Abstract
Fanconi anemia (FA) is a rare human genetic disease caused by mutations in any one of 13 known genes that encode proteins functioning in one common signaling pathway, the FA pathway, or in unknown genes. One characteristic of FA is an extremely high incidence of cancer, indicating the importance of the FA pathway in tumor suppression. However, the role of this pathway in the development and progression of human cancers in individuals who do not have FA has not been clearly determined. Here, we report that elevated expression of what we believe to be a novel splice variant of FA complementation group L (FANCL), which we identified and named FAVL, can impair the FA pathway in non-FA human tumor cells and act as a tumor promoting factor. FAVL expression was elevated in half of the human carcinoma cell lines and carcinoma tissue samples tested. Expression of FAVL resulted in decreased FANCL expression by sequestering FANCL to the cytoplasm and enhancing its degradation. Importantly, this impairment of the FA pathway by FAVL elevation provided human cancer cells with a growth advantage, caused chromosomal instability in vitro, and promoted tumor development in a xenograft mouse model. These data indicate that FAVL impairment of the FA pathway likely contributes to the development of non-FA human cancers and therefore add a challenging layer of complexity to the pathogenesis of human cancer. We further believe that these data will prove useful for developing additional tools for fighting human cancer.
Introduction
Fanconi anemia (FA) is a rare human genetic disease (1, 2), with severe congenital defects and an extremely high incidence of cancer (3). FA cells are hypersensitive to DNA crosslinking agents and display distinct chromosomal abnormalities. To date, 13 complementation groups of FA have been characterized. Each group can be accounted for by mutations in a specific FA gene. The same, or similar, phenotypes associated with each group, indicating that all FA proteins function in one common signaling pathway, termed the FA or FA-BRCA pathway, given that 3 breast cancer susceptibility gene products are FA proteins (4–10). The extremely high incidence of cancer in FA patients indicates the essential nature of this pathway in tumor suppression (7, 9, 11, 12); however, the role of this pathway in the development and progression of non-FA human cancer has not been adequately studied. Both DNA damage and DNA replication can activate this pathway, leading to monoubiquitination of FA complementation group D2 protein (FANCD2) (13) and its paralog FANCI (14, 15) through an E3 ubiquitin ligase complex comprising 8 known FA proteins (9), 2 FA-associated proteins (16, 17), and unknown proteins, with FANCL serving as a catalytic subunit (18, 19). The monoubiquitinated FANCD2 (13) and FANCI (14, 15) then function in concert with proteins such as FANCD1/BRCA2 (4), FANCN/PALB2 (10, 20), FANCJ/BRIP1/BACH1 (5, 6, 21), and others to repair damaged DNA. Moreover, FANCD2 and/or FANCI monoubiquitination appears to be a measure of the activation of the FA-BRCA pathway (11, 14). With discoveries that 2 FA proteins FANCD1 (4) and FANCN (10, 20) are BRCA2 and PALB2, respectively, and that 2 other FA proteins FANCJ (5, 6, 21) and FANCM (22) are a DNA helicase and translocase, respectively, FA has emerged as a unique genetic model to our knowledge to study mechanisms underlying genomic instability. Here, we report that elevation of what we believe to be a novel variant of FA complementation group L (FANCL), which we refer to herein as FAVL, expressed at high levels in 50% of carcinoma cell lines and carcinoma cases tested, can disrupt the FA signaling and cause formation and progression of xenograft tumors, representing an important and under investigated area that can help understand the pathogenesis of human cancer.
Results
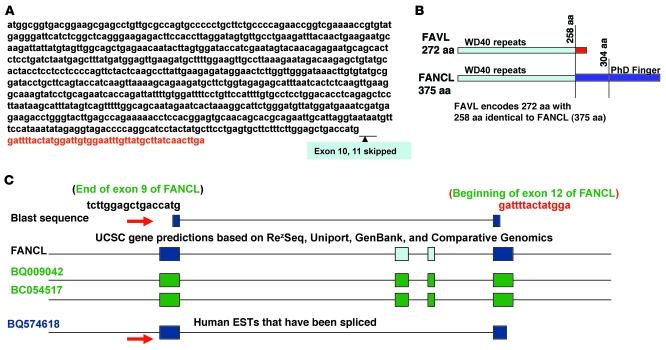
FAVL is identical to human expressed sequence tag BQ574618.
Chromosomal abnormalities are a hallmark of cancer (23). The crucial role of the FA-BRCA pathway in maintaining chromosomal stability prompted us to investigate how the FA-BRCA pathway is involved in non-FA human cancer. We found that an impaired FA-BRCA pathway associated with a lung cancer cell line might result from a greater expression of a splice variant of FANCL (24). We questioned whether this impaired FA-BRCA pathway, presumably triggered by this variant of FANCL, reflected a part of the essential role of an intact FA-BRCA pathway in suppressing non-FA human cancer. We found that the variant of FANCL named FAVL encoded 272 aa in total. Among the 272 aa, 258 were identical to those of FANCL, and 14 unique aa were translated from the RNA splice junction, which is the end of exon 9 fused with the beginning of exon 12 as a result of the exons 10 and 11 of FANCL being skipped (Figure 1, A and B). Further sequence analysis revealed that FAVL is identical to human expressed sequence tag (EST) BQ574618 (Figure 1C), which was identified from grade 2 chondrosarcoma, suggesting that FAVL may have a relevant role in development of non-FA human malignancy.
Figure 1. FAVL is a novel splice variant of FANCL to our knowledge and identical to human EST BQ574618.
(A) FAVL cDNA sequence. The cDNA sequence of FAVL is similar to the one of FANCL, with exons 10 and 11 of FANCL skipped. (B) Schematic protein structure comparison between FAVL and its cognate FANCL. FAVL has 272 aa, including 258 aa that are identical to FANCL and 14 unique aa that are translated from the junction region. (C) FAVL is identical to human EST BQ574816. Blast analysis of the fusion sequence, the end of exon 9 and the beginning of exon 12 of FANCL, shows that human EST BQ574618 matches the junction sequence (marked with red arrow), which is the unique part of FAVL (indicated with red arrow). Various colored boxes indicate exons.
FAVL is expressed at high levels in cancer cell lines and cancer tissue samples.
To investigate roles of FAVL in human tumorigenesis, we examined FAVL expression in lung cancer cell lines. Using FAVL antibodies that we generated (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI40908DS1), we found that FAVL protein was expressed at high levels in 8 out of 16 cancer cell lines tested compared with “normal” lung cells (Figure 2A and Supplemental Figure 2A). To validate FAVL protein expression, we examined FAVL mRNA expression in the cells harboring higher levels of FAVL protein by using a real-time quantitative RT-PCR (qRT-PCR). We found that levels of FAVL mRNA expression were consistent with levels of FAVL protein expression (Figure 2A). Thus, the high levels of expression of FAVL, only associated with carcinoma cells indicate, an important role of FAVL elevation in human tumorigenesis.
