Abstract
Increased levels of circulating saturated free fatty acids, such as palmitate, have been implicated in the etiology of type II diabetes and cancer. In addition to being a constituent of glycerolipids and a source of energy, palmitate also covalently attaches to numerous cellular proteins via a process named palmitoylation. Recognized for its roles in membrane tethering, cellular signaling, and protein trafficking, palmitoylation is also emerging as a potential regulator of metabolism. Indeed, we showed previously that the acylation of two mitochondrial proteins at their active site cysteine residues result in their inhibition. Herein, we sought to identify other palmitoylated proteins in mitochondria using a nonradioactive bio-orthogonal azido-palmitate analog that can be selectively derivatized with various tagged triarylphosphines. Our results show that, like palmitate, incorporation of azido-palmitate occurred on mitochondrial proteins via thioester bonds at sites that could be competed out by palmitoyl-CoA. Using this method, we identified 21 putative palmitoylated proteins in the rat liver mitochondrial matrix, a compartment not recognized for its content in palmitoylated proteins, and confirmed the palmitoylation of newly identified mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase. We postulate that covalent modification and perhaps inhibition of various mitochondrial enzymes by palmitoyl-CoA could lead to the metabolic impairments found in obesity-related diseases.
Keywords: azido-fatty acid, carbamoyl phosphate synthetase 1, Staudinger ligation, 3-hydroxy-3-methylglutaryl CoA synthase
For energy production via β-oxidation, or for lipid synthesis, long-chain fatty acids (LCFAs) must be activated to their coenzyme A (CoA) derivatives (LCFA-CoAs) by the fatty acyl-CoA synthetase enzyme. LCFA-CoAs are also the precursors for protein fatty acylation (myristoylation and palmitoylation), the covalent attachment of fatty acids to proteins. In myristoylation, the 14-carbon myristate is co-translationally (1, 2) or post-translationally (3-6) attached to the N-terminal glycine residue of a protein via a stable amide bond by N-myristoyltransferases (NMTs). Palmitoylation is characterized by the post-translational attachment of the 16-carbon fatty acid palmitate and other long-chain fatty acids to cysteine residues of a protein via labile thioester bond, although in some cases palmitate can also be attached to proteins through an amide linkage (7). Palmitate is transferred onto variably located cysteine residues of proteins, either enzymatically by a variety of enzymes known as protein fatty acyl transferases (PATs) (8-11) or spontaneously from palmitoyl-CoA (12-16).
Typically, fatty acids covalently attached to proteins serve as hydrophobic membrane anchors, although palmitoylation also plays a key role in cell signaling, protein localization, trafficking, and stability, as well as protein–protein interactions (7, 11, 17, 18). In addition to these well-established roles, palmitoylation is also emerging as a potential important regulator of metabolism. We and others have previously reported the occurrence of fatty acylation of mitochondrial proteins, with the majority of the fatty acylated proteins found in the matrix (15, 19). To date, only two mitochondrial proteins, methylmalonyl semialdehyde dehydrogenase (EC 1.2.1.27) (20, 21) and carbamoyl phosphate synthetase 1 (EC 6.3.4.16) (15), have had their palmitoylation confirmed, as well as the role of their palmitoylation characterized. In both cases, palmitoylation was shown to occur on an active site cysteine residue, which resulted in enzyme inhibition. Because of their implications in amino acid catabolism, we had postulated that palmitoylation of proteins in mitochondria could mediate a metabolic crosstalk between the pathways leading to the oxidation of fatty acids and amino acids (15, 20). The recent demonstration that the cytosolic glycolytic enzyme glyceraldehyde-3-P dehydrogenase (EC 1.2.1.12) (22) is also inhibited by fatty acylation confirms that physiological concentrations of LCFA-CoA can spontaneously acylate and inactivate certain enzymes involved in intermediary metabolism.
Increased levels of circulating saturated free LCFAs are known to result in an increase in intracellular LCFA-CoAs in cases of obesity and have been implicated in the etiology and development of type II diabetes (23-25). Because of the evidence that spontaneous palmitoylation can occur at physiological concentrations of palmitoyl-CoA and result in inhibition of the above-mentioned enzymes, we postulate that palmitoylation of mitochondrial metabolic enzymes may play a key role in metabolic regulation and energy homeostasis under normal physiological conditions and could very well contribute to maladaptative metabolic alterations found in obesity-related diseases.
Progress in the study of protein palmitoylation has long been impeded by the traditional long exposure time required to detect radioactive fatty acid label incorporation into proteins. In addition, methods that rely on incorporation of radioactive fatty acids into proteins are typically expensive, hazardous, and laborious and are often not sufficiently sensitive to allow the identification of new acylated proteins (26). To alleviate these encumbrances and further investigate the potential new role of protein fatty acylation as a metabolic regulator, we first sought to develop a nonradioactive, sensitive, and rapid method to detect and identify palmitoylated proteins to replace the traditional labeling methods using radioactive fatty acids. The alternative methodology reported here involves the incorporation of a synthetic azido-tetradecanoic acid, an isosteric analog of palmitate (henceforth referred to as azido-palmitate), from its coenzyme A derivative into proteins followed by chemoselective derivatization of the azido-fatty acylated proteins with a series of tagged (Myc, biotin, or fluorescein) triarylphosphines by the Staudinger ligation (27) (Fig. 1). The various tags used in this method are versatile and all offer same-day results, whereas traditional labeling methods require days or months of exposure time. This method is adapted from previous work on bio-orthogonal (synthetic analogues with functional handles, which are tolerated by enzymes) alkyl-azido glycoconjugates used to study glycosylated cell surface proteins (28). Importantly, alkyl azides are nontoxic and very stable at physiological temperature as well as under ambient or UV light (29), and they selectively react with modified triarylphosphines (Fig. 1). Our method takes advantage of these properties of azido-fatty acids as well as the fact that these analogues are readily activated by esterification to CoA (30), a prerequisite for the covalent attachment of fatty acids to proteins. Furthermore, a similar strategy was used to isolate and identify new farnesylated proteins using an azido-farnesyl analog (31).
Figure 1.

Schematic representation of the Staudinger ligation for the detection of palmitoylated proteins using a fatty acid analog. 1) Azido-palmitate is transferred to a protein from azido-palmitoyl-CoA, forming a thioester bond with a cysteine residue. 2) The azide moiety of the azido-palmitate reacts with the tagged triaryl-phosphine, forming an amide bond. Probe = Myc, biotin or fluorescein.
We utilized a strategy comparable to 2D electrophoresis to separate soluble mitochondrial proteins, first on the basis of their charge content then on the basis of their size. To do so we used a combination of native chromatography on a strong anion exchanger resin to separate the proteins by charge content and labeled each ion exchange fraction with [125I]iodopalmitoyl-CoA, followed by chemical ligation of the azido moiety to phosphine-biotin. The labeled fractions were then separated by size using SDS-PAGE. We could directly compare labeling of the proteins by the two methods and identify proteins from single bands using tandem mass spectrometry (MS). MS analysis of selected bands identified 21 (19 new) palmitoylated proteins, including the two previously characterized liver mitochondrial matrix proteins methylmalonyl semialdehyde dehydrogenase and carbamoyl phosphate synthetase 1 (15, 20), thereby validating our approach. The large number of fatty acylated mitochondrial proteins might represent a key, underappreciated phenomenon in mitochondrial physiology (See Supplemental Tables 1–3). We further sought to validate our findings by characterizing the acylation of the newly identified protein mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase (HMGCS) (EC 2.3.3.10). Recombinant HMGCS was used to further validate the use of azido-palmitate in detecting fatty acylated proteins compared with the established [125I]iodopalmitate analog (20, 32). We show that both fatty acid analogues are incorporated onto cysteine residues of proteins via neutral hydroxylamine and alkali sensitive thioester bonds, a major characteristic of naturally occurring protein palmitoylation.
