis, pathologic stress leads to limited mitochondrial biogenesis in adult myocardium (24). Since Myc has been reported to induce a number of nuclear-encoded mitochondrial genes (32–35), we determined its effect on mitochondrial biogenesis in the heart. Electron microscopy was performed on myocardial sections from NTg and MycER mice after OHT treatment to directly visualize mitochondria (Figure 6A). Mitochondria were more numerous and increased in size in the ventricles of Myc-activated mice compared with NTg control mice. Quantitative morphometric analysis confirmed that mitochondrial volume density was increased by 1.6-fold in Myc-activated hearts (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI38331DS1). Additionally, we examined total mitochondrial DNA (mtDNA) content as an indirect indicator of mitochondrial mass. mtDNA was isolated from ventricles of NTg and MycER mice treated with vehicle or 4-OHT for 7 days. As shown, Myc increased mtDNA, suggesting that mitochondrial biogenesis was induced (Figure 6B). To quantify the increase in mtDNA, we performed real-time PCR on total genomic and mtDNA isolated from treated NTg or MycER ventricles. DNA levels of the mitochondrial-encoded gene COX1 were compared with those of the nuclear-encoded gene PPRC (Figure 6C). Myc activation increased mtDNA 3.1-fold (P < 0.01 vs. untreated MycER). These observations corroborate the notion that Myc activation in adult heart induces mitochondrial biogenesis.
Figure 6. Myc stimulates mitochondrial biogenesis.
(A) Transmission electron microscopy performed on heart sections from NTg and MycER mice with OHT treatment. Scale bar: 1 μm. (B) Total mtDNA isolated from ventricles of NTg and MycER mice with or without 4-OHT treatment and loaded on ethidium bromide–stained agarose gel (1.2%). (C) Quantitative real-time PCR on the mitochondrial gene cytochrome c oxidase subunit I (COX1), along with the nuclear gene PPRC as an internal control from MycER mice with or without 4-OHT treatment. *P < 0.01 versus untreated MycER littermates; n = 3. (D) Representative polarograph of mitochondria isolated from treated NTg and MycER ventricles (n = 4/group) energized with complex I substrates in the presence of 2.5 mM Pi and ADP pulses, at the indicated concentrations. Mitochondria (mit) in both groups responded to ADP additions with transient dissipation of membrane potential (Dy) and acceleration of O2 consumption that slowed down after ADP was phosphorylated and Dy recovered. All data shown are representative of at least 3 independent experiments.
To clarify whether these new mitochondria in Myc-activated hearts functioned normally, we also assessed oxidative capacity in mitochondria isolated from the ventricles of 4-OHT–treated NTg and MycER mice. Myc induction in the adult heart had no significant effect on the respiratory control ratio (RCR; Supplemental Figure 1B), nor did Myc-induced mitochondrial biogenesis lead to significant changes in mitochondrial ability to phosphorylate ADP. Representative tracings (Figure 6D) demonstrate that energized mitochondria in both groups responded to ADP additions with membrane potential dissipation and acceleration of O2 consumption (state 3). As expected, membrane potential recovered, and O2 consumption returned to state 4 after ADP was phosphorylated. The time periods required for ADP phosphorylation were similar in the two genotypes (Supplemental Figure 1C), suggesting that ADP phosphorylating capacity did not change in Myc-activated hearts.
Myc directly activates nuclear genes involved in mitochondrial replication and biogenesis.
To determine the mechanism whereby Myc could be regulating mitochondrial biogenesis, we examined the expression of the critical mitochondrial transcription factor TFAM, mtDNA polymerase catalytic subunits POLG and POLG2, and mitochondrial protein import and assembly machinery subunits TOM20 and TOM70 in 4-OHT–treated MycER ventricles (Figure 7A). As shown, Myc activation led to upregulation of a panel of genes involved in mitochondrial replication and biogenesis. Myc also induced expression of TFAM in vitro. Adenoviral overexpression of Myc induced TFAM expression in NRVMs. Confocal microscopy of immunostained NRVMs revealed that while control or Ad-LacZ–infected cardiomyocytes demonstrated low-level TFAM expression (Supplemental Figure 2C), abundant TFAM was detected in cardiac myocytes after Ad-Myc infection (Supplemental Figure 2G).
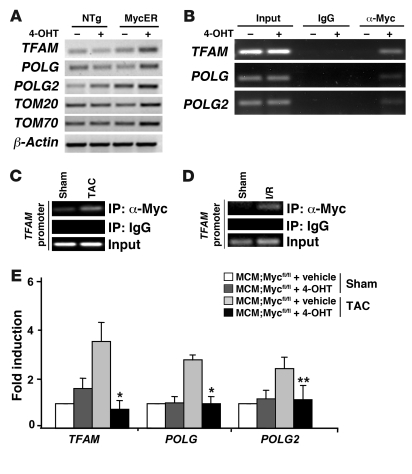
Figure 7. Myc directly regulates genes involved in mitochondrial replication and biogenesis.