Figure 2. FAVL is expressed at a high level in half of the tested carcinoma tissue samples and cancer cell lines.
(A) FAVL expression is elevated in 50% of lung cancer cell lines tested. Using specific FAVL antibody (Supplemental Figure 1), levels of FAVL protein were found to be higher in 8 out of 16 cancer cell lines tested (arrowheads) compared with 2 normal cell lines (the other normal cell line shown in Supplemental Figure 2A). TaqMan assay was used to confirm FAVL mRNA elevation in lung cancer cell lines carrying elevated FAVL protein levels. The VIC-FAVL splice junction (26mer, synthesized from ABI) was used as TaqMan probe, and 6FAM-actin (purchased from ABI) served as an internal control. Levels of FAVL mRNA are higher in lung cancer cells compared with normal lung cells (Beas2B) (relative expression ratio average ± SEM, with n = 3). (B) FAVL elevation also was found in nearly half of carcinoma tissue samples tested. Using IHC, FAVL shows focal or/and diffuse cytoplasmic staining on 13 out of 25 slides from lung cancer tissue, 30 out of 45 slides from prostate cancer tissue, and 10 out of 10 slides from osteosarcomas. But FAVL staining was negative in all normal samples tested (5 samples from lung tissue and 5 samples from prostate tissue) and in all normal tissue fields integrated in those malignant tissues, further indicating the specificity of FAVL expression in carcinoma cells (Supplemental Figure 2B). Images shown represent FAVL protein expression in lung cancer, prostate cancer, and osteosarcoma. NT, normal tissue.
Next we asked whether FAVL elevation was also closely associated with carcinoma tissues. Using immunohistochemistry (IHC), we found diffuse to focal cytoplasmic expressions of FAVL in nearly half of 80 cancer tissue samples tested across 3 types of cancer (Figure 2B and Supplemental Figure 2B) but not in any of corresponding nonmalignant tissue samples tested. To confirm the FAVL cytoplasmic stain, we examined FAVL protein levels in cytoplasmic and nuclear fractions prepared from Calu-6 cells and found that the level of FAVL protein was higher in the cytoplasm than in the nucleus (Supplemental Figure 2C), supporting the pattern of cytoplasmic stain displayed by IHC. We used qRT-PCR as an additional method to confirm FAVL elevation in tissue samples detected by IHC, and FAVL mRNA expression was consistent with FAVL protein expression detected by IHC (Supplemental Figure 2B and data not shown). Therefore, the elevation of FAVL levels in only carcinoma cells and tissues does not appear to be rare. In contrast, this elevation may play an important role in non-FA neoplastic transformation, given that FAVL is expressed at a high level in a considerable percentage of carcinoma cases tested (Figure 2B and Supplemental Figure 2B).
Elevated FAVL can compromise the FA-BRCA pathway.
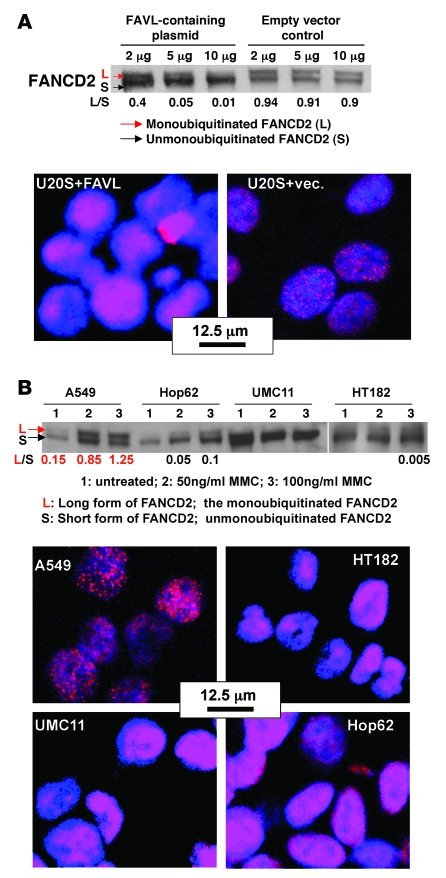
Many transcript variants are elevated during cancer development but not all of them have roles relevant to cancer development. FAVL, which we believe to be a novel splice variant of FANCL, was found to be expressed at a high level in nearly 50% of both cancer cell lines and cancer tissue samples detected, and it was thus necessary to determine whether FAVL elevation possessed any biological effects. Indeed, artificially increased FAVL expression levels seemed to impair the FA-BRCA pathway, as indicated by clearly reduced levels of monoubiquitinated FANCD2 and FANCD2 foci in cells transfected with a higher amount of FAVL cDNA–containing plasmid compared with control cells (Figure 3A and Supplemental Figure 3A). More importantly, including Calu-6 cells (24), there are additional lung cancer cell lines, UMC11, Hop62, and HT182, that harbor a noticeably compromised FA-BRCA pathway, measured by the compromised FANCD2 monoubiquitination and the failed focus formation of FANCD2 (Figure 3B and Supplemental Figure 3B). The other 4 cell lines expressing milder levels of FAVL demonstrated a compromised pathway to some extent (Supplemental Figure 3B). To determine whether endogenously elevated FAVL is a cause of an impaired FA-BRCA pathway, we investigated the possibility of restoring the integrity of the FA-BRCA pathway by downregulating FAVL expression. To this end, RNAi oligos specifically targeting FAVL (Supplemental Figure 1, C and D) were designed and used to treat Hop62 and HT182 cells. Subsequently, FANCD2 monoubiquitination and focus formation were analyzed in these cells after mitomycin C treatment. As shown in Supplemental Figure 3C, the level of FAVL in cells treated with RNAi oligos was lower than that in cells treated with nonspecific control RNAi oligos. Accordingly, the level of monoubiquitinated FANCD2 was relatively higher in cells treated with FAVL RNAi oligos compared with control cells, and FANCD2 focus formation was clearly detectable in cells treated with a higher dose of FAVL RNAi oligos (Supplemental Figure 3C). Therefore, similar to exogenous expression of FAVL, endogenously elevated FAVL in these cells is high enough to impair the FA-BRCA pathway. This statement may also apply to carcinoma tissues expressing FAVL at a comparable level to the one in cells harboring an impaired FA pathway (Supplemental Figure 3D), suggesting that the FA-BRCA pathway, which was disturbed by elevated FAVL levels, may have contributed to development of these carcinomas.
Figure 3. FAVL elevation compromises the FA-BRCA pathway.