MATERIALS AND METHODS
Materials
[125I]NaI (2.14 Ci/mmol) was from Amersham Biosciences (Arlington Heights, IL, USA). Mouse antimyc monoclonal antibody 9E10 was a gift from Dr. Steve Robbins (University of Calgary, Calgary, AB, Canada). Polyvinylidene difluoride (PVDF) membrane and centricon filters were from Millipore Corp. (Bedford, MA, USA). ECL Plus was from Amersham Biosciences. N-Ethylmaleimide (NEM) and hydroxylamine were from Sigma-Aldrich (St. Louis, MO, USA). Leupeptin and complete protease inhibitor cocktail were from Roche Molecular Biochemicals (Pleasanton, CA, USA). Bovine trypsin, dithiothreitol (DTT), iodoacetamide (IAA), α-cyano-4-hydroxy-cinnamic acid (4-HCCA), and trifluoroacetic acid (TFA) were all of highest available quality, purchased from Sigma-Aldrich (Oakville, ON, Canada). Solvents were all HPLC grade and purchased from Fisher Scientific (Ottawa, ON, Canada). All chemicals were used without further purification.
Plasmids
Mature cDNA encoding for human mHMG-CoA synthase was engineered with a C-terminal hexahistidine tag by polymerase chain reaction (PCR)-mediated mutagenesis using the appropriate primers and template cDNA (33) (a kind gift from Dr. John Capone, McMaster University, Hamilton, ON, Canada) and subsequently subcloned in the bacterial expression vector pET 19b (Novagen, Madison, WI, USA; see Supplemental Data for primer design).
Synthesis of azido-tetradecanoic acid (azido-palmitate) and tagged triaryl-phosphine probes
See Supplemental Data.
Preparation of radiolabeled and azido-palmitoyl-CoA
Radioiodination of iodopalmitate with [125I]NaI and the syntheses of azido-palmitoyl-CoA and [125I]palmitoyl-CoA derivatives were carried out using fatty acyl-CoA synthetase as reported previously (15, 32). Typical specific activity of [125I]iodopalmitate was 2 Ci/mmol. Stocks of azido-palmitoyl-CoA were made at 1 mM and kept frozen in aliquots at −80°C.
Metabolic labeling of primary culture of hepatocytes with [125I]iodopalmitate
Metabolic labeling of primary culture of hepatocytes was performed as described (34). Briefly, primary cultures of rat hepatocytes (a kind gift of Dr. Richard Lehner, University of Alberta, Edmonton, AB, Canada) were starved for 1 h by incubation with 10 ml of 10 μg/ml of fatty acid-free BSA in Dulbecco modified Eagle medium (DMEM; Life Technologies, Grand Island, NY, USA). The cells were incubated for 4 h in 3 ml DMEM containing 100 μCi of [125I]iodopalmitate conjugated to 30 μg BSA (6) and fractionated as described (20). Samples of the different fractions were analyzed by SDS-PAGE and autoradiography.
Mitochondrial isolation
Mitochondria were purified from the livers of Sprague-Dawley rats as described (35) with the following modifications: All buffers contained freshly added complete protease inhibitor cocktail; HEPES was used in place of N-Tris(hydroxymethyl)-methy-2-aminoethane sulfonic acid (TES); and the purified mitochondria were resuspended in a hypotonic lysis buffer (5 ml H2O plus complete protease inhibitor cocktail) for chromatography.
Labeling of proteins with [125I]palmitoyl-CoA and azido-palmitoyl-CoA
Incubations were performed in a final volume of 50–100 μl using 10 μg of lysed mitochondria or 1 μg purified proteins with 50 μM final concentration of [125I]palmitoyl-CoA or 100 μM azido-palmitoyl-CoA for 30 min at 25°C, as described in Corvi et al. (15). The radioactive reactions were stopped by the addition of 5× SDS-PAGE loading buffer (20), incubated for 2 min at 100°C and loaded onto a 10% SDS-polyacryl-amide gel. Radiolabel incorporation into proteins was visualized by autoradiography.
At the end of the incubation period, reactions containing the azido-palmitate analog were adjusted to 1% SDS and the various phosphine tags were added in a 2:1 mol:mol ratio to that of azido-palmitoyl-CoA. The reaction was allowed to proceed for another 2 h and was stopped and separated by electrophoresis as described above. For the palmitoyl-CoA competition experiments illustrated and explained in Fig. 6, purified HMGCS-His6 was preincubated for 45 min in various concentrations of palmitoyl-CoA prior to labeling as described above.
Figure 6.
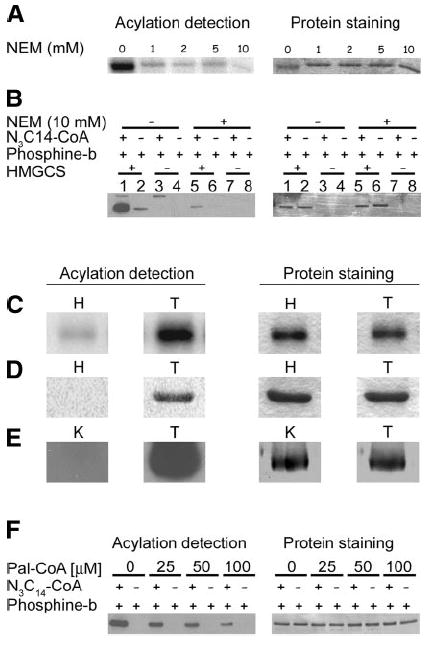
HMGCS-His6 is palmitoylated on a cysteine residue via a thioester bond. A) Purified HMGCS-His6 (1 μg) was incubated in buffer alone or the concentrations of NEM as shown prior to labeling with 50 μM (1 μCi) of [125I]iodopalmitoyl-CoA. Acylation of HMGCS-His6 was detected by autoradiography (left panel), and corresponding proteins were stained with Coomassie blue (right panel). B) Purified HMGCS-His6 (1 μg) was treated with 10 mM NEM or buffer alone prior to labeling with azido-palmitoyl-CoA (N3C14-CoA), incubation with phosphine-biotin, and SDS-PAGE. Azido-fatty acylated protein was detected by Western blot with Neutravidin-HRP/ECL. Corresponding protein was visualized by Coomassie blue staining of the membrane (right panel). Reactions were also carried out without HMGCS-His6 protein to detect the presence of fatty acyl-CoA synthetase (arrow) in the reaction solutions. C–E) To illustrate the versatility of our detection procedure, purified HMGCS-His6 samples (2 μg) were labeled with either [125I]iodopalmitoyl-CoA (C), azido-palmitoyl-CoA and phosphine-fluorescein (D), or azido-palmitoyl-CoA and phosphine-biotin (E), then subjected to SDS-PAGE (C, D) or subjected to SDS-PAGE and transferred to a PVDF membrane (E). The gels were soaked in 1 M hydroxylamine (H) or 1 M Tris (T) as a control prior to Coomassie blue staining, drying, and exposure to film (C) or scanning on a STORM 840 phosphorimager (D). The membranes in E were incubated in 0.1 M KOH, 90% methanol (K) or 0.1 M Tris 90% methanol (T) as a control, and protein azido-fatty acylation was detected by Western blotting with Neutravidin-HRP/ECL (left panel) prior to Coomassie blue staining (right panel). F) HMGCS-His6 (1 μg) was preincubated in the concentrations of palmitoyl-CoA as shown prior to labeling with azido-palmitoyl-CoA and phosphine-biotin. Acylation was detected by Western blot with neutravidin-HRP/ECL, and corresponding protein was visualized by Coomassie blue staining of the membrane.