(A) RT-PCR analysis performed for TFAM, POLG, POLG2, TOM20, and TOM70 on total RNA prepared from ventricular tissue from NTg and MycER mice with or without 4-OHT treatment. β-Actin was used as a loading control. (B) Myc binds to the TFAM, POLG, and POLG2 promoters in situ. ChIP analysis was performed with anti-Myc antibody and PCR primers to the TFAM, POLG, and POLG2 promoters. Ventricular tissue obtained from MycER mice 24 hours after vehicle or 4-OHT treatment was analyzed. Input lanes show PCR product derived from chromatin before immunoprecipitation to verify equal loading. (C and D) Endogenous Myc binds to the TFAM promoter in response to pathologic stress. Ventricular tissue obtained from wild-type mice subjected to sham operation or TAC (1 day) and after I/R injury (30 minutes of ischemia and 24 hours of reperfusion) were analyzed with anti-Myc antibody and PCR primers to the TFAM promoter. Input lanes show PCR product derived from chromatin before immunoprecipitation to verify equal loading. (E) Myc-null mice demonstrate attenuated stress-induced increase in mitochondrial replication and biogenesis genes. Total ventricular RNA from MCM;Mycfl/fl mice with or without 4-OHT treatment subjected to sham or TAC operation was assayed using quantitative real-time PCR for TFAM, POLG, and POLG2 (n = 3/group). *P < 0.05, **P < 0.001 for vehicle- versus 4-OHT–treated MCM;Mycfl/fl mice after TAC operation.
To determine whether these genes involved in mitochondrial biogenesis are direct targets of Myc, we performed ChIP analysis. Amplification of Myc-immunoprecipitated DNA complexes from 4-OHT–treated MycER ventricles revealed Myc binding to endogenous TFAM, POLG, and POLG2 promoters (Figure 7B). To confirm that endogenous Myc also bound these promoters in the heart after stress, we subjected wild-type mice to pressure overload for 24 hours (Figure 7C) or an ischemia for 30 minutes followed by 24 hours of reperfusion (Figure 7D). ChIP performed on protein-DNA complexes isolated from ventricular tissue from wild-type mice subjected to sham or TAC surgery revealed that binding of endogenous Myc to the TFAM promoter was enhanced by hypertrophic stimulus or pressure overload and after I/R injury (Figure 7, C and D).
To confirm that Myc was required for the upregulation of genes involved in mitochondrial replication and biogenesis after hypertrophic stimuli, we analyzed the total ventricular RNA from MCM;Mycfl/fl mice with or without 4-OHT treatment that underwent sham or TAC surgery. As shown in Figure 7E, TAC induced a 3.6-fold increase in TFAM (P < 0.05), 2.8-fold increase in POLG (P < 0.05), and 2.4-fold increase in POLG2 (P < 0.001) expression in vehicle-treated MCM;Mycfl/fl mice when compared with sham-operated mice. In contrast, Myc-deficient mice demonstrated attenuated expression of these genes after hemodynamic load. Thus, Myc directly activates nuclear genes involved in mitochondrial replication and biogenesis in response to hemodynamic stress in adult myocardium.
Myc activation attenuates ischemia-induced LV dysfunction and infarct size.
To determine whether the changes in gene expression and mitochondrial biogenesis we observed were adaptive or maladaptive, we examined LV function in response to global ischemia in isolated, Langendorff-perfused hearts from 4-OHT–treated NTg or MycER mice. Hearts were perfused with modified Krebs-Henseleit buffer, and after 30 minutes of stabilization, hearts were subjected to 30 minutes of global, normothermic ischemia followed by 60 minutes of reperfusion. LV function was assessed every 15 minutes during the recovery phase. Activation of Myc resulted in a 46.9% increase in cardiac mass normalized to body weight compared with treated NTg mice (Figure 8A; 6.13 ± 0.5 versus 4.17 ± 0.24 mg/g, P < 0.005). As shown in Figure 8B, although there was no significant increase in the maximal rate of contraction (+dP/dtmax) in Myc-activated hearts at baseline (P = 0.08), higher contractility was seen throughout the reperfusion period (P < 0.05 versus treated NTg). A detailed examination of hemodynamics at 1 hour after ischemia revealed that both +dP/dtmax and relaxation (–dP/dtmax) were improved (Table 1). Other parameters such as heart rate, LV end-diastolic pressure, and LV systolic pressure were not significantly different between genotypes.
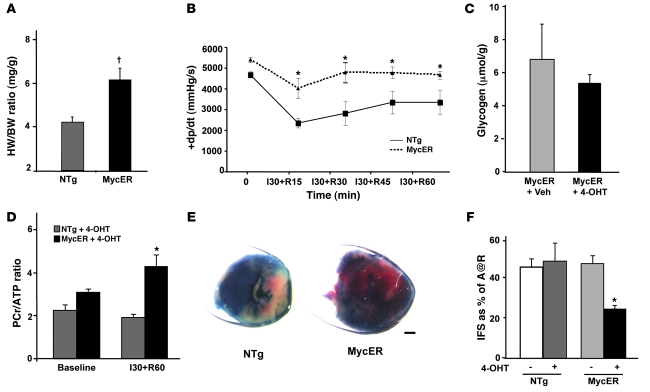
Figure 8. Myc-activated hearts display improved tolerance to I/R injury.