(A) FANCD2 activation is compromised in U2OS cells expressing Flag-FAVL. U2OS cells were transfected with various amounts of FAVL plasmid and treated with 40 ng/ml mitomycin C (MMC) 24 hours after transfection. The level of activated FANCD2 in these transfected cells (marked with the red arrow) decreased more when a higher amount of FAVL plasmid was transfected. The focus formation of FANCD2 is shown, derived from the same batch of cells transfected with 10 μg of FAVL cDNA–containing plasmid. The relevant controls and similar data obtained from the other cells are shown in Supplemental Figure 3A. (B) The FA-BRCA pathway is defective in 3 additional lung cancer cell lines, Hop62, UMC11, and HT182, expressing high levels of FAVL. Levels of monoubiquitinated FANCD2 were examined in all 17 lung cell lines following treatment with 25 ng/ml mitomycin C. Besides Calu-6 cells, FANCD2 activation was clearly compromised in Hop62, UMC11, and HT182 cells (data not shown) and compromised to some extent in A549, H650, H526, and H82 cells (Supplemental Figure 3B). FANCD2 activation in Hop62, UMC11, Calu-6, and HT182 cells remained defective after treatment with higher doses of mitomycin C (50 ng/ml and 100 ng/ml). Analysis of lysates from A549 cells, carrying an intact FA-BRCA pathway, is provided for comparison. FANCD2 foci were examined in mitomycin C–treated cells, at the dose of 100 ng/ml, and ubiquitinated FANCD2 was also examined by Western blotting in these corresponding cells (Supplemental Figure 3B). The L/S ratio in A549 cells (marked in red) is much higher than that in other cells (marked in black).
FAVL regulates FANCL at the protein level.
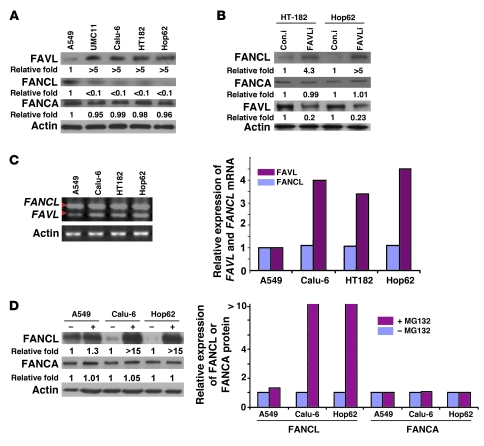
A compromised FA-BRCA pathway/FANCD2 monoubiquitination can result from any defective upstream regulators of FANCD2 monoubiquitination, including FANCL (25, 26). As FAVL was initially found in cells with lower levels of FANCL protein expression (24), we suspected that the effect of elevated FAVL levels might be mediated through lower levels of FANCL protein. To test this theory, we assessed levels of FANCL protein in those cancer cells carrying elevated levels of FAVL, with an impaired FA-BRCA pathway (Figure 3B). We found that, in contrast with other FA proteins tested, only levels of FANCL protein were low in UMC11, HT182, and Hop62 cells (Figure 4A), consistent with the finding in Calu-6 cells (24) and in cancer tissue samples (Supplemental Figure 4A). Importantly, ectopic expression of FANCL protein in these cells can partially restore FANCD2 activation (ref. 24 and Supplemental Figure 4B). This finding encouraged us to explore the possibility that silencing FAVL expression in these cells may restore levels of FANCL protein. Calu-6, UMC11, and HT182 cells transfected with FAVL-specific or -nonspecific RNAi oligos (indicated in Figure 4B, with FAVLi and Con.i, respectively) were collected 48 hours after transfection and analyzed by Western blotting for levels of FAVL and FANCL proteins. Indeed, FANCL protein was clearly detected in these cells treated with FAVL-specific RNAi oligos, especially in the nucleus (Figure 4B and Supplemental Figure 4, C and D). These results suggest that the function of elevated FAVL is at least partly mediated through lower levels of FANCL protein.
Figure 4. FAVL impairment of the FA-BRCA pathway is attributed to a low level of FANCL, resulting from FAVL regulation at posttranslational level.
(A) The level of FANCL protein, not that of FANCM (data not shown) or FANCA, is low in cells expressing high levels of FAVL compared with A459 cells. (B) Downregulating FAVL enhances levels of FANCL protein but not those of FANCM (data not shown) or FANCA. (C) Levels of FANCL mRNA remain similar among cells expressing low and high levels of FAVL. The ethidium bromide intensity of the high band (indicating the expression level of FANCL mRNA) in each reaction appears to be similar, but the intensity for the low band (indicating the expression level of FAVL mRNA) is stronger in HT182, Hop62, and Calu-6 cells than in A549 cells. (D) FANCL protein can be clearly detected in Calu-6, HT182, and Hop62 cells after MG132 treatment. Total cell lysates were prepared from Calu-6, HT182, and Hop62 cells, treated with or without 10 μM MG132 for 20 hours. Levels of FANCL, FANCM, and FANCA protein were analyzed. FANCL protein level was increased clearly in cells treated with MG132 compared with control cells but not too much for FANCM protein (data not shown) or FANCA protein. Relative expression levels of FA proteins tested were generated using the level of gray intensity of Western bands, measured using the NIH ImageJ program. (A, B, and D) Relative fold level increase is shown for each protein.
To define the manner in which FAVL affects the levels of FANCL protein, transcriptionally or posttranslationally, we examined levels of FAVL and FANCL mRNA in cells, with or without elevated FAVL. Using PCR primers bracketing the splice junction (Supplemental Figure 1C), we examined both FANCL and FAVL transcripts using the same RT-PCR reaction. We found levels of FAVL transcript were higher in cells carrying elevated FAVL protein levels than in cells harboring low levels of FAVL protein (Figure 4C). Levels of FANCL transcript, however, were similar among cells with different levels of FAVL expression (Figure 4C and Supplemental Figure 4E). Thus, FAVL did not appear to have a clear effect on levels of FANCL mRNA expression. Furthermore, FANCL protein was found to be increased in cells with elevated levels of FAVL after exposure to proteasome inhibitor when compared with mock-treated cells (Figure 4D). In contrast, the levels of other FA proteins detected (Figure 4D) or FAVL protein itself (data not shown) did not seem proportionally increased in the same batch of cells or in similarly treated A549 cells carrying a normal level of FANCL protein expression (Supplemental Figure 4F). These results suggest that low levels of FANCL protein are unlikely to result from a lower level of FANCL mRNA in cells expressing FAVL at a high level. Instead, FAVL appears to influence FANCL expression preferentially at the posttranslational level.
FAVL promotes FANCL degradation.