Detection of azido-palmitoylated proteins
When using phosphine-Myc or phosphine-biotin following SDS-PAGE and transfer to a PVDF membrane, membranes were probed for the detection of the Myc-tagged-azido-fatty acylated protein with mouse monoclonal antimyc (1:2000) and an horseradish peroxidase (HRP)-conjugated sheep anti-mouse IgG (1:5000). Detection of the biotinylated-azido-fatty acylated proteins was performed using a neutravidin-conjugated HRP (1:20,000; Pierce Biotechnology, Rockford, IL, USA). The azido-fatty acylated proteins labeled with phosphine-fluorescein were detected by direct scanning of the SDS-PAGE gel on a STORM 840 from Molecular Dynamics (Sunnyvale, CA, USA) utilized in the fluorescence mode.
Neutral hydroxylamine and alkali treatments
Lysed mitochondria (10 μg), carbamoyl phosphate syn-thetase 1, or HMGCS (1 μg) labeled with azido-palmitoyl-CoA and phosphine-Myc were subjected to 16 h treatments with 1 M Tris-HCl, pH 7.0; 1M NH2OH, pH 7.0; or a 1 h treatment with 0.2 M NaOH (final concentrations) in solution. The reactions were stopped and processed for detection as described above. For in gel hydroxylamine treatments the gels were fixed with 45% methanol and 10% acetic acid solution for 30 min and then incubated in 1 M Tris-HCl, pH 7.0, 50% isopropanol for the control or 1 M hydroxylamine pH 7.0, 50% isopropanol for 24 h. The gels were then washed for 48 h in 50% isopropanol, dried, stained, and exposed to film or directly scanned on a STORM 840. The phosphine-fluorescein treated protein-containing gels were scanned before and after the treatments without staining or drying. PVDF membranes were treated to 0.1 N NaOH in 90% methanol or soaked in 0.1 M Tris-HCl, pH 7.0, 90% methanol for 1 h.
N-ethylmaleimide treatment
All NEM treatments were performed with freshly prepared NEM in 100 mM HEPES, pH 7.4, or 50 mM MOPS, pH 7.4, for 30 min at room temperature in a final volume of 50–100 μl.
Anion exchange chromatography
Percoll-purified mitochondria (30 mg) were incubated on ice for 30 min in 5 ml hypotonic lysis buffer (H2O+protease inhibitors) followed by sonication. Following centrifugation at 12,000 g for 30 min at 4°C, the supernatant was added to 45 ml of degassed and filtered 20 mM Tris-HCl, pH 8.5, and 1 mM DTT (Tris-DTT buffer) and applied to a Source Q (GE Healthcare, Montreal, QC, Canada) anion-exchange column. The column was washed with 2 column volumes of TD buffer and eluted at a 1 ml/min flow rate with a gradient of 0–300 mM NaCl in 30 ml followed by a 30 ml gradient from 300 mM to 1 M NaCl. Fractions (2 ml) were collected, adjusted to 20% glycerol, and stored at −80°C.
Acylation/labeling of mitochondrial and purified proteins
Aliquots (80 μl) from the ion-exchange chromatography fractions were incubated for 30 min at room temperature with either 50 μM [125I]iodopalmitoyl-CoA or 100 μM azido-palmitoyl-CoA as described above.
Carbamoyl phosphate synthetase 1 purification
Carbamoyl phosphate synthetase 1 was purified from Sprague-Dawley rat livers as described previously (15).
Expression and purification of hexahistidine tagged HMGCS
pET19b plasmids containing cDNA encoding for mature hexahistidine tagged HMGCS were transformed into E. coli BL21 (DE3). Midlogarithmic 500 ml liquid broth (LB) -Amp cultures were induced with 0.1 mM isopropyl-β-d-thiogalactosid at 37°C for 4 h. HMGCS-His6 was purified by nickel-chelating chromatography (Qiagen, Valencia, CA, USA) followed by desalting on PD10 columns (GE Healthcare), ion-exchange chromatography, and finally ultrafiltration in a Centricon Plus-20 (Millipore) with a molecular weight cutoff of 30 kDa. Purified protein was analyzed for enzymatic activity as per (36) prior to other assays.
MS analysis of azido-palmitoylated proteins
A 160 μl aliquot from each fraction of the ion exchange column was labeled with azido-palmitoyl-CoA, followed by incubation with phosphine-biotin as described above. The samples were split into two sets and loaded on a 10% SDS gel for either Western blot detection (80 μl) or Coomassie blue staining (80 μl). Azido-fatty acid incorporation was visualized by Western blot after the biotinylation reaction as described above, and bands that were azido-palmitoylated and sufficiently separated from each other were cut out from the corresponding Biosafe Coomassie (Bio-Rad) stained gel and analyzed by MS as described in the Supplemental Data.
RESULTS
Mitochondrial protein fatty acylation is a process that occurs in primary tissue cultured cells
To assess the cellular localization of palmitoylated proteins and confirm that mitochondrial protein palmitoylation is a naturally occurring cellular process, we labeled freshly isolated primary rat hepatocytes with [125I]iodopalmitate followed by cell fractionation. As seen in Fig. 2A, the majority of [125I]iodopalmitate label incorporation occurs in proteins from the 10,000 g pellet fraction (P10), which is highly enriched in mitochondria. Of note, the abundant and heavily radiolabeled band present at ~165 kDa, previously identified as rat liver mitochondrial carbamoyl phosphate synthetase 1 enzyme (15), was also found enriched in the P10 fraction as expected.
Figure 2.

Detection of palmitoylated mitochondrial proteins by different methods in the presence or absence of NEM yields similar results. A) Freshly isolated rat hepatocytes were incubated with 100 μCi of [125I]iodopalmitate for 4 h, then subjected to cell fractionation by differential centrifugation. Of each fraction collected, 10% (v/v) was analyzed by SDS-PAGE/Coomassie staining (right panel) and autoradiography (left panel). S1 and P1 = supernatant and pellet of a 1000 g centrifugation; S10 and P10 = supernatant and pellet of a 10,000 g centrifugation; CPS 1 = carbamoyl phosphate synthetase 1. B) Lysed mitochondria (10 μg) labeled with azido-palmitoyl-CoA (N3C14-CoA) or buffer alone were incubated with phosphine-Myc or buffer alone, followed by SDS-PAGE. Azido-fatty acids were detected by Western blot with anti-Myc (left panel), and proteins were visualized by India ink staining of the membrane (right panel). C) Lysed mitochondria (10 μg) were treated with 10 mM NEM or buffer alone prior to labeling with azido-palmitoyl-CoA, incubation with phosphine-Myc, and SDS-PAGE. Azido-fatty acylated proteins were detected as described in B. D) Lysed mitochondria (10 μg) were treated with 10 mM NEM or buffer alone prior to labeling with azido-palmitoyl-CoA but reacted with phosphine-biotin prior to SDS-PAGE and analysis by Western blotting using Neutravidin-HRP/ECL (left panel). Proteins on the corresponding PVDF membrane were visualized by Coomassie blue staining (right panel). Reactions were also carried out without mitochondrial protein to detect the acylation of fatty acyl-CoA synthetase (arrow) present in the reaction solutions. E) Purified carbamoyl phosphate synthetase 1 (1 μg) was incubated with 10 mM N-ethylmaleimide or buffer alone prior to labeling with azido-palmitoyl-CoA prior to incubation with phosphine-Myc and SDS-PAGE analysis. Azido-fatty acylated carbamoyl phosphate synthetase 1 was detected by Western blot with anti-Myc (left panel), and proteins were visualized by India ink staining of the PVDF membrane (right panel).