(A) Activation of MycER increases cardiac mass. Measured heart weights (HW; mg) were normalized to body weight (BW; g); n = 3/group. †P < 0.005. (B) Contractile performance of hearts from MycER compared with NTg hearts after OHT treatment measured in Langendorff preparations. Hearts were subjected to 30 minutes of global normothermic ischemia (I) followed by the indicated periods of reperfusion (R). The rate of systolic pressure development (+dp/dt) measured at baseline and over time after reperfusion is shown. *P < 0.05 versus treated NTg littermates; n = 4/group. (C) Glycogen levels in μmol/g of tissue as determined by mass spectrometry analysis on ventricular tissue from MycER mice with or without 4-OHT treatment (n = 3/group). (D) PCr/ATP ratio at baseline and after ischemia for 30 minutes followed by 60 minutes of reperfusion as determined by 31P-NMR on ventricular tissue from NTg and MycER mice after 4-OHT treatment. *P < 0.05 versus treated NTg littermates; n = 4/group. (E) Representative 2,3,5-triphenyltetrazolium chloride–stained hearts from NTg and MycER mice treated with 4-OHT after 24 hours of reperfusion. (F) Mean infarct sizes from NTg and MycER mice with or without 4-OHT treatment after 24 hours of reperfusion are shown. A@R, area at risk. *P < 0.001 versus treated NTg littermates; n = 5/group.
Table 1 .
Detailed hemodynamic parameters of isolated perfused hearts at baseline and after 1 hour of reperfusion
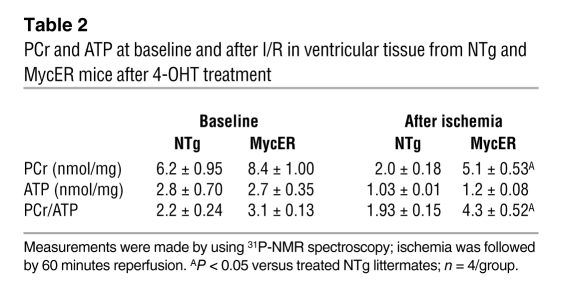
During ischemia, glycolysis assumes a more important role as the primary energy producing pathway for the heart, since FAO is restricted and glycogen supplies glucose for anaerobic glycolysis (36). Thus, to determine whether the improved tolerance to ischemia we saw in Myc-activated mice could be explained, at least in part, by beneficial alterations in myocardial energetic status, we examined hearts from 4-OHT–treated NTg or MycER mice after 30 minutes of ischemia followed by 15 minutes of reperfusion for glycogen levels. Although Myc-activated hearts displayed a 2-fold increase in glycogen compared with 4-OHT–treated NTg mice at baseline, Myc-activated hearts displayed preserved glycogen levels after 30 minutes of ischemia (5.3 ± 0.5 versus 6.8 ± 2.0 μmol/g, Figure 8C). To assess whether this resulted in improved high-energy phosphate content, we determined levels of phosphocreatine (PCr) and ATP at baseline and after ischemia followed by 60 minutes of reperfusion using 31P-NMR. Although PCr (6.2 ± 0.95 versus 8.4 ± 1.0 nmol/mg wet weight of tissue, Table 2) and PCr/ATP ratio (2.2 ± 0.24 versus 3.1 ± 0.13, Figure 8D and Table 2) were similar in baseline hearts of both the genotypes, activated MycER hearts had higher PCr (5.1 ± 0.53 versus 2.0 ± 0.18 nmol/mg wet weight of tissue, Table 2; P < 0.05) and PCr/ATP ratio (4.3 ± 0.5 versus 1.93 ± 0.15, Figure 8D; P < 0.05) after ischemia compared with NTg hearts. This suggests that improved energy supply secondary to glycogen utilization could account for at least part of the functional improvement in ischemic Myc transgenic hearts.
Table 2 .
PCr and ATP at baseline and after I/R in ventricular tissue from NTg and MycER mice after 4-OHT treatment
To determine whether the changes seen after Myc activation in the adult myocardium would also attenuate cardiac injury, we subjected hearts from 4-OHT–treated NTg or MycER mice to I/R injury and determined infarct size. Although the area at risk was indistinguishable between NTG and MycER hearts (Figure 8E), Myc activation led to a 50% reduction in infarct size when compared with vehicle-treated MycER mice or 4-OHT–treated NTg mice (24 ± 1.8 versus 46.7 ± 3.5 or 48 ± 8.4 infarct size as a percentage of area at risk, Figure 8F; P < 0.001).
