Abstract
The target of neutralizing antibodies that protect against influenza virus infection is the viral protein HA. Genetic and antigenic variation in HA has been used to classify influenza viruses into subtypes (H1–H16). The neutralizing antibody response to influenza virus is thought to be specific for a few antigenically related isolates within a given subtype. However, while heterosubtypic antibodies capable of neutralizing multiple influenza virus subtypes have been recently isolated from phage display libraries, it is not known whether such antibodies are produced in the course of an immune response to influenza virus infection or vaccine. Here we report that, following vaccination with seasonal influenza vaccine containing H1 and H3 influenza virus subtypes, some individuals produce antibodies that cross-react with H5 HA. By immortalizing IgG-expressing B cells from 4 individuals, we isolated 20 heterosubtypic mAbs that bound and neutralized viruses belonging to several HA subtypes (H1, H2, H5, H6, and H9), including the pandemic A/California/07/09 H1N1 isolate. The mAbs used different VH genes and carried a high frequency of somatic mutations. With the exception of a mAb that bound to the HA globular head, all heterosubtypic mAbs bound to acid-sensitive epitopes in the HA stem region. Four mAbs were evaluated in vivo and protected mice from challenge with influenza viruses representative of different subtypes. These findings reveal that seasonal influenza vaccination can induce polyclonal heterosubtypic neutralizing antibodies that cross-react with the swine-origin pandemic H1N1 influenza virus and with the highly pathogenic H5N1 virus.
Introduction
The HA is the major glycoprotein of influenza virus that mediates binding to cell surface sialic acid through the globular head domain (HA1) and the subsequent pH-dependent entry through endosomal fusion (1). Sixteen subtypes of HA that share between 40% and 60% amino acid sequence identity have been identified so far and have been clustered in 2 phylogenetic groups: group 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, and H16) and group 2 (H3, H4, H7, H10, H14, and H15) (2). The globular head is subject to continuing genetic evolution with amino acid substitutions in antibody-combining sites known as antigenic drift, while the structure of the stem region, which is primarily contributed by the HA2 domain, is more conserved (3, 4).
The globular head is the major target of neutralizing antibodies that inhibit virus binding to target cells and are classically detected by the hemagglutination inhibition assay (HAI). Distinct antigenic sites have been mapped mainly on the globular head using sequence information from naturally occurring and laboratory-selected antigenic variants (5–9). Less is known about the antigenic sites in the stem region. The first identified mAb (mAb C179) that binds to this region was isolated from an immunized mouse and showed a remarkable breadth of reactivity, being able to neutralize H1, H2, and H5 viruses by blocking membrane fusion (10–12). More recently, 2 groups described several heterosubtypic neutralizing mAbs isolated from phage display libraries that were either synthetic (13) or derived from immune donors (14). These mAbs utilize the VH1-69 germline sequence and bind to a conserved epitope in the HA stem region that is present in group 1 but not in group 2 influenza A subtypes. Crystallization studies revealed that the 2 most potent phage-derived mAb antibodies, CR6261 (15) and F10 (13), bind to a highly conserved helical region in the membrane proximal stem. Remarkably, the mAb contact residues are in the H chain CDR1 and CDR2, while the HCDR3 and the L chain do not contribute to antigen binding. The almost exclusive contribution of VH1-69 in its germline configuration to antibody binding is unprecedented and implies that a large fraction, up to 10%, of the human naive B cell repertoire (16) would be capable of responding to this conserved influenza epitope. This finding therefore raises the question of whether such antibodies are generated during the immune response to influenza virus infection or vaccination (17).
In this study, we investigated the human heterosubtypic antibody response following seasonal influenza vaccination. We report that some, but not all, individuals produced serum IgG antibodies that cross-reacted with the H5 hemagglutinin. By immortalizing memory B cells from these individuals, we isolated a panel of 20 heterosubtypic neutralizing mAbs that were characterized for their V gene usage, epitope specificity, and neutralizing activity in vitro and in vivo.
Results
Heterosubtypic neutralizing antibodies are produced in response to seasonal influenza vaccination.
Twenty-four healthy volunteers were immunized with trivalent inactivated influenza vaccine (6 in fall 2007, 9 in fall 2008, and 9 in the 2 consecutive seasons). The composition of the 2007 northern hemisphere vaccine included A/Solomon Islands/3/06 (H1N1), A/Wisconsin/67/05 (H3N2), and B/Malaysia/2506/04, while the 2008 vaccine contained A/Brisbane/59/07 (H1N1), A/Brisbane/10/07 (H3N2), and B/Florida/4/06. Plasma and mononuclear cells were collected before and 2 weeks after vaccination, a timing that corresponds to the peak of the expansion of vaccine-specific B cells (18).
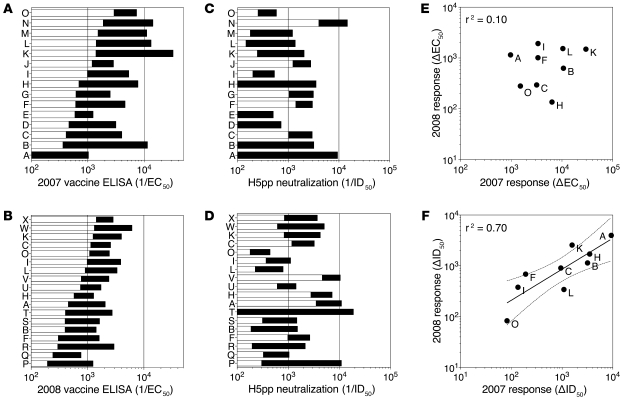
With a single exception, all donors had detectable ELISA titers of vaccine-specific IgG antibodies before vaccination, and the titers increased after vaccination, although to different extents in different individuals (Figure 1, A and B). The same plasma samples were also tested for their capacity to neutralize pseudoviruses expressing the HA from A/Viet Nam/1194/04 (H5N1) (Figure 1, C and D). Remarkably, H5-neutralizing activity was already detectable in some prevaccination sera and increased markedly following vaccination, reaching in some individuals ID50 titers as high as 1:20,000. All plasma samples failed to neutralize a VSV-G pseudovirus (ID50 <1:100; data not shown), indicating that the neutralizing response was H5 specific. The neutralizing activity was present in the IgG fraction and was lost following IgG depletion (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI41902DS1). Interestingly, in the 9 individuals who were vaccinated in the 2 consecutive seasons, there was a significant correlation (r2 = 0.70) in the magnitude of the H5-neutralizing antibody response measured in 2007 and 2008 (Figure 1F), while there was no correlation in the magnitude of the vaccine-specific response (Figure 1E). Although we cannot formally exclude priming of naive B cells, these findings strongly suggest that some individuals have memory B cells specific for epitopes shared by the seasonal vaccine and an unrelated H5 HA and that these B cells can be recalled by antigenically different vaccines.
Figure 1. Vaccine-binding and H5 pseudotype-neutralizing antibodies in plasma samples collected before and after seasonal influenza vaccination.