Bio-orthogonal azido-palmitate is readily incorporated into proteins in vitro
To further characterize the numerous palmitoylated proteins found in the mitochondrial fraction, we sought to develop an alternative approach to the traditional method of incorporating radioactive palmitate onto proteins. To establish our proof-of-principle, purified carbamoyl phosphate synthetase 1 (15) or rat liver mitochondrial lysates enriched in matrix proteins were labeled with or without azido-palmitoyl-CoA, followed by reaction in the presence or absence of phosphine-Myc. When azido-palmitate incorporation was visualized by Western blot using an anti-Myc antibody, we found that the Myc-label was only found attached to proteins when the samples were incubated in the presence of both azido-palmitoyl-CoA and phosphine-Myc (Fig. 2B). No detectable reaction products were visualized when only the azido-fatty acid or only the phosphine-Myc was used in the reactions.
Pretreatment of proteins with N-ethylmaleimide prevents the incorporation of azido-palmitate onto proteins from azido-palmitoyl-CoA
To test whether the azido-palmitoyl moieties are attached to proteins on cysteine residues, we pretreated the mitochondrial lysates with the cysteine alkylating agent NEM prior to incubation with azido-palmitoyl-CoA and either phosphine-Myc or phosphine-biotin for detection. We observed an almost total inhibition of azido-palmitate incorporation onto proteins pretreated with N-ethylmaleimide using either of the tagged phosphine reagents (Fig. 2C, D). Of interest, the NeutrAvidin-HRP reagent also detected a 75 kDa endogenous biotinylated protein, likely a decarboxylase (Fig. 2D, lanes 1, 2 and 5, 6). In addition, the labeling of purified carbamoyl phosphate synthetase 1, a mitochondrial protein known to be palmitoylated on a cysteine residue, was also completely prevented by pretreatment with NEM (Fig. 2E). These latter results are in accordance with our previous results obtained with [125I]iodopalmitoyl-CoA as a label (15). The arrow in Fig. 2D left of the gel shows the location of acyl-CoA synthetase, which is used to produce the azido-palmitoyl-CoA from azido-palmitate and CoA and is present in the reaction solutions containing the azido-palmitoyl-CoA.
Alkali or neutral hydroxylamine treatment removes the azido-palmitate label from proteins
To investigate the chemical nature of the bond linking the azido-palmitate to the various mitochondrial proteins or carbamoyl phosphate synthetase 1, we subjected protein samples preincubated with azido-palmitoyl-CoA followed by ligation with phosphine-Myc to either 1 M hydroxylamine, pH 7.0, for 16 h or 0.2 M NaOH for 60 min—treatments known to cleave thioester bonds (37-39). As shown in Fig. 3, alkali and neutral hydroxylamine treatments could remove virtually the entire label as compared to the 1 M Tris control treatment, suggesting that the azido-palmitate is attached to proteins via a thioester bond. Furthermore, this result also suggests that the azido-moiety does not affect the properties of the bond linking the azido-fatty acid to the proteins.
Figure 3.
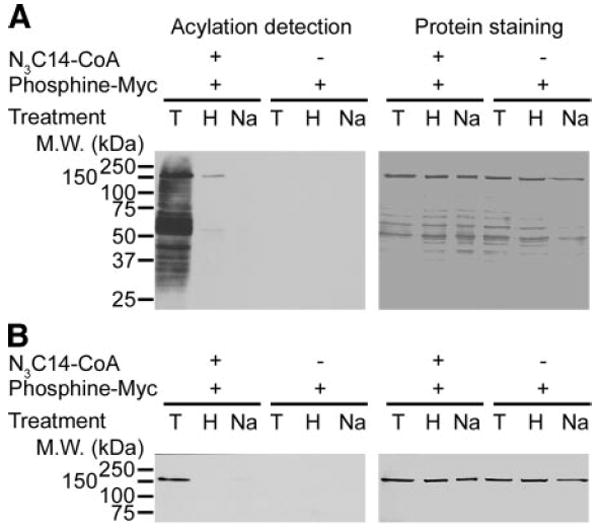
Azido-palmitate is bound to proteins via a hydroxylamine-sensitive thioester bond. Lysed mitochondria (10 μg) or purified carbamoyl phosphate synthetase 1 (1 μg) were labeled with azido-palmitoyl-CoA (N3C14-CoA), incubated with phosphine-Myc, and then treated either with 1 M Tris (T), 1 M NH2OH (H), or 0.2 N NaOH (Na), followed by SDS-PAGE. A) Azido-fatty acylated proteins from lysed mitochondria were detected by Western blotting with anti-Myc (left panel), and the corresponding membrane was stained with India ink (right panel). B) Aido-fatty acylated purified carbamoyl phosphate synthetase 1 was detected bt Western blot with anti-Myc (left panel), and the corresponding membrane was stained with by India ink (right panel).
Labeling and detection of palmitoylated proteins utilizing azido-palmitoyl-CoA and phosphine-biotin
To more accurately compare labeling methods and identify other mitochondrial fatty acylated proteins, we separated the soluble proteins from rat liver mitochondria by ion-exchange chromatography prior to labeling with [125I]iodopalmitoyl-CoA or azido-palmitoyl-CoA/phosphine-biotin (Fig. 4). We found that the electrophoretic protein banding pattern is comparable between the two labeling methods and that the detection of acylated proteins is higher (more sensitive) when using the azido-palmitoyl-CoA/phosphine-biotin labeling method followed by Western blotting with Neutravidin-HRP. The additional bands present in Fig. 4B but not in Fig. 4A at apparent molecular weights of 150 kDa and 75 kDa (marked by white asterisks) can be attributed to endogenously biotinylated proteins being detected by the NeutrAvidin-HRP. This finding is confirmed in Fig. 4C, D, in which the proteins were only labeled with phosphine-biotin (Fig. 4C) or left unlabeled (Fig. 4D) prior to separation and detection. Similarly, a ~60 kDa protein band present in all lanes in Fig. 4B, F, G (marked with an arrow at left edge of gel) is believed to be fatty acyl-CoA synthetase, which is used to produce the azido-palmitoyl-CoA from azido-palmitate and CoA and is present in the reaction solutions containing the azido-palmitoyl-CoA. The low level of reactivity in the samples incubated with phosphine-biotin alone (Fig. 4C), which is comparable to the nonlabeled proteins (Fig. 4D), confirms that the phosphine-biotin reagent reacts specifically with azido-palmitoylated proteins. In addition to its high degree of specificity, the new labeling protocol is significantly faster than the standard method of using radioisotope-labeled fatty acids, exposure times are often only a few seconds compared with typical overnight exposure with a phosphorimager screen when using [125I]iodopalmitoyl-CoA as label, or 15 to 30 days fluorographic exposure when using [3H]palmitoyl-CoA (data not shown) for these types of in vitro labeling reactions.
Figure 4.

Separation and labeling of mitochondrial protein for the identification of palmitoylated proteins. Soluble mitochondrial proteins from rat liver were separated by ion-exchange chromatography, and aliquots from each fraction were labeled with [125I]iodopalmitoyl-CoA (A), azido-palmitoyl-CoA (N3C14-CoA) and phosphine-biotin (B), or phosphine-biotin alone (C), or left unlabeled (D), followed by SDS-PAGE (A) or SDS-PAGE and Western blot analysis (B–D) as described in Materials and Methods. A) Autoradiogram. B–D) Western blot with Neutravidin-HRP/ECL. E–H) Corresponding Coomassie blue-stained gel and membranes (right panels). Asterisks show location of endogenously biotinylated proteins (B–D). Arrow shows the location of fatty acyl-CoA synthetase (B, F, G).