Discussion
We and others have shown that Myc and its downstream effectors play an important role in controlling hypertrophic and hyperplastic cardiac growth (21, 37–39). However, Myc is upregulated in the adult heart in response to many diverse environmental stresses not associated with growth, such as ischemia and preconditioning. The explanation and significance of this phenomenon were unknown, but our data imply a novel role for Myc in the heart; namely, regulating cardiac metabolism and mitochondrial biogenesis. Although is has been known that glucose utilization is enhanced after stress in the heart for some time (for review, see refs. 22, 40, 41), the mechanisms regulating this process are poorly understood. It has been assumed that HIF-1α played an important role in this process, at least after ischemic insults (42), but the transcriptional regulators in hypertrophy were unknown. HIF-1α cannot account for the changes in glucose metabolism we saw, as HIF-1α levels did not change with Myc activation (data not shown), and LDHA induction in response to hypertrophic agonists was attenuated when the Myc binding was impaired despite intact HIF-1α sites (Figure 4). Also, HIF-1α overexpression typically inhibits mitochondrial biogenesis (43). It is interesting to note that although mutating the Myc-binding site completely abrogated transactivation of LDHA-mE1 promoter by overexpression of Myc, induction of the LDHA-mE1 promoter was not completely abolished with the hypertrophic agonist ET-1. This observation suggests that factors in addition to Myc may be operational in regulating this process. The data presented here demonstrate that Myc induction leads to an increase in 18F-FDG uptake and alters the substrate oxidation pattern, with carbohydrates replacing fatty acids for oxidizing acetyl-CoA entering the citric acid cycle. Myc activation in the adult heart promotes glucose oxidation by inducing expression of genes involved in regulation of this pathway, with a parallel downregulation of expression of FAO enzymes. A number of key glycolytic enzymes in the pathway leading to either glucose oxidation or lactate production have been identified as direct Myc target genes. These include ENO-1α, HK-2, LDHA, and glucose transporter I, as well as PFK, defined as the rate-limiting enzyme for glycolysis in the hypertrophied heart (15, 27, 44, 45). Consistent with these findings our data demonstrated increased expression of ENO-1α, PFK-1, LDHA, and MCT-1 after Myc activation. Indeed, this was a direct effect of Myc activation in heart as revealed by Myc occupancy at the promoters of these key genes. Conversely, Myc-deficient hearts displayed attenuated expression of ENO-1α, PFK-1, LDHA, and HK-2 after hemodynamic load. Thus, Myc appears to be sufficient and necessary for regulating the expression of these genes in adult heart after stress.
Myc activation also resulted in the reduction in expression of FAO genes, although this was likely an indirect effect, mediated by reduced levels of the transcriptional cofactor PGC-1α. This reduction in protein was independent of its transcript levels. It has been reported that PGC-1α levels are also reduced in response to both hemodynamic load (46) and ischemia (22), although the mechanism(s) mediating this effect have been speculative. It has been proposed that PGC-1α levels are regulated posttranslationally (47, 48), and several plausible mechanisms for Myc-dependent regulation of PGC-1α levels exist that could explain our results. Recent studies have reported that PGC-1α is a short-lived protein that is rapidly degraded via the ubiquitin-proteasome system (49). Since Myc promotes ubiquitin-dependent proteolysis (50, 51), PGC-1α could be a target of Myc-dependent degradation by the ubiquitin-proteasome system. An alternate mechanism that could account for the reduction in PGC-1α is Myc induction of microRNAs, which recognize PGC-1α and preferentially inhibit translation, as has been proposed for other Myc-regulated transcriptional activators (52).
An unexpected finding of this study was the observation that Myc activation in adult myocardium triggers mitochondrial biogenesis, as current dogma holds that this process is PGC-1 dependent. Typically, mitochondrial biogenesis and FAO gene expression are regulated concomitantly in the heart with reductions in PGC-1α, leading to defects in both processes. However, our results demonstrate that Myc increased mitochondrial number while at the same time decreasing PGC-1α levels and FAO gene expression. This is the pattern typically seen in the early phases of the hypertrophic response when Myc is still expressed. Although it has been suggested that Myc can upregulate PGC-1β (43), which is also capable of mediating biogenesis (53), this does not appear to be true in the heart, as transcripts for all PGC-1 family members were unchanged in Myc-activated ventricles (Figure 5A). Our results suggest that Myc-induced mitochondrial biogenesis may be PGC-1 independent; however, further studies will be required to corroborate our findings, including assessing the effects of Myc activation in a PGC-1α/PGC-1β–deficient background. A novel Myc-dependent pathway for mitochondrial biogenesis is consistent with the relatively normal mitochondrial numbers in both PGC-1α– (12, 54) and PGC-1β–null (23) mice. More importantly, mitochondrial number and appearance were normal in fetal ventricles of combined PGC-1α– and PGC-1β–null mice at early developmental time points (23). Thus, PGC-1 expression cannot be an obligate requirement for mitochondrial biogenesis in the heart. Further studies will be required to resolve these findings with other recent in vitro studies suggesting that the PGC-1 coactivators play an absolutely essential but complementary role in differentiation-induced mitochondrial biogenesis in some tissues (55). The basis of Myc’s ability to induce mitochondrial biogenesis appears to be related to its ability to directly stimulate a wide panel of nuclear-encoded genes involved in this process (56, 57). Regardless, Myc-dependent mitochondrial biogenesis provides a rational mechanism to explain the early changes in mitochondrial number seen in the heart after pathologic stress at a time when PGC-1 levels are decreasing.