Volunteers (A to X) were immunized with seasonal influenza vaccine in 2 consecutive seasons. Plasma and PBMC samples were collected before and 2 weeks after vaccination. (A and B) Vaccine-specific IgG was measured in plasma by ELISA using the homologous vaccine as antigen. Reciprocal EC50 values before (white bars) and after vaccination (black bars) in 2007 (A) and 2008 seasons (B) are shown. (C and D) H5-specific neutralizing titers were measured in the same plasma samples by a pseudotyped neutralization assay against an H5 pseudovirus (A/VN/1194/04). Reciprocal ID50 values before (white bars) and after vaccination (black bars) in 2007 (C) and 2008 (D) are shown. (E and F) Correlation between the increase of vaccine binding titers (E) and H5-neutralizing titers (F) following vaccination in 2007 (x axis) and 2008 (y axis) in the 9 donors that received the seasonal influenza vaccine for the 2 consecutive years.
To estimate the frequency of vaccine-specific and H5–cross-reactive memory B cells, we polyclonally stimulated PBMCs collected before and after vaccination under limiting dilution conditions, as previously described (18), and tested the 10-day culture supernatants for the presence of vaccine binding and H5-neutralizing antibodies (Supplemental Table 1). In the 3 donors tested (B, F, and J), vaccine-specific B cells increased from 3,000–15,000 to 60,000–420,000 cells per million IgG+ B cells following vaccination. H5-neutralizing B cells could not be detected before vaccination but rose after vaccination to 320–16,680 cells per million IgG+ B cells. Taken together, these results indicate that a heterosubtypic response can be detected after influenza vaccination but it is generally small and is highly variable both in terms of serum antibodies and memory B cell frequencies.
Isolation and characterization of heterosubtypic neutralizing mAbs.
To characterize the heterosubtypic antibody response, we immortalized IgG+ memory B cells of the 4 donors that mounted the strongest H5-neutralizing response to the vaccine (donors A, B, H, and M) using EBV and CpG as described (19). The culture supernatants were screened using the H5-pseudotyped neutralization assay, and positive cultures were further tested for their ability to bind and neutralize infectious seasonal H1N1 virus. Cultures that scored positive for both H5 pseudovirus and H1N1 virus neutralization were subcloned, and 20 independent human mAbs (17 IgG1 and 3 IgG3) were isolated and their VH/VL genes sequenced.
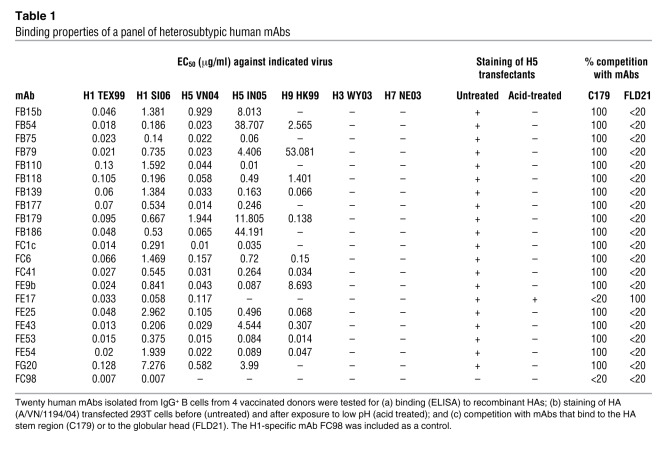
The mAbs were characterized for binding to recombinant HAs belonging to groups 1 and 2 (Table 1). All mAbs bound to H1 HAs (A/Texas/3/99 and A/Solomon Islands/3/06) as well as to H5 HA of A/Viet Nam/1203/04 with half-maximal binding concentration (EC50) values ranging from 0.01 to 7 μg/ml. In addition, some mAbs also bound to the clade 2.1 H5 HA of A/Indonesia/5/05 and the H9 HA of A/Hong Kong/1073/99, while none reacted with H3 and H7 HAs. We conclude that these antibodies react broadly with several unrelated HAs belonging to group 1, but not group 2, influenza A viruses.
Table 1 .
Binding properties of a panel of heterosubtypic human mAbs
In a preliminary attempt to determine whether the mAbs bind an epitope located on the HA1 or HA2 domains, we performed Western blot analysis with recombinant HA from the A/Viet Nam/1194/04 (H5N1) virus. None of the mAbs reacted in Western blot, suggesting that they may recognize a conformational epitope. We therefore evaluated mAb binding to HEK 293T cells transfected with the HA of A/Viet Nam/1194/04 before and after treatment at pH 5.0 that triggers an irreversible conformational change in the HA molecule. With the single exception of mAb FE17, all mAbs stained H5 HA–transfected cells, but failed to react with acid-treated cells (Table 1), indicating that they bind to a prefusion determinant that is lost upon acid treatment.
To further define whether the isolated mAbs recognize the globular head or the stem region of HA, we measured their capacity to inhibit HA binding of 2 mAbs of known epitope specificity: (a) C179, a mouse mAb that binds to an epitope in the HA stem region (10, 12); and (b) FLD21, a human mAb that binds to an epitope on the globular head of the H5 HA (20). ELISA plates were coated with recombinant H5 HA (A/Viet Nam/1203/04) and incubated with an excess of the isolated human mAbs followed by addition of C179 or FLD21 mAbs that were then detected using specific secondary reagents. With the single exception of mAb FE17, all the mAbs completely inhibited C179 but not FLD21 binding (Table 1). Reciprocally, FE17 inhibited FLD21 but not C179 binding. Among the heterosubtypic mAbs tested, FE17 was the only one with high HAI titers, but it reacted with 2 of the 4 H1N1 viruses tested. mAbs FE43 and FC41 did not inhibit hemagglutination, while FB110 showed modest HAI titers against all viruses that may be explained by the IgG3 isotype of this mAb (Supplemental Table 2). Taken together, these findings indicate that FE17 binds to an epitope on the globular head, while all the other heterosubtypic mAbs, such as FE43, bind to epitopes located in the HA stem region that overlap with the C179 epitope.
Titers and site specificity of heterosubtypic serum antibodies.
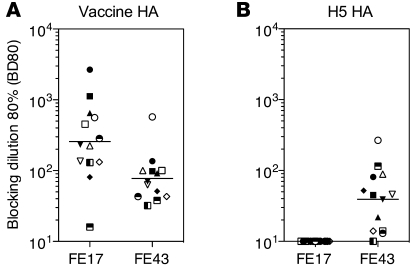
To gain insight into the relative proportion of the heterosubtypic component in the overall serum antibody response, we set up a competitive ELISA using 2 heterosubtypic mAbs that bind to the stem region (FE43) or to the globular head (FE17). ELISA plates coated with vaccine HA or with heterologous H5 HA were incubated with serial dilutions of postvaccination plasma followed by biotinylated FE43 and FE17 mAbs. The serum dilution that blocked mAb binding by 80% (defined as BD80) was determined. When tested on vaccine HA, all sera blocked both FE17 and FE43 binding, although to a different extent (Figure 2A). In contrast, when tested on H5 HA, none of the sera were able to block FE17 binding, while most blocked binding of FE43 (Figure 2B). These results indicate that heterosubtypic antibodies that bind to the stem region of the H5 HA can be readily detected in immune sera following seasonal vaccination, while heterosubtypic antibodies that bind to the globular head site defined by FE17 mAb cannot be detected. These results are consistent with the well-known conservation of the HA stem region as compared with the globular head.