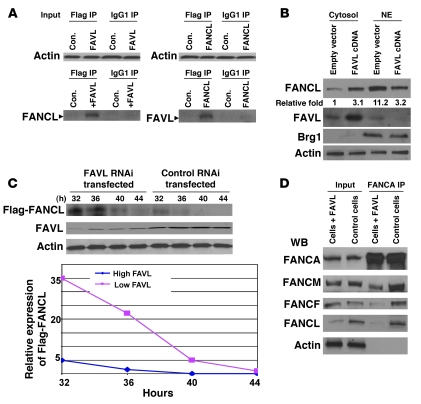
Because both FAVL and FANCL proteins contain WD40 repeats (Figure 1B), a protein domain conferring protein-protein interaction, we suspected that FAVL might interact with its cognate FANCL and that this interaction may ultimately lower levels of FANCL protein. To define the interaction between FANCL and FAVL, we performed IP-Western blot analysis of lysates of U2OS cells with overexpression of FANCL or FAVL protein. As shown in Figure 5A and Supplemental Figure 5A, FAVL and FANCL were detectable in the opposing pellets but in none of the few FA proteins tested, suggesting that both proteins may be strongly associated with each other. Next, we wished to examine the interaction between endogenous FAVL and FANCL proteins and performed IP-Western blot analysis of lysates of Calu-6 or A549 cells that carry relatively low levels of FANCL or FAVL protein, respectively. However, we could not detect a clear association between FAVL and FANCL (Supplemental Figure 5A and data not shown), and this might result from low levels of associated FANCL-FAVL. The clear detection of FANCL protein in Calu-6 cells treated with MG132 (Figure 4D) prompted us to perform IP-Western blot analysis of lysates of MG132-treated Calu-6 cells. As expected, FANCL, but not FANCM or FANCA, was detectable in the pellet of FAVL IP prepared from Calu-6 cells treated with MG132 (Supplemental Figure 5A and data not shown), supporting the interaction between FAVL and FANCL that was indicated by ectopically expressed proteins (Figure 5A).
Figure 5. FAVL sequesters FANCL protein in the cytoplasm and promotes its degradation.
(A) FAVL and FANCL can be coimmunoprecipitated. Lysates prepared from U2OS cells transfected with empty vector or Flag-tagged FANCL or FAVL were immunoprecipitated using an anti-Flag antibody (IgG1, control). FAVL and FANCL were coimmunoprecipitated with each other (similar results are shown in Supplemental Figure 5A). (B) Overexpression of FAVL enhances levels of FANCL in the cytoplasm. A549 cells were transfected with FAVL-containing plasmid or empty vector as the control. FANCL proteins were higher in the cytoplasm of cells overexpressing FAVL but lower in total cell lysates (Supplemental Figure 5B). Brg1 shows that nuclear proteins were mostly retained in the nuclear fractions we prepared. Relative fold level increase is shown for each protein. NE, nuclear extract. (C) Downregulating FAVL enhances the steady-state levels of FANCL protein. Calu-6 cells were first transfected with control RNAi oligos or FAVL RNAi oligos. Twelve hours later, both groups of cells were transfected with an equal amount of DNA mixture, containing 10 μg Flag-FANCL and 1 μg Renilla luciferase reporter (transfection efficiency control; Supplemental Figure 5C). Levels of FANCL protein were higher in cells with low levels of FAVL at time points tested compared with cells expressing a high level of FAVL. (D) Overexpression of FAVL leads to insufficient formation of the FA complex. About 1 mg nuclear extract, prepared from U2OS cells with or without ectopically expressed FAVL, was used for FANCA IP. Levels of FANCM, FANCF, and FANCL, but not FANCA, are lower in IP pellets prepared from FAVL-elevated U2OS cells.
Considering that FAVL protein was located predominantly in the cytoplasm (Figure 2B and Supplemental Figure 2C), we asked how the interaction between FAVL and FANCL influenced the levels of FANCL protein, which functions mainly in the nucleus in which it is located (Supplemental Figure 2C; refs. 18, 19). We reasoned that elevated FAVL, through its interaction with FANCL, may regulate the compartmentation of FANCL protein. We thus detected FAVL and FANCL proteins in both cytoplasmic and nuclear fractions prepared from A549 cells in which FAVL expression was low. We found that in cells transfected with FAVL cDNA, the FAVL signal was stronger in the cytoplasm and the FANCL signal was weak in the nucleus, and the opposite was the case for cells transfected with empty control vector (Figure 5B). However, nuclear FANCL protein could be restored when FANCL was ectopically expressed in Calu-6 cells in which FAVL was highly expressed (Supplemental Figure 5B). These results indicate that FAVL elevation can at least regulate the compartmentalization of FANCL protein, by sequestering FANCL in the cytoplasm, thus leading to a low level of FANCL protein in the nuclear compartment.
Above finding led us to reason that FAVL sequestration of FANCL in the cytoplasm might also contribute to a shorter half-life of FANCL protein. The steady-state levels of FANCL protein were examined in cells expressing high or low levels of FAVL protein. Flag-FANCL represented FANCL that was newly synthesized. We found that the levels of Flag-FANCL protein were higher and lasted longer in cells expressing lower levels of FAVL protein compared with cells harboring high levels of FAVL protein (Figure 5C and Supplemental Figure 5C). Clearly, FAVL elevation can decrease steady-state levels of FANCL protein; thus, FAVL sequestration of FANCL in the cytoplasm not only lowers its availability to FA core nuclear complex E3 but may also promote its turnover.
FAVL leads to a deficient formation of the FA complex.
Because FAVL elevation affects levels of FANCL protein, a pivotal component in FA core complex, we questioned whether the downregulated FANCL in turn affected the production of FA complex E3, essential for the activation of the FA-BRCA pathway. Therefore, established U2OS stable cell pairs (Supplemental Figure 5D), isogenic to an impaired FA pathway initiated by FAVL elevation, were used to isolate FA complexes. The production of FA complex in these cells was revealed by FANCA IP (18, 19) and subsequently by Western blotting for components of the FA complex. As shown in Figure 5D, lower levels of FA proteins detected in FA complexes isolated from U2OS cells with elevated levels of FAVL did not result from their IP inputs, in which levels of these FA proteins were the same as those in control U2OS cells carrying low levels of FAVL protein. In contrast, lower levels of FANCL protein detected in the input appeared to be the cause of the insufficient FA complex formation. A similar experiment was performed in A549, Calu-6, and Hop62 lung cancer cells, and results were consistent with those found in U2OS stable cell pairs (Supplemental Figure 5D). Therefore, high levels of FAVL in these lung cancer cells appeared to be responsible for a lower level of FANCL protein expression and thus a deficient FA complex (Figure 5D).
FAVL elevation promotes chromosomal instability.