It is interesting to note that the increased cardiac mass with increased glucose uptake and utilization that develops with Myc activation in MycER hearts is associated with normal LV contractility (39). Similar observations have been made in other transgenic models, where cardiac hypertrophy and increases in glucose uptake and glycolysis did not compromise cardiac function. Cardiac-specific ablated glucose transporter GLUT4 mice developed hypertrophy with an increase in glycolysis and displayed preserved +dP/dT and LV systolic pressure and contractile function (58). Also, insulin-dependent glucose transporter GLUT1–overexpressing mice demonstrated an increase in glycolysis that was associated with normal contractile performance under basal conditions (59). Furthermore, Liao et al. showed that an increase in glucose utilization in hypertrophied hearts protects against contractile dysfunction after chronic pressure overload. While more detailed metabolic studies are required, it is interesting to speculate that the metabolic changes we demonstrated could account for this benefit. Indeed, the cardioprotective effect of Myc activation was even more apparent when these hearts were subjected to ischemic stress; however, both ischemic reperfusion and mitochondrial functional studies were performed under conditions where pyruvate was the primary substrate, and it is possible that defects would have been seen if fatty acids were used as the substrate. Interestingly, despite use of a no-flow ischemia model, which is more susceptible to lactic acid accumulation from anaerobic glycolysis (60, 61), Myc-activated hearts demonstrated greater tolerance to I/R injury. In fact, it should be noted that creatine rephosphorylation rate, which is an in vivo assessment of mitochondrial function after ischemia (62), was superior in Myc-activated hearts. This is in contrast to previous studies wherein hypertrophied hearts were more sensitive to ischemic injury and more dependent on glycolytic substrates than nonhypertrophied hearts (63–67). The cellular mechanisms responsible for the observed cardioprotective effect in Myc-activated hearts are speculative; however, Myc’s ability to induce mitochondrial biogenesis with normally functioning mitochondria in the setting of increased glucose metabolism might explain this effect. This result is reminiscent of the protection against ischemic injury seen in GLUT1-overexpressing transgenic mice, which also demonstrated increases in glucose uptake (68), although Myc-activated mice demonstrated increased glucose uptake and utilization as well as increased numbers of functional mitochondria. In fact, there is suggestive evidence that shifting the substrate preference of the heart away from fatty acids toward carbohydrate oxidation can improve LV function and slow the progression of heart failure (for review, see refs. 7, 41, 69– 71).
In summary, we have demonstrated that Myc activation induced a coordinated metabolic alteration in the adult heart, similar to that seen after hemodynamic or ischemic stress, that was associated with increase in the number of functional mitochondria. Furthermore, our data suggest that Myc might mediate a novel PGC-1–independent pathway for mitochondrial biogenesis. These Myc-dependent metabolic alterations serve an adaptive role, equipping the heart with the enhanced ability to respond to ischemic insults.
Methods
Transgenic mice and animal studies.
Inducible α–myosin heavy chain (α-MHC) MycER transgenic mice have been described previously (39). Myc-floxed mice (Mycfl/fl) were provided by F. Alt (Harvard University, Boston, Massachusetts, USA) and genotyped as described (72). The tamoxifen-inducible MCM mice were generated by J. Molkentin (Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA) under the control of the α-MHC promoter (73). To activate Myc or Cre in these transgenic mice, 1 mg 4-OHT (Sigma-Aldrich) dispersed in peanut oil by sonication was injected intraperitoneally daily. All mice were maintained on a C57BL/6 (MycER) or FVB (MCM;Mycfl/fl) background. All animal studies were conducted in strict accordance with the NIH guidelines for humane treatment of animals and were approved by the Animal Research Committee at the University of California, Los Angeles. The TAC, myocardial I/R, and delayed preconditioning of the heart models have been previously described (39, 74).
Metabolic and mitochondrial studies.
For 13C-NMR and isotopomer analyses, fractional contributions of metabolic substrates were determined in NTg and Myc-induced mice induced for 3 days as previously published and detailed in Supplemental Methods (28). Infarct size was determined after 30 minutes of ischemia followed by 24 hours of reperfusion as we have described (75). The perfused mouse heart model is described in detail in Supplemental Methods. For microPET analyses of cardiac 18F-FDG uptake, we examined 3 littermates each from NTg and MycER mice with or without OHT treatment. Mice were imaged for 10 minutes in a Siemens microPET Focus220 (Siemens Preclinical Solutions) using intraperitoneal injection of FDG (~220 μCi) and 60 minutes uptake with a single reconstructed image volume.
Cell culture and plasmids.
NRVMs were prepared (76) and infected with adenoviruses 36 hours before use. Construction of AdMyc (77) has been previously described. For gene expression studies, NRVMs were transfected with wild-type LDHA, LDHA mutant E-box (E1) plasmid (gift from C.V. Dang, Johns Hopkins University, Baltimore, Maryland, USA), and pCMV-β-galactosidase. Twenty-four hours after transfection, cells were treated with 10–8 M endothelin-1 (Sigma-Aldrich) for an additional 24 hours and then subjected to β-galactosidase assay and luciferase activity assay (Promega). Luciferase activity was normalized to β-galactosidase activity and presented as the mean ± SEM of 3 independent experiments performed in triplicate.
Transmission electron microscopy.