Figure 2. Detection of heterosubtypic antibodies in plasma samples following seasonal vaccination.
ELISA plates were coated with vaccine or H5 HAs and incubated with serial plasma dilutions, followed by biotinylated FE17 or FE43 mAbs, which were then detected using enzyme-conjugated streptavidin. Shown is the reciprocal plasma dilution that blocks 80% binding (BD80) of heterosubtypic mAbs FE17 and FE43 to homologous HA (A) or heterologous H5 HA (B). Each symbol represents a different individual from Figure 1, A and C. Lines indicate geometric mean values.
Epitope mapping and V gene usage of heterosubtypic neutralizing mAbs.
To identify the target epitopes of the isolated heterosubtypic mAbs, we attempted to isolate virus escape mutants. Seven escape mutants isolated in the presence of FE17 all carried a single-point mutation in the globular head of the HA in the Ca2 antigenic site (8, 21) (S to N in position 145, according to H3 numbering). This finding is consistent with the competition data above and with the failure of this antibody to bind to the HA of A/Indonesia/5/05 that carries an S to P substitution in the same position. The epitope recognized by FC98, a control mAb that binds to some H1 HAs, was also mapped using escape mutants to the globular head of the HA in the Sb antigenic site (8, 21) (A to T in position 193, H3 numbering). In spite of repeated attempts, we failed to isolate escape mutants using the stem-specific mAbs FE43 and FB110. These findings suggest that in contrast to the neutralizing epitopes of the globular head, the conserved epitopes on the HA stem region recognized by heterosubtypic antibodies are less prone to mutate without loss of viral fitness.
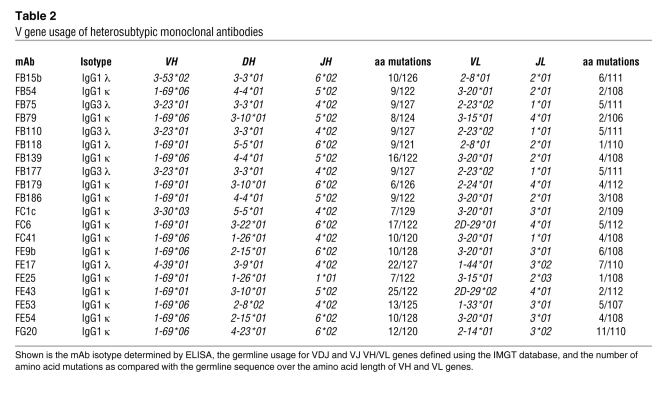
The VH/VL sequence information of the 20 mAbs is summarized in Table 2. Fourteen mAbs used VH1-69, which was rearranged with different D and J segments and paired with different VL genes, indicating an independent clonal origin. Three mAbs (all IgG3) that were independently isolated from donor B carried identical rearrangements on H and L chain genes (VH3-23/D3-3/JH4 paired with VL2-23/JL1) and therefore represent an in vivo expanded clone. The remaining 3 mAbs used yet different VH genes: VH3-30, VH3-53, and VH4-39. The number of somatic mutations was higher in the VH (6 to 25 amino acid substitutions) as compared with the VL (1 to 11 amino acid substitutions). These findings are consistent with a preferential although not exclusive usage of VH1-69 in the heterosubtypic response to influenza HA and demonstrate that this response is polyclonal within and among different individuals. In addition, the high level of somatic mutations observed in several clones (up to 25 amino acid substitutions in FE43 H chain) is consistent with a germinal center origin of these B cell clones.
Table 2 .
V gene usage of heterosubtypic monoclonal antibodies
Breadth of neutralization of influenza virus subtypes.
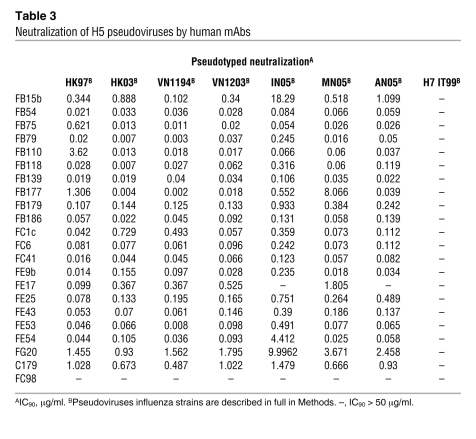
The neutralizing activity of the isolated mAbs was first characterized using a pseudotype neutralization assay (Table 3). Most mAbs showed a broad pattern of neutralization against different H5 pseudoviruses representative of the antigenically divergent clades 0, 1, 2.1, 2.2, and 2.3, while they failed to neutralize a pseudovirus expressing an avian influenza H7 HA. Some antibodies, such as FB54, FB110, FC6, FC41, and FE43, were more potent than others since they neutralized pseudoviruses with IC90 values lower than 0.4 μg/ml. In contrast, FE17 neutralized only some of the H5 pseudoviruses and failed to neutralize pseudoviruses expressing HAs derived from A/Indonesia/5/05 and A/Anhui/1/05, a finding that is consistent with the data obtained with escape mutants.
Table 3 .
Neutralization of H5 pseudoviruses by human mAbs
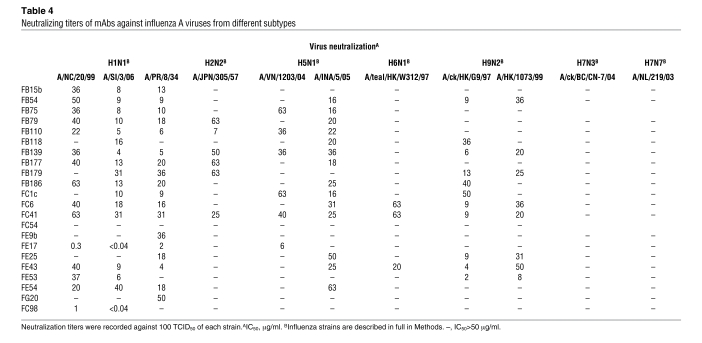
We next tested the mAbs for their capacity to neutralize human and avian infectious influenza A viruses belonging to group 1 and group 2 subtypes (22) (Table 4). Most antibodies showed considerable breadth of viral neutralization, being able to neutralize viruses of several subtypes belonging to group 1, namely H1N1, H2N2, H5N1, H6N1, and H9N2, although they failed to neutralize group 2 H7N3 and H7N7 viruses. In particular, the pattern of reactivity against group 1 viruses was distinct for each antibody. Notably, the 2 antibodies that bound to the globular head of HA (FE17 and FC98) neutralized viruses with low IC50 values (<0.04–6 μg/ml), comparable to those found using H5 pseudoviruses. In contrast, the HA stem–specific mAbs neutralized infectious viruses with IC50 values ranging from 4 to 63 μg/ml that were 100- to 1,000-fold higher than the IC90 values determined with pseudoviruses.