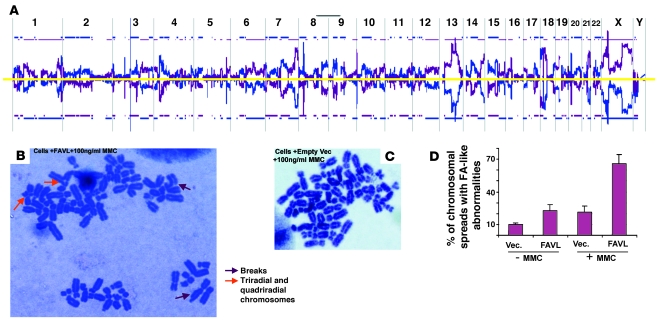
To address the question of whether the impaired pathway resulting from FAVL elevation would possess an effect on chromosome stability similar to that caused by any defective FA protein, we attempted to use normal lung cells for revealing effect of FAVL elevation on chromosomal instability. However, the cytotoxicity of the transfection reagent challenged us to obtain normal stable cell pairs isogenic to an impaired FA pathway, triggered by high-level FAVL expression. All osteosarcoma cases tested, however, appeared to harbor a higher expression of FAVL (Figure 2 and Supplemental Figure 2B), thus U2OS cells derived from osteosarcoma with mild malignancy appeared to be proper parental cells for establishing stable cell pairs, isogenic to FAVL overexpression and thus to FA complex stability (Figure 5D and Supplemental Figure 5D). Subsequently, U2OS stable cell pairs were used for comparative genomic hybridization (CGH), and we found that FAVL elevation contributed to an extreme degree of chromosomal abnormality (Figure 6A and Supplemental Figure 6A), which was consistent with the increased number of FA-like cytogenetic changes found in the same stable cells as well as in endogenous lung cancer cells with elevated levels of FAVL (Figure 6B and Supplemental Figure 6B). Therefore, like any defective FA gene, FAVL impairment of the FA-BRCA pathway can promote chromosomal instability.
Figure 6. Overexpression of FAVL leads to chromosomal instability.
(A) Genomic DNA was isolated from control U2OS cells and U2OS cells with elevated levels of FAVL, respectively. Both DNA samples were sent to MOgene LC for array CGH analysis through the dye swap approach. Chromosomal alterations occurred on nearly all chromosomes of cells with elevated levels of FAVL, standardized by control cells (blue line) and elevated levels of FAVL (purple line). Blue and purple lines appearing above or below the yellow line stand for DNA amplification and deletion, respectively. Representative raw data for chromosome 1 and also for chromosome Y (essentially negative, consistent with the origin of U2OS cells) are provided (Supplemental Figure 6A). (B–D) The chromosomal spreads were prepared by using U2OS stable cell pairs, and typical FA-like chromosomal abnormalities were highly associated with U2OS cells expressing FAVL at a high level. The population of U2OS cells overexpressing FAVL (B) has a higher percentage of FA-like chromosomal changes compared with control cells with empty vectors (C), with or without mitomycin C treatment. There are broken, triradial, and quadriradial chromosomes in FAVL-overexpressed cells (B), which are cytogenetic features of FA cells. These tumor cells carry a considerable amount of aneuploidy spreads, which remained similar between 2 types of cells. Original magnification, ×1,000. (D) Average percentage of FA-like chromosomal spreads ± SEM (n = 3).
FAVL triggers growth advantages.
To reveal a direct link of a malfunctioning FA-BRCA pathway to the development of human cancer, we investigated whether FAVL impairment of the FA-BRCA pathway can affect tumor development. We found that cells with high-level FAVL expression had a faster growth rate compared with control cells with empty vectors (Figure 7A), indicating FAVL overexpression can provide a growth advantage for host cells. The growth advantage initiated from overexpressed FAVL was further tested using colony formation assay in soft agar. FAVL elevation consistently promoted the colony formation in soft agar (Figure 7A). The inactivation of the FA-BRCA pathway by FAVL elevation ultimately promotes chromosomal abnormalities and tumor cell growth advantage in vitro. However, it remains unclear whether the growth advantage triggered by FAVL elevation can contribute to the development of human tumors in vivo.
Figure 7. FAVL elevation initiates growth advantages.
(A) Overexpression of FAVL promotes cell proliferation rate. A growth curve made from average cell number ± SEM, with n = 3, is shown. Cells overexpressing FAVL have a faster growth rate (pink line) than control cells (black line). Elevated FAVL leads to colony formation in soft agar. There was not a single colony (>50 cells) that could be scored from dishes plated with U2OS control cells carrying empty vector. Ten fields per one culture dish were viewed under a light microscope, at an original magnification of ×40. A pair of typical views was shown. (B) U2OS cells overexpressing FAVL form xenograft tumors in nude mice. The left side of the mouse was injected with U2OS cells, which had been pool selected and overexpressed FAVL, and the right side of the same mouse was injected with control U2OS cells, containing empty vector that had been subjected to the same selection process. One of four mice in the group is shown here, and the other mice and growth curves of xenograft tumors are shown in Supplemental Figure 7A. (C) U2OS xenograft tumors expressing high levels of FAVL are high-grade, poorly differentiated osteosarcomas. H&E images show a high number of mitotic figures (red arrows) and large areas of coagulative tumor cell necrosis (marked with yellow lines) and nuclear polymorphism. Morphologic differentiation is shown in Supplemental Figure 7C. Original magnification, ×400.
U2OS cells by themselves are incapable of forming xenograft tumors in nude mice (described by ATCC). Therefore, these cells appear to be suitable for a proper xenograft model system for studying the contribution of FAVL impairment of the FA-BRCA pathway to tumor development in vivo. As shown in Figure 7B and Supplemental Figure 7A, cells expressing FAVL at a higher level possessed a strong growth potential that permits host cells to develop tumors; in contrast, most control cells did not survive beyond the time when the second or the third image was taken, because the photon counts of these images were dramatically decreased compared with initial ones. Moreover, a lower level of FANCL protein expression also promoted the growth of xenograft tumors (Supplemental Figure 7B), further supporting the mechanistic theory that the function of FAVL elevation is partly mediated through lowering FANCL protein. Collectively, these results demonstrate that FAVL elevation can provide substantial growth advantages that can lead to the development of non-FA human tumors.