To prepare myocardial tissue for electron microscopy, hearts were arrested in diastole by CdCl2 and fixed in 2% glutaraldehyde, 2% paraformaldehyde at room temperature for 2 hours, followed by 24 hours at 4°C. Specimens were rinsed in 0.1 M phosphate buffer (PB), pH 7.4, postfixed in 1% osmium tetroxide in PB (pH 7.4) at 4°C for 1 hour, dehydrated in graded ethanol followed by propylene oxide, and embedded in Eponate 812 (Ted Pella Inc.). Ultrathin sections (around 60–70 nm) from longitudinal parts were cut and contrasted with uranyl acetate followed by lead citrate and examined in a JEOL 10000X transmission electron microscope at 80 kV.
Protein and RNA analysis.
Western blots were performed on protein extracts from whole ventricles according to established protocols (76). Total RNA was extracted from frozen tissue samples using Tri Reagent (Sigma-Aldrich) and real-time quantitative PCR or ribonuclease protection assay (RPA) performed and analyzed as described. Complete details on methodology and primers are provided in Supplemental Methods.
ChIP analysis.
ChIP analysis of ventricular tissue has been previously described (78). Ventricular tissues obtained from hearts were treated with 1% formaldehyde, and chromatin was sheared by sonication on ice with nuclear lysis buffer. DNA-protein complexes were immunoprecipitated with anti-Myc antibody or control rabbit IgG. After immunoprecipitation, precipitated DNA was analyzed by the PCR primers listed in Supplemental Table 1.
Statistics.
All data are presented as mean ± SEM. Results were compared by ANOVA and the Fisher protected least-significant difference tests, with significance at a P value less than 0.05.
Supplementary Material
Acknowledgments
We thank S. Chan and D. Liem for technical assistance and Angélica Keller, University of Paris, for the gift of enolase-1α antibody. We also thank Marianne Cilluffo for expert assistance with electron microscopy and David Stout for his assistance with animal imaging. This work was supported by gifts from the Laubisch Fund (W.R. MacLellan), as well as NIH grants P01 HL080111, R01 HL70748, and HL09491 to W.R. MacLellan. Work performed in the Portman laboratory was supported by a subcontract to R01CA106650-06 (David Hockenbery, Fred Hutchinson Cancer Research Center).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(5):1494–1505. doi:10.1172/JCI38331.
References
- 1.Lopaschuk GD, Spafford MA. Energy substrate utilization by isolated working hearts from newborn rabbits. Am J Physiol. 1990;258(5 Pt 2):H1274–H1280. doi: 10.1152/ajpheart.1990.258.5.H1274. [DOI] [PubMed] [Google Scholar]
- 2.Lopaschuk GD, Collins-Nakai RL, Itoi T. Developmental changes in energy substrate use by the heart. Cardiovasc Res. 1992;26(12):1172–1180. doi: 10.1093/cvr/26.12.1172. [DOI] [PubMed] [Google Scholar]
- 3.Spitkovsky D, et al. Activity of complex III of the mitochondrial electron transport chain is essential for early heart muscle cell differentiation. FASEB J. 2004;18(11):1300–1302. doi: 10.1096/fj.03-0520fje. [DOI] [PubMed] [Google Scholar]
- 4.Nadal-Ginard B, Mahdavi V. Molecular basis of cardiac performance. Plasticity of the myocardium generated through protein isoform switches. J Clin Invest. 1989;84(6):1693–1700. doi: 10.1172/JCI114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz K, Boheler KR, de la Bastie D, Lompre AM, Mercadier JJ. Switches in cardiac muscle gene expression as a result of pressure and volume overload. Am J Physiol. 1992;262(3 pt 2):R364–R369. doi: 10.1152/ajpregu.1992.262.3.R364. [DOI] [PubMed] [Google Scholar]
- 6.Buttrick PM, Kaplan M, Leinwand LA, Scheuer J. Alterations in gene expression in the rat heart after chronic pathological and physiological loads. J Mol Cell Cardiol. 1994;26(1):61–67. doi: 10.1006/jmcc.1994.1008. [DOI] [PubMed] [Google Scholar]
- 7.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85(3):1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 8.Rosano GM, Fini M, Caminiti G, Barbaro G. Cardiac metabolism in myocardial ischemia. Curr Pharm Des. 2008;14(25):2551–2562. doi: 10.2174/138161208786071317. [DOI] [PubMed] [Google Scholar]
- 9.de Jonge R, de Jong JW. Ischemic preconditioning and glucose metabolism during low-flow ischemia: role of the adenosine A1 receptor. Cardiovasc Res. 1999;43(4):909–918. doi: 10.1016/S0008-6363(99)00137-6. [DOI] [PubMed] [Google Scholar]
- 10.Lehman JJ, Kelly DP. Gene regulatory mechanisms governing energy metabolism during cardiac hypertrophic growth. Heart Fail Rev. 2002;7(2):175–185. doi: 10.1023/A:1015332726303. [DOI] [PubMed] [Google Scholar]
- 11.Lee SS, et al. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15(6):3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3(4):e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillies RJ, Gatenby RA. Adaptive landscapes and emergent phenotypes: why do cancers have high glycolysis? J Bioenerg Biomembr. 2007;39(3):251–257. doi: 10.1007/s10863-007-9085-y. [DOI] [PubMed] [Google Scholar]
- 14.Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis — the seventh hallmark of cancer. Cell Mol Life Sci. 2008;65(24):3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yalcin A, Telang S, Clem B, Chesney J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp Mol Pathol. 2009;86(3):174–179. doi: 10.1016/j.yexmp.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Evan GI, Littlewood TD. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993;3(1):44–49. doi: 10.1016/S0959-437X(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 17.Brand T, et al. Proto-oncogene expression in porcine myocardium subjected to ischemia and reperfusion. Circ Res. 1992;71(6):1351–1360. doi: 10.1161/01.res.71.6.1351. [DOI] [PubMed] [Google Scholar]