Table 4 .
Neutralizing titers of mAbs against influenza A viruses from different subtypes
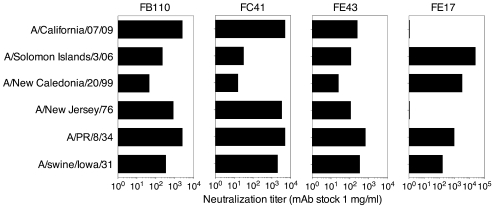
Three selected HA stem–binding mAbs (FE43, FC41, and FB110) and the globular head–specific mAb (FE17) were also tested against a panel of human and animal H1N1 viruses spanning more than 70 years of antigenic evolution and including the newly emerged pandemic H1N1 virus A/California/07/09 (H1N1) (Figure 3). While FE17 neutralized only 4 of the 6 viruses, FB110, FC41, and FE43 neutralized all 6 H1N1 viruses, including the swine-origin influenza A virus A/California/07/09 isolate. It is worth noting that while all the mAbs were elicited by a vaccine containing A/Solomon Islands/3/06, the stem-binding mAbs cross-neutralized other viruses more potently. Instead, FE17 that binds to the globular head showed a higher potency against the immunizing virus.
Figure 3. Broad neutralization of H1N1 viruses including A/California/07/09 by selected human mAbs.
Serial dilutions of the indicated mAbs (starting at 1 mg/ml) were tested for the capacity to neutralize a panel of H1N1 viruses spanning more than 70 years of antigenic evolution. Shown is the dilution providing 50% protection (ID50).
Prophylactic efficacy of the human mAbs in vivo.
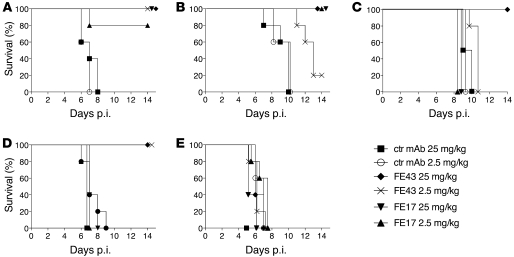
To determine whether the in vitro neutralizing activity displayed by the isolated human mAbs would be predictive of their prophylactic efficacy in vivo, BALB/c mice were passively inoculated i.p. with 25 or 2.5 mg/kg of FE17, FE43, or a control mAb and were challenged 24 hours later with 50 MLD50 (50% mouse lethal doses) of the following influenza viruses: A/Puerto Rico/8/34 (PR8, H1N1), A/teal/Hong Kong/W312/97 (H6N1), A/Viet Nam/1203/04 (H5N1), A/Indonesia/5/05 (H5N1), or A/Netherlands/219/03 (H7N7). MAb FE43 protected mice from lethal challenge with PR8 at either concentration tested, while FE17 conferred protection in a dose-dependent manner, affording 100% protection in animals that received 25 mg/kg of the antibody and 80% protection in animals that received 2.5 mg/kg (Figure 4A). Interestingly, despite the absence of detectable in vitro neutralizing activity (Table 4), FE43 protected all the mice from lethal infection with the clade 1 H5N1 A/Viet Nam/1203/04 virus at 25 mg/kg and conferred partial protection at 2.5 mg/kg (Figure 4B). Both doses of FE17 protected mice from lethal A/Viet Nam/1203/04 (H5N1) virus challenge as predicted by its neutralizing activity in vitro. After challenge with the clade 2 H5N1 A/Indonesia/5/05 virus, protection was observed only with the highest dose of FE43 (Figure 4C). Consistent with the in vitro neutralizing activity, FE43 fully protected mice against challenge with lethal dose of A/teal/Hong Kong/W312/97 (H6N1) virus at both doses tested, whereas mice injected with FE17 were not protected (Figure 4D). As expected, none of the mAbs conferred protection against lethal challenge with the group 2 H7N7 virus A/Netherlands/219/03 (Figure 4E).
Figure 4. Prophylactic efficacy of human mAbs in vivo.
5 BALB/c mice per group were passively injected i.p. with 25 or 2.5 mg/kg of mAbs FE17 or FE43 or a control (ctr) mAb and challenged 24 hours later with 50 MLD50 of the following influenza viruses: (A) A/PR/8/34(H1N1); (B) A/VN/1203/04 (H5N1); (C) A/INA/5/05(H5N1); (D) A/teal/Hong Kong/W312/97 (H6N1); or (E) A/Netherlands/219/03 (H7N7). mAb efficacy was measured as percentage survival after 14 days.
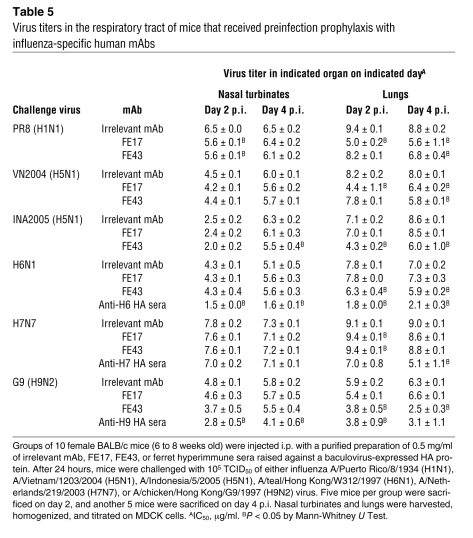
To gain further insights into the kinetics of viral neutralization in vivo and to measure the neutralizing activity against nonlethal viruses (such as H9N2 A/chicken/Hong Kong/G9/97 virus), we measured viral replication in the upper and lower respiratory tracts (Table 5). Mice passively immunized with FE17 or FE43 were challenged 24 hours later with 105 TCID50 of PR8 (H1N1), A/Viet Nam/1203/04 (H5N1), A/Indonesia/5/05 (H5N1), A/teal/Hong Kong/W312/97 (H6N1), A/chicken/Hong Kong/G9/97 (H9N2), or A/Netherlands/219/03 (H7N7). The mice were sacrificed on days 2 and 4 post infection (p.i.), and the level of virus replication was determined in nasal turbinates and in the lungs. Both FE17 and FE43 significantly reduced virus titers in the lungs of mice infected with PR8 by about 1,000- and 100-fold, respectively, on day 4 p.i. Both mAbs also significantly reduced the virus burden in the lungs of mice infected with A/Viet Nam/1203/04 on day 4 p.i., but their impact on the kinetics of virus replication differed. A significant reduction in pulmonary virus titers was noted as early as day 2 p.i. in mice that received FE17 while mice that received FE43 only showed a significant reduction in pulmonary virus titers on day 4. Consistent with its neutralizing activity profile in vitro, mice that were immunized with FE43 displayed significantly reduced virus replication in the lungs after infection with either the H5N1 A/Indonesia/5/05 virus or the H6N1 or H9N2 viruses, while no significant difference was observed in the titers of these challenge viruses in the lungs of mice that received either the irrelevant mAb or FE17. Finally, mice immunized with either FE17 or FE43 did not show any reduction in virus titers in the respiratory tract after infection with the H7N7 virus. Taken together, these results indicate that the mAbs significantly reduced the virus burden in the lungs to levels that were associated with survival.