To reevaluate the status of the FA-BRCA pathway and levels of FAVL expression in U2OS xenograft tumors, we examined levels of FAVL protein and monoubiquitinated FANCD2 protein in primary xenograft tumor cells (27). We found these xenograft tumor cells continued to express high levels of FAVL protein and harbor an impaired FA-BRCA pathway compared with control cells. Thus, it was the elevated FAVL that caused U2OS cells to develop tumors in nude mice. Moreover, we examined histology of these U2OS xenograft tumors and found that these tumors were not moderately differentiated osteosarcomas, from which U2OS cells were derived. Instead, these xenograft tumors were poorly differentiated, high-grade osteosarcomas (Figure 7C and Supplemental Figure 7C). Therefore, an FAVL-triggered impaired FA-BRCA pathway not only promoted the formation of U2OS xenograft tumors but also caused tumor progression. These data demonstrate that FAVL impairment of the FA-BRCA pathway harbored in non-FA human tumors possesses a strong potential to promote transformation to malignancy in humans, representing an important and under investigated area of cancer research that can increase our understanding of human cancer development.
Discussion
Although initial studies implicating a link between FA and human cancers can be traced back to 1971 (1), progress has not been made in this aspect of FA research until recently. Some studies have demonstrated a relationship between germline FA gene mutations in cancer patients who do not have FA and the development of a subset of cancers associated with these mutations (10, 28, 29). However, cancer as a genetic disease is believed to be caused by a series of mutations occurring in both germline and somatic cells (23, 30). Understanding the role of not only inherited mutations but also somatic changes in the FA-BRCA pathway during tumor development is of great importance, although to date it has been largely understudied.
The extremely high incidence of cancer formation in FA patients prompted us to investigate how the FA-BRCA pathway is involved in the development of non-FA human tumors. Interestingly, we found that in a particular lung cancer cell line, Calu-6, the activation of the FA pathway upon mitomycin C treatment was compromised and, furthermore, that this impairment of the FA pathway was a result of reduced levels of FANCL (24). Then, we discovered that FAVL, a variant of FANCL, is at least in part responsible for this reduced FANCL expression. FAVL expression was elevated in nearly 50% of cancer cell lines and multiple types of cancer tissue samples that we tested compared with corresponding normal tissue samples (Figure 2 and Supplemental Figure 2). Moreover, we determined that high-level expression of FAVL compromised the FA-BRCA pathway activation (Figure 3 and Supplemental Figure 3), promoted chromosomal instability (Figure 6 and Supplemental Figure 6), and conferred substantial growth advantages both in vitro and in vivo (Figure 7 and Supplemental Figure 7). These results suggest that FAVL may act as a potent oncogenic factor in promoting tumor development by targeting the FA-BRCA tumor suppressor pathway. Indeed, as we carefully compared levels of FAVL expression in tumor samples with cells known to harbor a higher level of FAVL expression and thus an impaired FA-BRCA tumor suppressor pathway (Supplemental Figure 3D), nearly 45% of tumor samples tested carry a compromised FA-BRCA pathway, which might have contributed to the development of human caner.
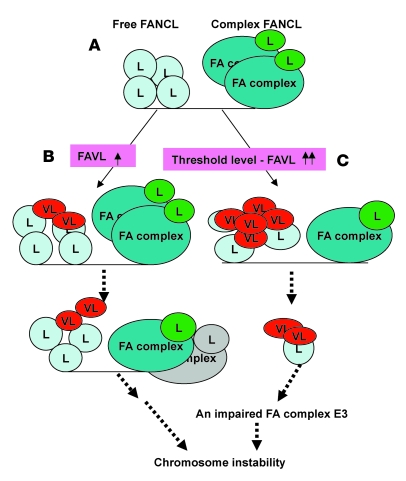
Cancer results from the accumulation of genetic and epigenetic alterations. Identification of alterations distinguishing cancer cells from normal cells will ultimately help us determine the nature of and understand the pathologic behavior of cancer cells. We have shown that the status of the FA-BRCA pathway can be impaired or restored when FAVL is overexpressed or silenced, respectively, in cells harboring lower or higher levels of FAVL expression (Figure 3 and Supplemental Figure 3). These results suggest that increased expression of FAVL beyond a certain level — the threshold level (as proposed in Figure 8) — might lead to negative effects on genomic stability, especially in human tumors. Indeed, about 25% of lung carcinomas tested and possibly more than 50% of osteosarcomas tested (Figure 2 and Supplemental Figure 2) can be attributed in part to an impaired FA-BRCA pathway initiated from FAVL elevation, because FAVL expression levels in these carcinomas are comparable with those in cells harboring an impaired FA-BRCA pathway. Many variants have important roles in human tumorigenesis, for example, MDM2 variants (31) and pyruvate kinase M2 variant (32). Detailed characterization of FAVL may prove crucial to our understanding of malignant transformation. As illustrated in Figure 8, FAVL transcript was found to be expressed at a high level only in tumor cells and tumor tissues, and its protein product could interact with and sequester its cognate FANCL protein, thus lowering levels of FANCL protein (Figure 5). Consequently, FAVL elevation could lead to genomic instability and tumor development (Figures 6 and 7 and Supplemental Figures 6 and 7). All of these points are important in the understanding of tumorigenesis, because the high level of FAVL expression was restricted to tumors (Figure 2, Supplemental Figure 2, and Supplemental Figure 4A). Future studies that continue to characterize FAVL, asking questions such as what the exact threshold level of FAVL is (Figure 8) and whether the missing carboxyl terminal of FANCL (Figure 1B) is responsible for its cytoplasmic location, may provide further insights into the contribution of FAVL elevation to the development of non-FA human cancer.
Figure 8. FAVL elevation confers genomic instability.
(A) In cells expressing a low level of FAVL (VL), there is perhaps a dynamic balance between 2 versions of FANCL protein (L): the free form and the complex form, activating the FA-BRCA tumor suppressor pathway and maintaining chromosomal stability. (B and C) In cells overexpressing FAVL, this balance may be disturbed, as a result of FAVL sequestration of FANCL protein and its enhancement of FANCL turnover. (B) The production of the FA complex E3 will be slightly (or not) affected when levels of FAVL are high but not beyond a threshold level. (C) However, the production of the FA complex E3 will be affected substantially when levels of FAVL exceed the threshold level. Both slightly and substantially compromised complex E3 will be able to restrain the activation of the FA-BRCA tumor suppressor pathway and, ultimately, contribute to chromosomal instability and cancer development within a relatively longer or shorter period of time, respectively.
The existence of FAVL-EST (Figure 1C) in malignant tissue also supports an important role of FAVL involved in the development of non-FA human cancer. Given the fact that FAVL was expressed at a high level in half of 80 tumor samples tested, across 3 types of cancer (Figure 2B and Supplemental Figure 2B), FAVL elevation in non-FA human cancer appears to be an important hidden factor, capable of contributing to the development of non-FA human cancer. The cause of the aberrant FANCL splicing and whether FAVL elevation in carcinomas was a result and/or a cause of tumor development need to be investigated, since both are as important as characterizing FAVL in understanding of tumorigenesis. Our ongoing studies (data not shown) suggest that elevated variant transcripts, including FAVL, may result from malfunctioned RNA biogenesis machinery that, at least in part, is caused by malfunctioning p53 tumor suppressor. The insufficient RNA biogenesis machinery will then lead to an aberrant transcriptome in which elevated variants of M2, MDM2, and others were already reported to have oncogenic activity (31, 32).