Incorporation of bio-orthogonal azido-palmitate analog onto proteins facilitates the identification of palmitoylated mitochondrial proteins from rat liver
Using azido-palmitoylated bands shown in Fig. 4 as a guide, we cored 31 gel slices corresponding to the labeled bands from a duplicate gel (Supplemental Fig. 1) and subjected these proteins to mass spectrometry analysis. From these, we could identify 21 putative palmitoylated mitochondrial proteins (Supplemental Tables 1–3). Of note, 19 of the 21 bands contained peptides corresponding to a single protein, which suggests that our separation methodology is highly efficient. In the case of fraction 40, band 6 (see Supplemental Fig. 1), 2 bands are clearly labeled, which were too close together to be excised separately. Importantly, carbamoyl phosphate synthetase 1 and methylmalonyl semialdehyde dehydrogenase—two previously characterized palmitoylated proteins—were identified as palmitoylated in this screen, thereby further validating our method. All proteins were confidently identified by tryptic in-gel digestion of the excised bands followed by matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF). With external calibration, a mass accuracy <100 ppm (i.e., <0.1 Da for a 1000 Da peptide) was achieved. The obtained peptide maps allowed protein identification with a significance level of better than 5% (P<0.05). All molecular weights (MW) agreed well with the MW of the identified proteins on the gel. For bands that contained more than one detectable protein, hypothetical proteins or protein bands that yielded 5 or fewer significant peptides, we obtained further confirmation of the protein identification by electrospray mass spectrometric fragmentation (ESI MS/MS) of peptides with protein-specific sequences. Details on all procedures and results of the fragmentation analysis are listed in the Supplemental Data. Many of the enzymes for β-oxidation as well as a large number of dehydrogenases were identified in this screen and are listed in Supplemental Tables 1–3.
Human HMGCS is palmitoylated on a cysteine residue via an alkali and neutral hydroxylamine sensitive thioester bond
Because of its importance in ketone body synthesis, the apparent acylation of mitochondrial HMG-CoA synthase (HMGCS) was characterized further. To do so, we subcloned an hexahistidine-tagged version of the human HMG-CoA synthase cDNA into the pET19b bacterial expression vector, and using a combination of nickel-chelating, anion-exchange chromatography and ultrafiltration, we could purify the recombinant HMGCS-His6 from E. coli lysates to apparent homogeneity as judged by Coomassie blue staining of a sample containing 10 μg of protein (Fig. 5A). The recombinant protein was catalytically active and had a specific activity of 0.11 U/mg protein. The enzymatic activity of our recombinant HMGCS-His6 was inhibited by palmitoyl-CoA with an apparent IC50 of 50 μM (data not shown). This finding contrasts with the original report by Reed et al. (40), who reported palmitoyl-CoA as a potent inhibitor of avian mitochondrial HMGCS with a Ki of ~0.5 μM.
Figure 5.

Expression, purification, and labeling of recombinant hexahistidine-tagged HMGCS. A) Protein (10 μg) from each purification step was separated on a 10% SDS-PAGE as described in Materials and Methods. Lane 1: total bacterial lysate. Lane 2: S10 supernatant of a 10,000 g centrifugation. Lane 3: elution from the Ni-chelating column. Lane 4: elution from anion exchange column. Lane 5: concentrated protein after ultrafiltration with Centricon Plus-20. B) Recombinant HMGCS-His6 (1 μg) labeled with 1 μCi of [125I]iodopalmitoyl-CoA ([125I]IC16-CoA). Acylation was detected by autoradiography (left panel), and proteins by staining with Coomassie blue (right panel). C) HMGCS-His6 samples (1 μg) were labeled with azido-palmitoyl-CoA (N3C14-CoA) with or without reaction with phosphine-Myc. Acylated protein was detected by Western blot with anti-Myc antibody (left panel), and corresponding proteins were stained with Coomassie blue (right panel). D) HMGCS-His6 samples (1 μg) were labeled with azido-palmitoyl-CoA (N3C14-CoA) and reacted with or without phosphine-biotin. Acylated protein was detected by Western blotting with Neutravidin-HRP/ECL (left panel) and corresponding proteins were stained with Coomassie blue (right panel).
To confirm whether palmitoylation of HMGCS occurs in the recombinant human enzyme as well as determine the fatty acyl acceptor amino acid and the type of bond involved in the linkage of the fatty acid, we labeled HMGCS-His6 with [125I]iodopalmitoyl-CoA (50 μM) or azido-palmitoyl-CoA (100 μM) followed by appropriate separation and detection. As shown in Fig. 5, recombinant HMGCS-His6 incorporated the radioactive fatty acid analog (Fig. 5B), the azido-palmitate/phosphine-Myc or azido-palmitate/phosphine-biotin derivatives (Fig. 5C, D) in an apparent covalent manner. Because the apparent molecular masses of HMGCS-His6 and fatty acyl-CoA synthetase are close, we included a control devoid of HMGCS-His6 to eliminate the possibility that traces of fatty acyl-CoA synthetase in the labeling reaction were responsible for the signal (Fig. 5B). The fact that our method can detect the covalent intermediate formed by fatty acyl-CoA synthetase during catalysis attests to the sensitivity of our method.
Treatment of HMGCS-His6, with increasing concentrations of NEM prior to the labeling reaction with [125I]iodopalmitoyl-CoA, showed a progressive reduction in labeling with almost complete blocking at 10 mM NEM (Fig. 6A), a concentration that lowered labeling with azido-palmitate/phosphine-biotin to background levels (Fig. 6B). When an SDS-polyacryl-amide gel containing radiolabeled or azido-palmitoylated HMGCS-His6 was treated with neutral hydroxylamine, the majority of the incorporated radiolabel and virtually all the azido-palmitate/phosphine-fluorescein label were removed, which therefore suggests that these fatty acids were linked to HMGCS-His6 via a thioester bond (Figs. 6C, D). The phosphine-fluorescein labeling allowed us to directly compare the ingel hydroxylamine treatments with the radiolabeling method. The same results were achieved when PVDF membranes containing the azido-palmitate/phosphine-biotin labeled HMGCS-His6 were treated with methanol-KOH (Fig. 6E).
To further assess the binding specificity of our azido-palmitoyl-CoA analog to HMGCS-His6 and ensure that the azido-palmitate modification was occurring at the same cysteine residue(s) as palmitate itself, we performed a competition experiment by preincubating HMGCS-His6 with increasing concentrations of palmitoyl-CoA prior to labeling with azido-palmitoyl-CoA. Figure 6F shows that palmitoyl-CoA incubation can compete for the incorporation of azido-palmitate from azido-palmitoyl-CoA in a concentration-dependent manner. Taken together these results provide strong evidence that HMGCS-His6 is palmitoylated on a cysteine residue via an alkali and hydroxylamine-sensitive thioester bond and that azido-palmitate is incorporated into the same sites as palmitate.
DISCUSSION
To circumvent the long exposure times often associated with detection of radioactive fatty acid incorporated into proteins, we developed a two-step chemical method to detect palmitoylated proteins (Fig. 1). In the first step, a bio-orthogonal isosteric azido-palmitate analog (azido-tetradecanoate) is converted into its CoA-derivative by fatty acyl-CoA synthetase and incubated with mitochondrial proteins. In the second step, the azido-palmitoylated protein is reacted with a variety of tagged triaryl-phosphines (e.g., Myc, biotin, or fluorescein) for detection. The detection achieved with the various triarylphosphine tags was not only rapid and highly sensitive but also highly specific.