Table 5 .
Virus titers in the respiratory tract of mice that received preinfection prophylaxis with influenza-specific human mAbs
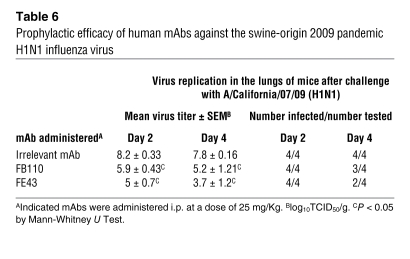
Finally, we measured the capacity of mAbs FE43 and FB110 to restrict viral replication of the swine origin H1N1 A/California/07/09 virus, which is not lethal in mice (Table 6). Mice were injected with mAbs at 25 mg/kg and challenged 24 hours later with 105 TCID50 of the A/California/07/09 (H1N1) virus. Viral titers in the lungs on day 4 were reduced on average 10,000-fold in mice that received FE43 and 400-fold in mice that received FB110. In addition, infectious virus was below the limit of detection in 2 out of 4 FE43-treated mice and in 1 out of 4 FB110-treated mice.
Table 6 .
Prophylactic efficacy of human mAbs against the swine-origin 2009 pandemic H1N1 influenza virus
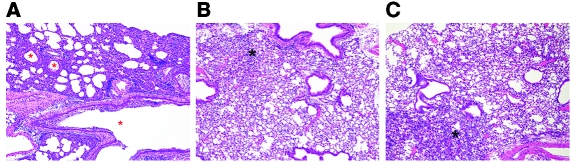
Histopathological analysis of the lungs of mice challenged with A/California/07/09 (H1N1) showed diffuse necrotizing bronchiolitis with various degrees of necrosis and inflammation of the surrounding interstitium (Figure 5A). This pathology was markedly decreased in animals that received either mAb FE43 or mAb FB110. On 4 day p.i., some of these animals showed focal bronchiolitis and inflammation of the interstitium, mononuclear perivascular cuffs, congestion, and minimal edema (Figure 5, B and C). Antigen, as demonstrated by immunohistochemistry (IHC), was not detected in animals receiving FB110 (data not shown), while in other groups antigen was detected in the bronchial and bronchiolar epithelium as well as in alveolar macrophages and alveolar epithelial cells.
Figure 5. Histologic sections (H&E) of lungs infected with 2009 H1N1.
Original magnification, ×10. (A) Prominent necrotizing bronchointerstitial pneumonia in the lungs of control animals; red asterisks denote necrotic airways. (B and C) Lungs of animals treated with either FB110 (B) or FE43 (C) show focal areas of interstitial inflammation, edema, and congestion (black asterisks).
These results demonstrate that heterosubtypic neutralizing mAbs derived from immune donors can confer protection against lethality and/or pulmonary virus replication in the mouse model following challenge with a variety of influenza virus subtypes, including the new swine origin influenza A H1N1 pandemic virus.
Discussion
The main finding of this study is that seasonal influenza vaccines can elicit heterosubtypic IgG antibodies that broadly neutralize group 1 influenza A virus subtypes both in vitro and in vivo. The extent of the heterosubtypic response varied considerably from donor to donor and in the same donor could be triggered by antigenically divergent vaccines. These findings illustrate a special case of original antigenic sin (OAS) (23). While in the typical case of OAS, memory B cell clones specific for the highly variable loops of the HA globular head are recalled only by closely related viruses, memory B cell clones specific for the stem region will be stimulated even by widely divergent group 1 viruses encountered over the lifetime of the individual.
Our results are consistent with the well-known heterogeneity in the antibody response to influenza virus both in terms of titer and epitope specificity (24). A recent report described IgG antibodies in H5N1 convalescent sera that bind to a variety of conserved peptides in the stem region (20). Another study reported on H5N1-neutralizing activity in sera of some individuals vaccinated with seasonal vaccine, although the authors did not investigate the specificity of the serum antibodies (25). To dissect the serologic response, we developed a quantitative mAb-blocking assay to determine the titers of antibodies specific for different regions of H1 or H5 HA. We found that antibodies that bind to epitopes overlapping FE43 in the stem region are present at relatively low concentrations as compared with antibodies binding to epitopes overlapping FE17 in the globular head (Ca2 antigenic site). However, while the former cross-react extensively with the H5 HA, the latter, although more abundant, do not cross-react with H5 HA and appeared to be genuinely subtype specific. These findings are also consistent with the notion that FE17 represents a rare heterosubtypic specificity that is not readily detected in serum.
Two heterosubtypic mAbs derived from phage libraries were recently characterized in great detail. CR6261 was isolated from a library prepared from IgM memory B cells (14), while F10 was isolated from a random human Ig library (13). The crystal structure of these antibodies bound to the target antigen revealed that binding is mediated only by the HCDR1 and HCDR2 encoded by the VH1-69 germline sequence, while HCDR3 and the light chain do not contribute significantly to antigen binding. These findings suggest that the naive repertoire of most individuals (who possess different copies of VH1-69 genes; ref. 16) contains a very high frequency, up to 8%, of B cells capable of responding to group 1 influenza A viruses, while the rare individuals lacking VH1-69 (16) may be unable to generate such a response. Our findings, which are based on the analysis of IgG antibodies and IgG memory B cell responses, confirm and extend these observations in several aspects. First, while most of the heterosubtypic mAbs isolated in this study use VH1-69, others use different VH, such as VH3-53, VH3-23, and VH3-30. Second, the VH genes of heterosubtypic mAbs carry different loads of somatic mutations, indicating that these cells have undergone germinal center reaction, although VH1-69 mAbs retain critical hydrophobic residues at positions 53 and 54, which have been shown to make a major contribution to binding (13, 15). Third, different B cell clones and different V gene usage could be found within the same individual, indicating that the response is polyclonal.
Considering the conservation of the target epitope, the prominent use of VH germline genes, and the polyclonal nature of the response, it is surprising that only some individuals generate a robust heterosubtypic neutralizing response. At present, we can only speculate that factors such as accessibility of the epitope to B cells, antigen presentation, and the individual history of influenza virus exposure may be limiting in this type of response. It will be interesting to investigate the feasibility of selectively priming a heterosubtypic response using vaccines where the stem region of the HA is engineered to achieve optimal presentation to B cells. It is also interesting that all stem-reactive heterosubtypic antibodies described so far, including those in the present study, react with group 1, but not group 2, viruses, a fact that may be attributed to the presence of an additional glycosylation site (N38 of HA1) in group 2 viruses. Repeated attempts to isolate heterosubtypic neutralizing antibodies specific for group 2 viruses by screening for binding to group 2 H7 HA were unsuccessful, suggesting that heterosubtypic responses are indeed directed primarily against group 1 viruses (D. Corti, unpublished data).