Splice variants found predominantly in tumors may have diagnostic value. Many alternative splice variants have turned out to be valuable tools for the diagnosis of human cancers (31). Because FAVL has been expressed at a high level in 3 types of cancer that we tested, it may be developed into a biomarker to assist in multiple types of cancer diagnosis in the future. On the other hand, these splice variants, with high expression only in tumors, may also be considered potential drug targets. FAVL might be of benefit to cancer treatment, especially with crosslinking chemotherapeutic agents. Tumor cells harboring a deficient FA-BRCA pathway, due to elevated FAVL, may act like FA cells that are hypersensitive to DNA crosslinking agents (8, 9). In fact, our studies (ref. 24 and Supplemental Figure 8) and those of others (25, 33) have revealed an association between the impaired FA-BRCA pathway and cancer cell sensitivity to DNA crosslinking agents in vitro. Future studies supporting this association in vivo will have profound effects on current types of chemotherapy relevant to DNA crosslinking agents. For example, by detecting the status of the FA-BRCA pathway to predict outcomes of types of chemotherapy, we might be able to choose a more effective therapy before tumors develop resistance and/or to improve drug sensitivity by targeting the FA-BRCA pathway. As the FA-BRCA pathway appears to be involved with multiple DNA repair mechanisms (9, 26), the status of this FA-BRCA pathway may hold promise of a better therapeutic marker for cancer therapies, using many types of DNA damage agents in addition to DNA crosslinking agents.
In conclusion, over the past few years, the field of FA research has become exciting, progressing from the study of a relatively obscure chromosomal breakage disorder to determining the role of FA proteins in DNA damage repair and tumorigenesis. The recent identification of new FA genes, along with detailed studies on FA proteins, has helped advance our understanding of complexities of the FA-BRCA tumor suppressor pathway substantially. However, to date, the role of this tumor suppressor pathway in development of non-FA human tumors has not been addressed. We believe our study is among the first to investigate how the FA-BRCA tumor suppressor pathway is involved in development of non-FA human tumors, thus demonstrating a new, challenging layer of complexity in the pathogenesis of human cancer. Moreover, our use of FA as a unique genetic model system to gain insights into the FA-BRCA pathway and the development and progression of non-FA tumors indicates the value of studying cancer susceptibility syndromes to advance multiple aspects of cancer research, including those directly and indirectly related to diseases at hand (8, 9, 34–38).
Methods
Cell lines, antibodies, chemicals, and RNAi oligos.
All cell lines, with an exception of WI-38 from Coriell, were purchased from ATCC. Anti-FANCD2 antibody was purchased from NOVUS (catalog no. N100-182). Anti-Flag (catalog no. F3165) and anti–β-actin (catalog no. 5316) antibodies as well as mitomycin C and cisplatin were from Sigma-Aldrich. d-luciferin was purchased from Xenogen. Coelenterazine was bought from Biotium Inc. FAVL siRNA, cugaccauggauuuuacuaug, and nonspecific control siRNA, LacZ-siRNA, gugaccagcgaauaccugu, were synthesized from Dharmacon.
Oligo or plasmid transfection, Western blot analysis, IHC, qRT-PCR, and imaging reporter assay.
All methods were performed as previously described (18, 39, 40). Primary antibodies were used at a dilution ratio of 1:1,000 for FANCD2, Brg1, and FANCL and 1:5,000 for β-actin. IHC was performed by using FAVL antibody, with 1:50 dilution ratio, for primary incubation, followed by use of the ImmPRESS Reagent Kit (catalog no. MP-7401; Vector). IHC of CKAE1/AE3, S100, and myogenin was conducted through standard services provided at the Mayo Clinic, Rochester, Minnesota, USA. VIC-FAVL, VIC-FANCL, and 6FAM-actin internal controls were purchased from Applied Biosystems. The imaging reporter assay was conducted by using the Xenogen IVIS 200.
Protein fraction preparation.
Both cytoplasmic and nuclear fractions were prepared essentially as described in protocols provided by the manufacturer (Pierce). The NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (catalog 78833; Pierce) was used.
IP.
Essentially as described previously (22), the whole cell lysates, prepared from cells transfected with Flag-FANCL or Flag-FAVL, and cells with empty vectors, used as the experimental controls accordingly, were mixed with 1 μg Flag antibody and 20 μl of flurry IgA beads (Invitrogen) for rotating overnight at 4°C. The eluate from IP-pellets was Western blotted for FAVL or FANCL, accordingly. For the FA complex IP, FANCA antibody was used to pull down the complex from nuclear extracts prepared from cells with or without overexpressed FAVL. Control antibodies, including the isotype mouse IgG1 and pooled rabbit IgG (Sigma-Aldrich), were used simultaneously when IPs were performed.
Cell proliferation.
An equal number of cells with or without overexpressed FAVL were plated at day 0, and total cell numbers were recounted at day 3 and day 7. The cell growth curve was generated by plotting cell numbers counted (mean ± SEM) (n = 3) from 3 independent experiments, averaged from triplicate samples for each individual experiment.
Soft agar colony formation.
Colony formation on soft agar was conducted essentially as described previously (41). Briefly, 10,000 cells of each kind were suspended in 1.5 ml of 0.3% low-melting gel with 1X DMEM/F-12, plated on top of 1.5 ml of 0.6% agar with 1X medium solidified in the 35-mm dishes, and incubated in humidity chambers for 2 weeks. The colonies (>50 cells) were scored by randomly counting 10 fields per 1 dish.
Xenograft formation.
Nude mice (4–6 weeks old), purchased from NCI Frederick, were injected with 300 μl Geltrex flurry (Invitrogen) (prepared at a 1:1 ratio with 1× PBS), containing 3 million cells constitutively expressing firefly luciferase. The luciferase activity was measured by intraperitoneally injecting d-luciferin into anesthetized mice and examining live images, using Xenogen IVIS as described previously (39).
Generation of primary cells from xenograft tumors.
Xenograft tissue (~0.5 cm3) was chopped into pieces (approximately <1 mm3) under sterile conditions, and the whole mixture then was placed in a 10-cm cell culture dish, containing 6 ml DMEM/F-12 medium with 20% FBS, 100 IU/ml penicillin, and 100 μg/ml of streptomycin. Cells were trypsinized at day 10 and maintained with regular DMEM culture medium.
Animals.