- 18.Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci U S A. 1988;85(2):339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci U S A. 1999;96(23):13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baena E, et al. c-Myc regulates cell size and ploidy but is not essential for postnatal proliferation in liver. Proc Natl Acad Sci U S A. 2005;102(20):7286–7291. doi: 10.1073/pnas.0409260102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong W, et al. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J. 2006;25(16):3869–3879. doi: 10.1038/sj.emboj.7601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: a question of balance. J Clin Invest. 2005;115(3):547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai L, et al. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22(14):1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106(7):847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finck BN, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) regulatory cascade in cardiac physiology and disease. Circulation. 2007;115(19):2540–2548. doi: 10.1161/CIRCULATIONAHA.107.670588. [DOI] [PubMed] [Google Scholar]
- 26.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18(4):357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 27.Osthus RC, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275(29):21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 28.Hyyti OM, et al. Cardioselective dominant-negative thyroid hormone receptor (Delta337T) modulates myocardial metabolism and contractile efficiency. Am J Physiol Endocrinol Metab. 2008;295(2):E420–E427. doi: 10.1152/ajpendo.90329.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kammermeier H, Rose H. Are isolated cardiomyocytes a suitable experimental model in all lines of investigation in basic cardiology? Basic Res Cardiol. 1988;83(4):343–349. doi: 10.1007/BF02005819. [DOI] [PubMed] [Google Scholar]
- 30.Semenza GL, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 31.Shim H, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94(13):6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci U S A. 2002;99(9):6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ. Identifying genes regulated in a Myc-dependent manner. J Biol Chem. 2002;277(40):36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- 34.Wonsey DR, Zeller KI, Dang CV. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc Natl Acad Sci U S A. 2002;99(10):6649–6654. doi: 10.1073/pnas.102523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orian A, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17(9):1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Depre C, Vanoverschelde JL, Taegtmeyer H. Glucose for the heart. Circulation. 1999;99(4):578–588. doi: 10.1161/01.cir.99.4.578. [DOI] [PubMed] [Google Scholar]
- 37.Jackson T, Allard MF, Sreenan CM, Doss LK, Bishop SP, Swain JL. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol Cell Biol. 1990;10(7):3709–3716. doi: 10.1128/mcb.10.7.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson T, Allard MF, Sreenan CM, Doss LK, Bishop SP, Swain JL. Transgenic animals as a tool for studying the effect of the c-myc proto-oncogene on cardiac development. Mol Cell Biochem. 1991;104(1–2):15–19. doi: 10.1007/BF00229798. [DOI] [PubMed] [Google Scholar]
- 39.Xiao G, et al. Inducible activation of c-Myc in adult myocardium in vivo provokes cardiac myocyte hypertrophy and reactivation of DNA synthesis. Circ Res. 2001;89(12):1122–1129. doi: 10.1161/hh2401.100742. [DOI] [PubMed] [Google Scholar]
- 40.Taha M, Lopaschuk GD. Alterations in energy metabolism in cardiomyopathies. Ann Med. 2007;39(8):594–607. doi: 10.1080/07853890701618305. [DOI] [PubMed] [Google Scholar]
- 41.Ingwall JS. Energy metabolism in heart failure and remodelling. Cardiovasc Res. 2009;81(3):412–419. doi: 10.1093/cvr/cvn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seagroves TN, et al. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol. 2001;21(10):3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Kim JW, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24(13):5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nascimben L, et al. Mechanisms for increased glycolysis in the hypertrophied rat heart. Hypertension. 2004;44(5):662–667. doi: 10.1161/01.HYP.0000144292.69599.0c. [DOI] [PubMed] [Google Scholar]
- 46.Barger PM, Brandt JM, Leone TC, Weinheimer CJ, Kelly DP. Deactivation of peroxisome proliferator-activated receptor-alpha during cardiac hypertrophic growth. J Clin Invest. 2000;105(12):1723–1730. doi: 10.1172/JCI9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29(4):339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 48.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 49.Sano M, et al. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor gamma coactivator 1alpha. J Biol Chem. 2007;282(35):25970–25980. doi: 10.1074/jbc.M703634200. [DOI] [PubMed] [Google Scholar]
- 50.O’Hagan RC, et al. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000;14(17):2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von der Lehr N, Johansson S, Larsson LG. Implication of the ubiquitin/proteasome system in Myc-regulated transcription. Cell Cycle. 2003;2(5):403–407. [PubMed] [Google Scholar]