Together with previous reports, the present study emphasizes crucial differences between antibodies that bind to the HA stem region and those that bind to the HA globular head as to their neutralizing potency and selection of escape mutants. The globular head–specific antibodies are generally isolate/subtype specific, but may include also some heterosubtypic antibodies such as FE17 (this study), human mAbs isolated from H5N1 survivors (26), and the mouse mAb S139/1 (27). These mAbs show high neutralizing potency with IC50 below 1 μg/ml and show comparable titers in neutralization of infectious viruses and pseudoviruses. In contrast, the stem-reactive heterosubtypic mAbs potently neutralized H5 pseudoviruses at concentrations as low as 1 ng/ml, but show 100-1,000 lower neutralizing potency against infectious viruses. This difference is also apparent in the sera of boosted individuals where the heterosubtypic neutralizing antibodies can be easily detected using pseudoviruses (Figure 1) but not using H5N1 infectious virus (A.L. Suguitan Jr., unpublished data). The different neutralizing potencies of heterosubtypic antibodies measured with the 2 assays has already been observed in another study (13) and would be consistent with the notion that accessibility to the target epitope is limited by the tight packing of HA molecules in intact viruses as compared with pseudoviruses. It should be noted that in vitro viral neutralization does not necessarily predict the capacity of antibodies to protect mice from virus challenge. Indeed, it has been shown that antibodies that neutralize poorly in vitro can be very effective in vivo through recruitment of effector mechanisms such as complement and FcR+ cells (28–30). A striking example is the FE43 mAb described in the present study that did not neutralize A/Viet Nam/1203/04 H5N1 virus in vitro at 50 μg/ml but protected mice in vivo when administered at 25 mg/kg. It is possible that in vivo, the stem-reactive antibodies may efficiently bind to virus-infected cells expressing the HA molecule, thus signaling these cells for destruction by complement or FcR+ cells. Another major difference between classical and stem-reactive heterosubtypic mAbs relates to the mechanisms of viral escape. While escape mutants are readily selected using antibodies specific for the globular head (8), this is not the case for mAbs that bind to the stem region (13). Indeed, despite several attempts, we were unable to isolate influenza virus escape mutants using HA stem–specific mAbs such as FE43 and FB110. Further in vivo studies in relevant animal models and eventually in humans will be required to determine the potential of these antibodies in prevention and treatment of influenza virus infection.
Given the continuous antigenic drift of influenza viruses (31) and the unpredictability of new pandemics (32), it would be important to identify strategies of active and passive vaccination that target the most conserved regions of the HA. Our results show a potential pathway toward this goal, but also raise a question as to the relative role of the heterosubtypic versus isolate/subtype neutralizing antibody response, since even in high responder individuals, heterosubtypic antibodies hardly reach effective neutralizing concentration in the serum. It will be important to establish their protective concentration in vivo and to determine whether such concentration could be achieved by vaccination or by passive serotherapy.
Methods
Viruses.
The viruses used in this study were A/Puerto Rico/8/1934 (H1N1) (PR8), A/Solomon Islands/3/2006 (H1N1) (SI), A/swine/Iowa/1931 (H1N1), A/New Jersey/1976 (H1N1), A/California/07/2009 (H1N1) (CAL2009), A/Viet Nam/1203/2004 (H5N1) (VN2004), A/Indonesia/5/2005 (H5N1) (INA2005), A/teal/Hong Kong/W312/1997 (H6N1), A/Netherlands/219/2003 (H7N7), and A/chicken/Hong Kong/G9/1997 (H9N2). Replication-incompetent viruses pseudotyped with the HA genes were produced as previously described (33). Briefly, HEK293T/17 were cotransfected with HA-expressing plasmids and with a complementing viral-genome reporter gene vector, pNL4-3.Luc+.E-R+ (provided by John R. Mascola, Vaccine Research Center, NIAID, NIH, Bethesday, Maryland, USA) in the presence of 0.1 U/ml recombinant neuraminidase from Clostridium perfringens (Sigma-Aldrich). The pseudoviruses used were A/Hong Kong/156/97 (H5N1) (HK97), A/Hong Kong/213/03 (H5N1) (HK03), A/Viet Nam/1194/04 (H5N1) (VN1194), A/Viet Nam/1203/04 (H5N1) (VN1203), A/Indonesia/5/05 (H5N1) (IN05), A/Anhui/1/05 (H5N1) (AN05), A/whooper swan/Mongolia/244/05 (H5N1) (MN05), and A/chicken/Italy/13474/99 (H7N1) (IT99). All H5 HA encoding genes were provided by Barbara Capecchi (Novartis Vaccines and Diagnostic, Siena, Italy).
Samples collection.
Fresh PBMCs were obtained from healthy donors before and 2 weeks after vaccination with the seasonal influenza vaccine. All donors gave written informed consent for research use of blood samples, following approval by the Cantonal Ethical Committee of Cantone Ticino, Switzerland.
ELISA.
Half-area ELISA plates (Corning) were coated with appropriate concentrations of baculovirus-expressed recombinant HA (H5N1 A/Viet Nam/1203/04, H9N2 A/Hong Kong/1073/99, H7N7 A/Netherlands/219/03; Protein Sciences Corp.), purified inactivated virus (H1N1 A/Solomon Islands/3/06 and A/Texas/36/91; Prospec-Tany Technogene), or vaccine preparations (Inflexal, Berna Biotech, or Influvac; Solvay Pharma).
Quantitation of site-specific serum antibodies.
mAbs were purified on protein A or G columns (GE Healthcare) and biotinylated using the EZ-Link NHS-PEO solid-phase biotinylation kit (Pierce). The competition between polyclonal serum antibodies and biotinylated mAbs for binding to immobilized antigens was measured by ELISA. Briefly, plasma samples were added to HA-coated plates at different dilutions. After 1 hour, biotinylated mAbs were added at a concentration corresponding to 70%–80% of the maximal OD level, and the mixture was incubated at room temperature for 1 hour. Plates were then washed and bound biotinylated mAb was detected using AP-labeled streptavidin (Jackson Immunoresearch). The percentage of inhibition was tested in duplicates and calculated as follows: (1 – [(ODsample – ODneg ctr)/(ODpos ctr – ODneg ctr)]) × 100. BD80 value was calculated by interpolation of curves fitted with a 4-parameter nonlinear regression.
Virus neutralization.