Our alternative methodology reduced the exposure times from days or months with autoradiographic/fluorographic methods, using [125I]iodopalmitate or [3H]palmitate as label, to seconds, with ECL or fluorescence imaging. Recently, Hang et al. (41) also demonstrated that a variety of azido-fatty acid analogues can be used as chemical probes to monitor protein fatty acylation using the Staudinger ligation, therefore establishing a proof-of-principle for such a methodology. Of relevance to our study, they also showed that 14-azidododecanoate (azido-palmitate herein) is preferentially incorporated into proteins via thioester bonds. In the present study, we established the proof of principle with greater detail and versatility and exploited this new technique to demonstrate the existence of several palmitoylated proteins in rat liver mitochondria. Furthermore, using anio-exchange chromatography, in vitro labeling, separation by SDS-PAGE, and mass spectrometry, we identified 21 of these palmitoylated proteins in a compartment not yet recognized for its content in palmitoylated proteins. Primary hepatocyte labeling with radioactive palmitate (Fig. 2A) and labeling of mitochondria in vitro (Figs. 2-4) demonstrate that mitochondrial palmitoylated proteins are numerous and attest to the importance of investigating mitochondrial protein palmitoylation further. Of the bands identified, 19 of 21 contained peptides with masses that corresponded to only one protein, thus illustrating the efficiency of our strategy to identify these palmitoylated proteins.
The fact that two previously confirmed mitochondrial palmitoylated proteins (carbamoyl phosphate synthetase 1 and methylmalonyl semialdehyde dehydrogenase) were identified in this screen further validates our methodology (15, 20). Utilizing a recombinant HMGCS-His6, we further compared the labeling properties and efficiencies of traditional radiolabeling to azido-palmitate labeling in vitro. The newly identified palmitoylated protein, mitochondrial HMGCS-His6, incorporated the traditional radiolabeled fatty acid analog. It also incorporated the azido-palmitate followed by tagged phosphine detection on a cysteine residue via a thioester bond (Figs. 5 and 6) at the same sites since azido-palmitate incorporation could be competed out by palmitoyl-CoA preincubation (Fig. 6).
HMGCS is the rate-limiting enzyme in ketogenesis, the process of converting acetyl-CoA, derived mainly from β-oxidation of fatty acids, into ketone bodies. Interestingly, we and others have demonstrated that palmitoyl-CoA inhibits HMG-CoA synthase (data not shown; also, see ref. 40). Because the concentrations of LCFA-CoAs are increased in the liver mitochondria when ketone production is necessary, e.g., during fasting or starvation (42, 43), we do not expect that inhibition of the enzyme by palmitoylation is the main function for this type of modification. Hence, further investigation should focus on possible roles for its palmitoylation.
A large group of the identified palmitoylated proteins (9 of 21) are dehydrogenases (Supplemental Tables 1 and 3). We believe, as proposed earlier (22), that the internal NAD(H) binding site of these dehydrogenases could help facilitate acylation of the enzymes through the binding of the ADP-ribose moiety of acyl-CoA, which brings the thioester bond into close proximity to a nucleophilic cysteine residue. The previous demonstration of the palmitoylation of bovine liver glutamate dehydrogenase (EC 1.4.1.2) (20) and methylmalonyl semialdehyde dehydrogenase (20) also suggests that a significant subset of mitochondrial dehydrogenases could be potential substrates for palmitoylation. This is also corroborated by the identification of a palmitoylation site in the cytosolic glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (22).
Palmitoylation of these dehydrogenases and of the other identified mitochondrial enzymes appeared to require only the coenzyme A form of long-chain fatty acid and purified enzyme, thereby suggesting that the reaction occurs in a spontaneous fashion. Further to this point, fatty acylation of the recombinant HMGCS-His6 purified from bacteria required only the presence of LCFA-CoA analogues, which thus ruled out that contamination by an “acylating mitochondrial transferase” is required for the acylation of mitochondrial proteins in vitro.
The alpha and beta subunits of the electron-transferring flavoprotein (ETF) as well as 3 of their electron donating dehydrogenases—sarcosine dehydrogenase, isovaleryl-CoA dehydrogenase, and dimethylglycine dehydrogenase (44, 45)—were identified as palmitoylated. This finding suggests that palmitoylation is a possible requirement for their membrane localization and subcompartmentalization in close vicinity to one another and to the ETF:ubiquinone oxidoreductase (ETF-QO) and complex III of the electron transport chain, their downstream electron acceptors (46-49).
The ETF also serves as the electron acceptor for the various β-oxidation acyl-CoA dehydrogenases (50-54). We also show that the β-oxidation enzymes 3-ketoacyl-CoA thiolase (EC 2.3.1.16), hydroxyacyl-CoA dehydrogenase, enoyl-CoA hydratase (EC 2.3.1.16), and long-chain specific acyl-CoA dehydrogenase appear to be palmitoylated on a cysteine residue. Of note, short-chain specific acyl-CoA dehydrogenase (EC 1.3.99.2) was identified in a previous screen using [125I]-iodo-palmitoyl-CoA as a label (data not shown). One could argue that these enzymes may form covalent acyl-enzyme intermediates during catalysis, like we have seen with fatty acyl-CoA synthetase (Figs. 2 and 4-6). Arguing in favor of this possibility is the fact that 3-ketoacyl-CoA thiolase is known to form a thioester intermediate (55, 53), but arguing against it are the facts that enoyl-CoA hydratase, hydroxyacyl-CoA hydratase, and the acyl-CoA dehydrogenases are not reported to use cysteine residues during catalysis nor to form thioester intermediates (54-56, 58). We believe that palmitoylation of these enzymes might provide a membrane anchor and offer a plausible mechanistic explanation for the known localization of these enzymes and of others (59-62) to the inner mitochondrial membrane.
Also detected as putative palmitoylated proteins in this screen were the key enzymes of the malate aspartate shuttle, the malate dehydrogenase (EC 1.1.1.37), and the aspartate aminotransferase (EC 2.6.1.1). In vitro binding experiments have revealed that the addition of palmitoyl-CoA enhances the binding of glutamate dehydrogenase to aspartate aminotransferase or malate dehydrogenase (63, 64) and that ternary complexes of all three enzymes could be formed (65). In contrast, addition of glutamate dehydrogenase could prevent the binding of citrate synthase to malate dehydrogenase in the presence of palmitoyl-CoA (63). From these observations and since glutamate dehydrogenase can be palmitoylated in vitro (20), we can speculate that selective palmitoylation of these enzymes may function as a general mechanism to promote or stabilize protein–protein interactions and possibly perform a regulatory role in the directional flow of the malate-aspartate shuttle. During conditions of high palmitoyl-CoA as seen in obesity, disregulation of the malate-aspartate shuttle could contribute to the increased gluconeogenesis commonly found in type II diabetes (23).
We also identified alanine-glyoxylate aminotransferase 2 (EC 2.6.1.44 and EC 2.6.1.40) as a putative palmitoylated protein. This enzyme catalyzes the reversible transamination between alanine and pyruvate, utilizing either glycine and glyoxylate or glutamate and α-ketoglutarate as substrates (EC 2.6.1.44) or methylmalonate semialdehyde and 3-amino-isobutyrate as substrates (EC 2.6.1.40). Note that alanine glyoxylate aminotransferase competes with methylmalonyl semialdehyde dehydrogenase for a common substrate, methylmalonate semialdehyde, a product of valine catabolism, and that methylmalonyl semialdehyde dehydrogenase is inhibited by fatty acylation (20). It is unknown how palmitoylation affects alanine glyoxylate aminotransferase, but under conditions of high fatty acyl-CoA the inhibition of methylmalonyl semialdehyde dehydrogenase would free up the methylmalonate semialdehyde to be used for the conversion of alanine to pyruvate by alanine glyoxylate aminotransferase. Should palmitoylation have a positive affect on the activity of alanine glyoxylate aminotransferase, then increased levels of LCFA-CoA in mitochondria could result in the disregulation of pyruvate production favoring gluconeogenesis over glycolysis.