- 52.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 53.Liesa M, et al. Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS ONE. 2008;3(10):e3613. doi: 10.1371/journal.pone.0003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arany Z, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1(4):259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 55.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3(5):333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17(2):240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li F, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25(14):6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abel ED, et al. Cardiac hypertrophy with preserved contractile function after selective deletion of GLUT4 from the heart. J Clin Invest. 1999;104(12):1703–1714. doi: 10.1172/JCI7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao R, et al. Cardiac-specific overexpression of GLUT1 prevents the development of heart failure attributable to pressure overload in mice. Circulation. 2002;106(16):2125–2131. doi: 10.1161/01.CIR.0000034049.61181.F3. [DOI] [PubMed] [Google Scholar]
- 60.Dyck JR, Lopaschuk GD. Glucose metabolism, H+ production and Na+/H+-exchanger mRNA levels in ischemic hearts from diabetic rats. Mol Cell Biochem. 1998;180(1–2):85–93. doi: 10.1023/A:1006891007014. [DOI] [PubMed] [Google Scholar]
- 61.Liu Q, Docherty JC, Rendell JC, Clanachan AS, Lopaschuk GD. High levels of fatty acids delay the recovery of intracellular pH and cardiac efficiency in post-ischemic hearts by inhibiting glucose oxidation. J Am Coll Cardiol. 2002;39(4):718–725. doi: 10.1016/S0735-1097(01)01803-4. [DOI] [PubMed] [Google Scholar]
- 62.Portman MA, Standaert TA, Ning XH. Relation of myocardial oxygen consumption and function to high energy phosphate utilization during graded hypoxia and reoxygenation in sheep in vivo. J Clin Invest. 1995;95(5):2134–2142. doi: 10.1172/JCI117902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lorell BH, Wexler LF, Momomura S, Weinberg E, Apstein CS. The influence of pressure overload left ventricular hypertrophy on diastolic properties during hypoxia in isovolumically contracting rat hearts. Circ Res. 1986;58(5):653–663. doi: 10.1161/01.res.58.5.653. [DOI] [PubMed] [Google Scholar]
- 64.Wexler LF, Lorell BH, Momomura S, Weinberg EO, Ingwall JS, Apstein CS. Enhanced sensitivity to hypoxia-induced diastolic dysfunction in pressure-overload left ventricular hypertrophy in the rat: role of high-energy phosphate depletion. Circ Res. 1988;62(4):766–775. doi: 10.1161/01.res.62.4.766. [DOI] [PubMed] [Google Scholar]
- 65.Cunningham MJ, Apstein CS, Weinberg EO, Vogel WM, Lorell BH. Influence of glucose and insulin on the exaggerated diastolic and systolic dysfunction of hypertrophied rat hearts during hypoxia. Circ Res. 1990;66(2):406–415. doi: 10.1161/01.res.66.2.406. [DOI] [PubMed] [Google Scholar]
- 66.Eberli FR, Apstein CS, Ngoy S, Lorell BH. Exacerbation of left ventricular ischemic diastolic dysfunction by pressure-overload hypertrophy. Modification by specific inhibition of cardiac angiotensin converting enzyme. Circ Res. 1992;70(5):931–943. doi: 10.1161/01.res.70.5.931. [DOI] [PubMed] [Google Scholar]
- 67.Kagaya Y, Weinberg EO, Ito N, Mochizuki T, Barry WH, Lorell BH. Glycolytic inhibition: effects on diastolic relaxation and intracellular calcium handling in hypertrophied rat ventricular myocytes. . J Clin Invest. 1995;95(6):2766–2776. doi: 10.1172/JCI117980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luptak I, Yan J, Cui L, Jain M, Liao R, Tian R. Long-term effects of increased glucose entry on mouse hearts during normal aging and ischemic stress. Circulation. 2007;116(8):901–909. doi: 10.1161/CIRCULATIONAHA.107.691253. [DOI] [PubMed] [Google Scholar]
- 69.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95(6):568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 70.Fragasso G. Inhibition of free fatty acids metabolism as a therapeutic target in patients with heart failure. Int J Clin Pract. 2007;61(4):603–610. doi: 10.1111/j.1742-1241.2006.01280.x. [DOI] [PubMed] [Google Scholar]
- 71.Kodde IF, van der Stok J, Smolenski RT, de Jong JW. Metabolic and genetic regulation of cardiac energy substrate preference. Comp Biochem Physiol A Mol Integr Physiol. 2007;146(1):26–39. doi: 10.1016/j.cbpa.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 72.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14(1):45–55. doi: 10.1016/S1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 73.Sohal DS, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89(1):20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 74.Wang G, et al. Nitric oxide donors protect murine myocardium against infarction via modulation of mitochondrial permeability transition. Am J Physiol Heart Circ Physiol. 2005;288(3):H1290–H1295. doi: 10.1152/ajpheart.00796.2004. [DOI] [PubMed] [Google Scholar]
- 75.Liem DA, et al. Cyclin-dependent kinase 2 signaling regulates myocardial ischemia/reperfusion injury. . J Mol Cell Cardiol. 2008;45(5):610–616. doi: 10.1016/j.yjmcc.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol Cell Biol. 2000;20(23):8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitchell KO, El-Deiry WS. Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ. 1999;10(4):223–230. [PubMed] [Google Scholar]
- 78.MacLellan WR, et al. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol Cell Biol. 2005;25(6):2486–2497. doi: 10.1128/MCB.25.6.2486-2497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.