Influenza microneutralization assay was performed using Madin-Darby canine kidney (MDCK) cells and 100 TCID50 (50% tissue culture infectious doses) of influenza viruses essentially as previously described (34). Briefly, purified mAbs were serially diluted 1:2 prior to mixing with 100 TCID50 of virus for 1 hour at room temperature prior to addition to monolayers of MDCK cells that were then incubated for a further 3–4 days and examined for cytopathic effect (CPE). Neutralizing titer was defined as the reciprocal of the lowest antibody concentration at which the infectivity of 100 TCID50 of the appropriate WT virus for MDCK cells was completely neutralized in 50% of the wells. Infectivity was identified by the presence of CPE on day 4, and the titer was calculated by the Reed-Muench method.
Pseudotyped neutralization assay.
A single-cycle infectivity assay was used to measure the neutralization of luciferase-encoding virions pseudotyped with the desired influenza HA proteins, as previously described (33, 35). Briefly, appropriate dilutions of the virion-containing culture supernatants were preincubated at 37°C for 1 hour with mAbs at various concentrations. The virus-mAb mixtures were added to HEK 293T/17 cells and incubated for 3 days at 37°C. The cells were then lysed with Britelite reagent (Perkin-Elmer), and the RLU in the cell lysates were determined on a luminometer microplate reader (Veritas; Turner Biosystems). The reduction of infectivity was determined by comparing the RLU in the presence and absence of mAbs and expressed as percentage of neutralization. The IC90 was defined as the sample concentration at which RLU were reduced 90% compared with virus control wells after subtraction of background RLU in cell control wells.
Isolation of human mAbs.
PBMCs were stained with directly labeled antibodies to CD22 (Pharmingen) and to immunoglobulin IgM, IgD, and IgA. CD22+IgM–IgD–IgA– B cells were isolated using FACSAria, pulsed with EBV (50% B958 supernatant) and seeded at 5 cells/well in replicate cultures in medium supplemented with CpG 2006 and irradiated allogeneic PBMCs, as previously described (19). Culture supernatants were screened for the ability to neutralize the H5 VN1194 pseudovirus, and positive cultures were further tested for their ability to neutralize infectious seasonal H1N1 virus. Cultures that scored positive for both H5 pseudovirus and H1N1 virus neutralization were subcloned by limiting dilution. The usage of VH and VL gene segments was determined by sequencing, and analysis for homology to known human V, D, and J genes was performed using the IMGT database.
ELISA competition assay.
Plates were coated with H5 HA, and competition was performed between unlabeled human mAbs and the murine C179 mAb (Takara Bio Inc.) or biotinylated FLD 21. Briefly, unlabeled competitor mAbs were added to H5-coated plates at 10 μg/ml for 1 hour, followed by the addition of the C179 or FLD 21 mAbs at limiting concentration that was chosen to give a net optical density in the linear part of the titration curve (corresponding to 70%–80% of the maximal OD level). After incubation for 1 hour, the plates were washed and the amount of C179 or FLD 21 mAb bound was detected using anti-mouse alkaline-phosphatase–conjugated antibodies (Southern Biotechnologies) or alkaline-phosphatase–conjugated streptavidin.
Sensitivity to acid treatment.
HEK293T/17 cells were transfected with an expression plasmid expressing the H5 HA of A/VN/1194/04. Two days after transfection, cells were washed and incubated for 2 minutes at 37°C with an acid buffer (10 mM HEPES, 10 mM 2-(N-morpholino) ethanesulfonic acid [MES] in PBS, pH 5.0). Untreated or acid-treated HA-transfected cells were then stained with 10 μg/ml of each mAb, and the binding to HA was detected using a secondary antibody by flow cytometry.
Isolation of escape mutants.
A fixed amount of A/Solomon Islands/3/06 H1N1 virus containing 3 × 108 TCID50 was incubated with a constant amount (0.5 mg/ml) of antibody FE17, FC98, FE43, or FB110 for 1 hour at 37°C and 5% CO2. The incubated mixture was then adsorbed by MDCK cells for 4 hours at 10,000 TCID50/well in 96 multi-well plates. Infected cells were then washed twice with PBS and replenished with TPCK-containing fresh medium containing 50 μg/ml of selecting mAbs. CPE of these infected cells was monitored 48–72 hours p.i. Supernatants from CPE-positive wells were harvested for a subsequent round of infection. The HA sequences of these escape variants were examined by routine sequencing techniques.
HAI.
Antibodies and receptor-destroying enzyme–treated control sera were tested for hemagglutination-inhibiting activity by standard methods using 4 HA units of influenza virus in V-bottom 96-well microtiter plates with 0.5% v/v washed turkey erythrocytes (for H1N1 viruses) or 1.0% v/v washed horse erythrocytes (for H5N1 VN 2004).
Evaluation of the efficacy of prophylaxis with the human mAbs in mice.
To evaluate prophylactic efficacy, groups of 6- to 8-week-old female BALB/c mice were injected i.p. with 500 or 50 μg (corresponding to 25 and 2.5 mg/kg) purified mAbs (FE17, FE43, or FB110 and a control mAb specific for HCV). When available, ferret hyperimmune antisera raised against recombinant baculovirus-expressed hemagglutinins (H6 HA of H6N1, H7 HA of H7N7, and H9 HA) were employed as positive controls (provided by Hong Jin, MedImmune). For CAL2009, p.i. antiserum (provided by the Centers for Disease Control and Prevention) was used as a positive control. Twenty-four hours after i.p. injection, the mice were bled to determine the serum-neutralizing antibody titer achieved by passive transfer and were challenged i.n. with 10 MLD50 of either PR8, H6N1, VN2004, INA2005, or the H7N7 virus. The mice were monitored daily for survival and weight loss until day 14 p.i. Animals that lost more than 25% of their initial body weight were euthanized in accordance with our animal study protocol.
To evaluate the influence of human mAbs on viral replication, mice were challenged with 105 TCID50 of PR8, H6N1, VN2004, INA2005, G9, CAL2009, or H7N7 virus 24 hours after i.p. injection of 1.0 ml of 0.5 mg/ml preparation of FE17, FE43, the irrelevant mAb, or ferret hyperimmune sera. Mice were sacrificed on days 2 and 4 p.i., and the nasal turbinates, lungs, and brains were collected aseptically. For mice challenged with CAL2009, only lungs were collected. These tissues were homogenized in Leibovitz L-15 medium (Invitrogen) supplemented with an antibiotic-antimycotic solution (Invitrogen) to achieve either a 5% or 10% w/v organ suspension. The organ homogenates were titrated on MDCK cells, and virus titers were determined as reported previously (36). All experiments involving highly pathogenic avian influenza viruses were conducted in a BSL3 containment facility that was approved for use by the US Department of Agriculture and Centers for Disease Control and Prevention. Animal experiments were approved by the NIH Animal Care and Use Committee.
Tissue staining.