Other than providing possible membrane tethering, we also found proteins for which we could not offer simple alternative explanations for possible palmitoylation-related functions. These are alpha-methylacyl-CoA racemase (EC 5.1.99.4), sulfite oxidase (EC 1.8.3.1), and—surprisingly—the heat-shock protein 75 kDa chaperone. In addition to the above, we identified the hypothetical protein LOC365699, with homology to sugar, NAD, and sphingosine kinases, as potentially palmitoylated.
The chemical detection of palmitoylated protein using a bio-orthogonal azido-palmitate analog has multiple advantages. Since azido-fatty acids are abiotic and nontoxic to cells or animals and are readily converted into acyl-CoAs by fatty acyl-CoA synthetase (30), they could eventually be used to monitor protein palmitoylation in vivo and to perform activity based protein profiling studies (66). Furthermore, the potential use of a phosphine-biotin tag could allow for wider scale proteomics analyses using avidin-based technology. On a smaller scale herein, we demonstrated the existence of many palmitoylated proteins in rat liver mitochondria, identified several of these proteins, and postulated that mitochondrial protein palmitoylation could represent an active regulator of intermediary metabolism.
Supplementary Material
Acknowledgments
We thank Gareth Lambkin for excellent technical assistance in the preparation of the [125I] iodo-palmitate and azido-palmitoyl-CoA. L.G.B. is a senior scholar of the Alberta Heritage Foundation for Medical Research. This work was initiated by funding from CIHR grant MOP 64256 and completed by funding from CIHR grant MOP 81248 to L.G.B. M.A.K. held a Province of Alberta Graduate Scholarship. M.M.C held a Ph.D. scholarship from Alberta Heritage Foundation for Medical Research. B.O.K. thanks University of British Columbia’s Child & Family Research Institute for an Establishment Award. We thank Sheila Innis, Roger Dyer, and Suzanne Perry for access to and help with the mass spectrometric equipment, which is funded through grants from the Canadian Foundation for Innovation and the Michael Smith Foundation for Health Research. We thank Erin K. Lee for help with the protein identification. C.R.B. thanks the U.S. National Institutes of Health (NIH; GM58867) for financial support. J.R.F. thanks the Robert A. Welch Foundation and NIH (GM31278) for financial support.
References
- 1.Deichaite I, Casson LP, Ling HP, Resh MD. In vitro synthesis of pp60v-src: myristylation in a cell-free system. Mol Cell Biol. 1988;8:4295–4301. doi: 10.1128/mcb.8.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilcox C, Hu JS, Olson EN. Acylation of proteins with myristic acid occurs cotranslationally. Science. 1987;238:1275–1278. doi: 10.1126/science.3685978. [DOI] [PubMed] [Google Scholar]
- 3.Utsumi T, Sakurai N, Nakano K, Ishisaka R. C-terminal 15 kDa fragment of cytoskeletal actin is posttranslationally N-myristoylated upon caspase-mediated cleavage and targeted to mitochondria. FEBS Lett. 2003;539:37–44. doi: 10.1016/s0014-5793(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 4.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 5.Warden SM, Richardson C, O’Donnell J, Jr, Stapleton D, Kemp BE, Witters LA. Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem J. 2001;354:275–283. doi: 10.1042/0264-6021:3540275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilas GL, Corvi MM, Plummer GJ, Seime AM, Lambkin GR, Berthiaume LG. Posttranslational myristoylation of caspase-activated p21-activated protein kinase 2 (PAK2) potentiates late apoptotic events. Proc Natl Acad Sci U S A. 2006;103:6542–6547. doi: 10.1073/pnas.0600824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 8.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Linder ME, Deschenes RJ. New insights into the mechanisms of protein palmitoylation. Biochemistry. 2003;42:4311–4320. doi: 10.1021/bi034159a. [DOI] [PubMed] [Google Scholar]
- 10.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 11.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE 2006. 2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 12.Bizzozero OA, Bixler HA, Pastuszyn A. Structural determinants influencing the reaction of cysteine-containing peptides with palmitoyl-coenzyme A and other thioesters. Biochim Biophys Acta. 2001;1545:278–288. doi: 10.1016/s0167-4838(00)00291-0. [DOI] [PubMed] [Google Scholar]
- 13.Duncan JA, Gilman AG. Autoacylation of G protein alpha subunits. J Biol Chem. 1996;271:23594–23600. doi: 10.1074/jbc.271.38.23594. [DOI] [PubMed] [Google Scholar]
- 14.Bano MC, Jackson CS, Magee AI. Pseudoenzymatic S-acylation of a myristoylated yes protein tyrosine kinase peptide in vitro may reflect non-enzymatic S-acylation in vivo. Biochem J. 1998;330(Pt 2):723–731. doi: 10.1042/bj3300723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corvi MM, Soltys CL, Berthiaume LG. Regulation of mitochondrial carbamoyl-phosphate synthetase 1 activity by active site fatty acylation. J Biol Chem. 2001;276:45704–45712. doi: 10.1074/jbc.M102766200. [DOI] [PubMed] [Google Scholar]
- 16.Veit M. Palmitoylation of the 25-kDa synaptosomal protein (SNAP-25) in vitro occurs in the absence of an enzyme, but is stimulated by binding to syntaxin. Biochem J. 2000;345(Pt 1):145–151. [PMC free article] [PubMed] [Google Scholar]
- 17.Bijlmakers MJ, Marsh M. The on-off story of protein palmitoylation. Trends Cell Biol. 2003;13:32–42. doi: 10.1016/s0962-8924(02)00008-9. [DOI] [PubMed] [Google Scholar]
- 18.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 19.Stucki JW, Lehmann LH, Siegel E. Acylation of proteins by myristic acid in isolated mitochondria. J Biol Chem. 1989;264:6376–6380. [PubMed] [Google Scholar]
- 20.Berthiaume L, Deichaite I, Peseckis S, Resh MD. Regulation of enzymatic activity by active site fatty acylation. A new role for long chain fatty acid acylation of proteins. J Biol Chem. 1994;269:6498–6505. [PubMed] [Google Scholar]
- 21.Deichaite I, Berthiaume L, Peseckis SM, Patton WF, Resh MD. Novel use of an iodo-myristyl-CoA analog identifies a semialdehyde dehydrogenase in bovine liver. J Biol Chem. 1993;268:13738–13747. [PubMed] [Google Scholar]
- 22.Yang J, Gibson B, Snider J, Jenkins CM, Han X, Gross RW. Submicromolar concentrations of palmitoyl-CoA specifically thioesterify cysteine 244 in glyceraldehyde-3-phosphate dehydrogenase inhibiting enzyme activity: a novel mechanism potentially underlying fatty acid induced insulin resistance. Biochemistry. 2005;44:11903–11912. doi: 10.1021/bi0508082. [DOI] [PubMed] [Google Scholar]
- 23.Lam TK, Carpentier A, Lewis GF, van de Werve G, Fantus IG, Giacca A. Mechanisms of the free fatty acid-induced increase in hepatic glucose production. Am J Physiol Endocrinol Metab. 2003;284:E863–873. doi: 10.1152/ajpendo.00033.2003. [DOI] [PubMed] [Google Scholar]