Five- to six-week-old male nude mice were obtained from NCI Frederick. The mice were euthanized at the end of experiments, and all animal experiments were performed using an Institutional Animal Care and Use Committee protocol approved by the Mayo Clinic, Rochester, which followed the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. Xenograft tumors were excised out and part of the tumor tissue was used for primary tumor cell culture. The rest was fixed in 4% paraformaldehyde overnight at 4°C and paraffin embedded.
Array-CGH.
Array-CGH was performed using MOgene LC through the dye swap approach with human 105K Chip.
Immunofluorescence.
Immunofluorescence was performed according to standard procedures. Briefly, cells were first fixed using 1% paraformaldehyde, permeabilized with 0.1% Triton X-100, and then blocked with 3% goat serum in 1X PBS and incubated with primary antibody (FANCD2 antibody, 1:1,000 dilution in 1X PBS containing 0.05% goat serum).
Biospecimens.
All experiments using human tissue samples in this study were performed using an IRB protocol approved by the Mayo Clinic, Rochester.
Statistics.
A 2 × 3 contingency table was used for statistical analysis. P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Alan D’Andrea (Harvard Medical School) for offering his expertise on FA cancer research, Weidong Wang (NIH) for providing FANCL cDNA and Brg1 and FANCL antibodies, Maureen Hoatlin (University of Oregon) for the FANCA antibody, and the Fanconi Anemia Research Foundation for FANCA, FANCM, and FANCF antibodies. We also think Liang Wang for his statistical support (Mayo Clinic, Rochester, Minnesota, USA). This study is partly supported by NIH grant (CA1R01CA136532-01A2 to P. Fei), an award to P. Fei from Minnesota Ovarian Cancer Alliance, a career development award to P. Fei from Mayo Prostate Cancer Spore (5P50CA091956-08 to D.J. Tindall), Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, and Mayo Clinic Cancer Center.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(5):1524–1534. doi:10.1172/JCI40908.
Deping Zhao’s present address is: Department of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China.
Hong Wang’s present address is: Department of Gastroenterology, First Municipal People’s Hospital of Guangzhou, the First Affiliated Hospital Sun Yat-Sen University, Guangzhou, China.
References
- 1.Swift M. Fanconi’s anaemia in the genetics of neoplasia. Nature. 1971;230(5293):370–373. doi: 10.1038/230370a0. [DOI] [PubMed] [Google Scholar]
- 2.Bagby GC, Alter BP. Fanconi anemia. Semin Hematol. 2006;43(3):147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Alter BP, Greene MH, Velazquez I, Rosenberg PS. Cancer in Fanconi anemia. Blood. 2003;101(5):2072. doi: 10.1182/blood-2002-11-3597. [DOI] [PubMed] [Google Scholar]
- 4.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297(5581):606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 5.Litman R, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8(3):255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Levitus M, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37(9):934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 7.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3(1):23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 8.Fei P, Yin J, Wang W. New advances in the DNA damage response network of Fanconi anemia and BRCA proteins. FAAP95 replaces BRCA2 as the true FANCB protein. Cell Cycle. 2005;4(1):80–86. doi: 10.4161/cc.4.1.1358. [DOI] [PubMed] [Google Scholar]
- 9.Wang W. Emergence of a DNA-damage response network consisting of Fanconi aneamia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 10.Rahman N, et al. PALB2, which encodes a BRCA2–interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niedzwiedz W, Patel KJ. “Dub”bing a tumor suppressor pathway. Cancer Cell. 2005;7(2):114–115. doi: 10.1016/j.ccr.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Bagby GC., Jr Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10(1):68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Taniguchi T, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109(4):459–472. doi: 10.1016/S0092-8674(02)00747-X. [DOI] [PubMed] [Google Scholar]
- 14.Grompe M, van de Vrugt H. The Fanconi family adds a fraternal twin. Dev Cell. 2007;12(5):661–662. doi: 10.1016/j.devcel.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129(2):289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciccia A, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25(3):331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Ling C, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26(8):2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meetei AR, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35(2):165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 19.Gurtan AM, Stuckert P, D’Andrea AD. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J Biol Chem. 2006;281(16):10896–10905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- 20.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA–N and predispose to childhood cancer. Nat Genet. 2007;39(2):142–143. doi: 10.1038/ng0207-142. [DOI] [PubMed] [Google Scholar]
- 21.Cantor SB, Andreassen PR. Assessing the link between BACH1 and BRCA1 in the FA pathway. Cell Cycle. 2006;5(2):164–167. doi: 10.4161/cc.5.2.2338. [DOI] [PubMed] [Google Scholar]
- 22.Meetei AR, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37(9):958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Wang X, Lin CJ, Couch FJ, Fei P. Altered Expression of FANCL confers mitomycin C sensitivity in Calu–6 lung cancer cells. Cancer Biol Ther. 2006;5(12):1632–1636. doi: 10.4161/cbt.5.12.3351. [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107(11):4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19(24):2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 27.Masuda N, et al. Establishment of tumor cell lines as an independent prognostic factor for survival time in patients with small-cell lung cancer. J Natl Cancer Inst. 1991;83(23):1743–1748. doi: 10.1093/jnci/83.23.1743. [DOI] [PubMed] [Google Scholar]
- 28.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 29.Pal T, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104(12):2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 30.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 31.Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell. 2002;2(1):9–15. doi: 10.1016/S1535-6108(02)00091-0. [DOI] [PubMed] [Google Scholar]
- 32.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi T, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9(5):568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 34.D’Andrea AD. The Fanconi road to cancer. Genes Dev. 2003;17(16):1933–1936. doi: 10.1101/gad.1128303. [DOI] [PubMed] [Google Scholar]
- 35.Franchitto A, Pichierri P. Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle. 2004;3(10):1331–1339. doi: 10.4161/cc.3.10.1185. [DOI] [PubMed] [Google Scholar]
- 36.Lee JW, Harrigan J, Opresko PL, Bohr VA. Pathways and functions of the Werner syndrome protein. Mech Ageing Dev. 2005;126(1):79–86. doi: 10.1016/j.mad.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Monnat RJ., Jr Werner syndrome: molecular genetics and mechanistic hypotheses. Exp Gerontol. 1992;27(4):447–453. doi: 10.1016/0531-5565(92)90080-j. [DOI] [PubMed] [Google Scholar]
- 38.Kudlow BA, Kennedy BK, Monnat RJ., Jr Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat Rev Mol Cell Biol. 2007;8(5):394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- 39.Fei P, et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6(6):597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 40.Fei P, Bernhard EJ, El-Deiry WS. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62(24):7316–7327. [PubMed] [Google Scholar]
- 41.Yoshioka N, Wang L, Kishimoto K, Tsuji T, Hu GF. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc Natl Acad Sci U S A. 2006;103(39):14519–14524. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.