Tissue sections (4–7 μm) from formalin-fixed paraffin-embedded lungs were stained with H&E for histopathological evaluation or processed for IHC. Endogenous peroxidase activity was quenched for 10 minutes in a peroxide solution followed by blocking for 10 minutes with 10% normal rabbit serum. Antigen detection for H1N1, H7N7, and H9N2 was achieved using a goat polyclonal antibody against influenza A virus strain USSR (H1N1) (Serotec) at a 1:300 dilution for 60 minutes at room temperature. H5N1 antigen was detected using a goat polyclonal antibody at a 1:1,000 dilution under similar conditions. Following incubation with the primary antibody, a sequential application of biotinylated antibody (rabbit anti-goat BA-5000; Vector Laboratories), avidin-biotinylated horseradish peroxidase macromolecular complex (R.T.U. Vectastain PK-7100; Vector Laboratories), and diaminobenzidine (Betazoid DAB, BDB2004L; Biocare Medical) were applied. Sections were counterstained with modified Harris hematoxylin, dehydrated, and mounted using permanent media. Antibody-negative controls for each slide included replacing the primary antibody with normal goat serum at protein concentrations comparable to those of the primary antibody.
Statistics.
Linear regression analysis shown in Figure 1, E and F, was performed using the least squares method. In the in vivo challenge experiments, log-transformed viral titers were compared using the Mann-Whitney U test. Data were analyzed using GraphPad Prism software.
Supplementary Material
Acknowledgments
We thank David Jarrossay for cell sorting; Isabella Giacchetto-Sasselli for antibody sequence analysis; Jadon Jackson, Leatrice Vogel, and the staff of the Comparative Medicine Branch, NIAID, for assistance with the mouse studies; Lawrence Faucette and Katherine Shea for assistance with pathology studies and IHC; and Barbara Cappechi (Novartis) for providing HA H5 genes. We thank the National Institute of Hygiene and Epidemiology (NIHE) in Hanoi, Vietnam, for the A/Viet Nam/1194/2004 HA gene. We thank Hong Jin of MedImmune for providing the ferret hyperimmune sera that were used as controls in the animal studies. This work was supported in part by the Swiss National Science Foundation (31003A-126027), the European Commission FP6 programme (IMECS, 201169), the Human Frontier Science Program (RGP9/2007), and the Intramural Research Program of National Institute of Allergy and Infection Diseases, NIH. A. Lanzavecchia is supported by the Helmut Horten Foundation. N.J. Temperton and R.A. Weiss are supported by the UK Medical Research Council.
Footnotes
Conflict of interest: A. Lanzavecchia is the scientific founder of Humabs LLC, a company that develops human antibodies for treatment of infectious diseases. D. Corti and F. Vanzetta are currently employees of Humabs. A. Lanzavecchia and F. Sallusto hold shares in Humabs, and R.A. Weiss is consultant.
Citation for this article: J Clin Invest. 2010;120(5):1663–1673. doi:10.1172/JCI41902.
References
- 1.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Russell RJ, Burns WH, White DO, Anders EM, Ward CW, Jackson DC. Antigenic determinants of influenza virus hemagglutinin. III. Competitive binding of antibodies directed against “common” and “strain-specific” antigenic determinants of A/Memphis/72 hemagglutinin. J Immunol. 1979;123(2):825–832. [PubMed] [Google Scholar]
- 3.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouchier RA, et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79(5):2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 6.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 7.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119(1):1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caton A, Brockman-Schneider CJ, Yewdell J, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 9.Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol. 1990;8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 10.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67(5):2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuno Y, Matsumoto K, Isegawa Y, Ueda S. Protection against the mouse-adapted A/FM/1/47 strain of influenza A virus in mice by a monoclonal antibody with cross-neutralizing activity among H1 and H2 strains. J Virol. 1994;68(1):517–520. doi: 10.1128/jvi.68.1.517-520.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnov Y, et al. An epitope shared by the hemagglutinins of H1, H2, H5, and H6 subtypes of influenza A virus. Acta Virol. 1999;43(4):237–244. [PubMed] [Google Scholar]
- 13.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16(3):265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasso EH, Johnson T, Kipps TJ. Expression of the immunoglobulin VH gene 51p1 is proportional to its germline gene copy number. J Clin Invest. 1996;97(9):2074–2080. doi: 10.1172/JCI118644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen GL, Subbarao K. Attacking the flu: neutralizing antibodies may lead to ‘universal’ vaccine. Nat Med. 2009;15(11):1251–1252. doi: 10.1038/nm1109-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinna D, Corti D, Jarrossay D, Sallusto F, Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur J Immunol. 2009;39(5):1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10(8):871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khurana S, et al. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 2009;6(4):e1000049. doi: 10.1371/journal.pmed.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brownlee GG, Fodor E. The predicted antigenicity of the haemagglutinin of the 1918 Spanish influenza pandemic suggests an avian origin. Philos Trans R Soc Lond B Biol Sci. 2001;356(1416):1871–1876. doi: 10.1098/rstb.2001.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell RJ, et al. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology. 2004;325(2):287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 23.Fazekas de St G, Webster RG. Disquisitions of original antigenic sin. I. Evidence in man. J Exp Med. 1966;124(3):331–345. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang ML, Skehel JJ, Wiley DC. Comparative analyses of the specificities of anti-influenza hemagglutinin antibodies in human sera. J Virol. 1986;57(1):124–128. doi: 10.1128/jvi.57.1.124-128.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gioia C, et al. Cross-subtype Immunity against Avian Influenza in Persons Recently Vaccinated for Influenza. Emerg Infect Dis. 2008;14(1):121–128. doi: 10.3201/eid1401.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashyap A, et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A. 2008;105(16):5986–5991. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida R, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5(3):e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng J, Mozdzanowska K, Gerhard W. Complement component C1q enhances the biological activity of influenza virus hemagglutinin-specific antibodies depending on their fine antigen specificity and heavy-chain isotype. J Virol. 2002;76(3):1369–1378. doi: 10.1128/JVI.76.3.1369-1378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mozdzanowska K, Feng J, Eid M, Zharikova D, Gerhard W. Enhancement of neutralizing activity of influenza virus-specific antibodies by serum components. Virology. 2006;352(2):418–426. doi: 10.1016/j.virol.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Hessell A, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449(7158):101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 31.Smith D, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 32.Capua I, Kajaste-Rudnitski A, Bertoli E, Vicenzi E. Pandemic vaccine preparedness--have we left something behind? PLoS Pathog. 2009;5(6):e1000482. doi: 10.1371/journal.ppat.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Temperton N, et al. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza and Other Respiratory Viruses. 2007;1(3):105–112. doi: 10.1111/j.1750-2659.2007.00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowe T, et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol. 1999;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alberini I, et al. Pseudoparticle neutralization is a reliable assay to measure immunity and cross-reactivity to H5N1 influenza viruses. Vaccine. 2009;27(43):5998–6003. doi: 10.1016/j.vaccine.2009.07.079. [DOI] [PubMed] [Google Scholar]
- 36.Suguitan A, et al. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 2006;3(9):e360. doi: 10.1371/journal.pmed.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.