- 24.Lewis GF, Carpentier A, Adeli K, Giacca A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr Rev. 2002;23:201–229. doi: 10.1210/edrv.23.2.0461. [DOI] [PubMed] [Google Scholar]
- 25.Unger RH. Lipotoxicity in the pathogenesis of obesity-dependent NIDDM. Genetic and clinical implications. Diabetes. 1995;44:863–870. doi: 10.2337/diab.44.8.863. [DOI] [PubMed] [Google Scholar]
- 26.Resh MD. Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods. 2006;40:191–197. doi: 10.1016/j.ymeth.2006.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 28.Saxon E, Luchansky SJ, Hang HC, Yu C, Lee SC, Bertozzi CR. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J Am Chem Soc. 2002;124:14893–14902. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- 29.Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 30.Devadas B, Lu T, Katoh A, Kishore NS, Wade AC, Mehta PP, Rudnick DA, Bryant ML, Adams SP, Li Q, Gokel GW, Gordon JI. Substrate specificity of Saccharomyces cerevisiae myristoyl-CoA: protein N-myristoyltransferase. Analysis of fatty acid analogs containing carbonyl groups, nitrogen heteroatoms, and nitrogen heterocycles in an in vitro enzyme assay and subsequent identification of inhibitors of human immunodeficiency virus I replication. J Biol Chem. 1992;267:7224–7239. [PubMed] [Google Scholar]
- 31.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci U S A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berthiaume L, Peseckis SM, Resh MD. Synthesis and use of iodo-fatty acid analogs. Methods Enzymol. 1995;250:454–466. doi: 10.1016/0076-6879(95)50090-1. [DOI] [PubMed] [Google Scholar]
- 33.Meertens LM, Miyata KS, Cechetto JD, Rachubinski RA, Capone JP. A mitochondrial ketogenic enzyme regulates its gene expression by association with the nuclear hormone receptor PPARalpha. EMBO J. 1998;17:6972–6978. doi: 10.1093/emboj/17.23.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, Lobo S, Dong X, Ault AD, Deschenes RJ. Erf4p and Erf2p form an endoplasmic reticulum-associated complex involved in the plasma membrane localization of yeast Ras proteins. J Biol Chem. 2002;277:49352–49359. doi: 10.1074/jbc.M209760200. [DOI] [PubMed] [Google Scholar]
- 35.Susin SA, Larochette N, Geuskens M, Kroemer G. Purification of mitochondria for apoptosis assays. Methods Enzymol. 2000;322:205–208. doi: 10.1016/s0076-6879(00)22020-x. [DOI] [PubMed] [Google Scholar]
- 36.Lowe DM, Tubbs PK. 3-Hydroxy-3-methylglutaryl-coenzyme A synthase from ox liver. Purification, molecular and catalytic properties. Biochem J. 1985;227:591–599. doi: 10.1042/bj2270591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magee AI, Koyama AH, Malfer C, Wen D, Schlesinger MJ. Release of fatty acids from virus glycoproteins by hydroxylamine. Biochim Biophys Acta. 1984;798:156–166. doi: 10.1016/0304-4165(84)90298-8. [DOI] [PubMed] [Google Scholar]
- 38.Ross NW, Braun PE. Acylation in vitro of the myelin proteolipid protein and comparison with acylation in vivo: acylation of a cysteine occurs nonenzymatically. J Neurosci Res. 1988;21:35–44. doi: 10.1002/jnr.490210106. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt MF, Lambrecht B. On the structure of the acyl linkage and the function of fatty acyl chains in the influenza virus haemagglutinin and the glycoproteins of Semliki Forest virus. J Gen Virol. 1985;66(Pt 12):2635–2647. doi: 10.1099/0022-1317-66-12-2635. [DOI] [PubMed] [Google Scholar]
- 40.Reed WD, Clinkenbeard D, Lane MD. Molecular and catalytic properties of mitochondrial (ketogenic) 3-hydroxy-3-methylglutaryl coenzyme A synthase of liver. J Biol Chem. 1975;250:3117–3123. [PubMed] [Google Scholar]
- 41.Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. Chemical probes for the rapid detection of fatty-acylated proteins in mammalian cells. J Am Chem Soc. 2007;129:2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 42.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323(Pt 1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegardt FG. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem J. 1999;338(Pt 3):569–582. [PMC free article] [PubMed] [Google Scholar]
- 44.Frisell WR, Mackenzie CG. Separation and purification of sarcosine dehydrogenase and dimethylglycine dehydrogenase. J Biol Chem. 1962;237:94–98. [PubMed] [Google Scholar]
- 45.Ikeda Y, Tanaka K. Purification and characterization of isovaleryl coenzyme A dehydrogenase from rat liver mitochondria. J Biol Chem. 1983;258:1077–1085. [PubMed] [Google Scholar]
- 46.Beckmann JD, Frerman FE. Electron-transfer flavoprotein-ubiquinone oxidoreductase from pig liver: purification and molecular, redox, and catalytic properties. Biochemistry. 1985;24:3913–3921. doi: 10.1021/bi00336a016. [DOI] [PubMed] [Google Scholar]
- 47.Beckmann JD, Frerman FE. Reaction of electron-transfer flavoprotein with electron-transfer flavoprotein-ubiquinone oxidoreductase. Biochemistry. 1985;24:3922–3925. doi: 10.1021/bi00336a017. [DOI] [PubMed] [Google Scholar]
- 48.Ruzicka FJ, Beinert H. A new membrane iron-sulfur flavoprotein of the mitochondrial electron transfer system. The entrance point of the fatty acyl dehydrogenation pathway? Biochem Biophys Res Commun. 1975;66:622–631. doi: 10.1016/0006-291x(75)90555-0. [DOI] [PubMed] [Google Scholar]
- 49.Ruzicka FJ, Beinert H. A new iron-sulfur flavoprotein of the respiratory chain. A component of the fatty acid beta oxidation pathway. J Biol Chem. 1977;252:8440–8445. [PubMed] [Google Scholar]
- 50.Eder M, Krautle F, Dong Y, Vock P, Kieweg V, Kim JJ, Strauss AW, Ghisla S. Characterization of human and pig kidney long-chain-acyl-CoA dehydrogenases and their role in beta-oxidation. Eur J Biochem. 1997;245:600–607. doi: 10.1111/j.1432-1033.1997.00600.x. [DOI] [PubMed] [Google Scholar]
- 51.Hall CL, Lambeth JD. Studies on electron transfer from general acyl-CoA dehydrogenase to electron transfer flavoprotein. J Biol Chem. 1980;255:3591–3595. [PubMed] [Google Scholar]
- 52.Ikeda Y, Okamura-Ikeda K, Tanaka K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J Biol Chem. 1985;260:1311–1325. [PubMed] [Google Scholar]
- 53.Izai K, Uchida Y, Orii T, Yamamoto S, Hashimoto T. Novel fatty acid beta-oxidation enzymes in rat liver mitochondria. I Purification and properties of very-long-chain acyl-coenzyme A dehydrogenase. J Biol Chem. 1992;267:1027–1033. [PubMed] [Google Scholar]
- 54.Williamson G, Engel PC. Butyryl-CoA dehydrogenase from Megasphaera elsdenii. Specificity of the catalytic reaction. Biochem J. 1984;218:521–529. doi: 10.1042/bj2180521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert HF, Lennox BJ, Mossman CD, Carle WC. The relation of acyl transfer to the overall reaction of thiolase I from porcine heart. J Biol Chem. 1981;256:7371–7377. [PubMed] [Google Scholar]
- 56.Agnihotri G, Liu HW. Enoyl-CoA hydratase. Reaction, mechanism, and inhibition. Bioorg Med Chem. 2003;11:9–20. doi: 10.1016/s0968-0896(02)00333-4. [DOI] [PubMed] [Google Scholar]
- 57.Ghisla S, Thorpe C. Acyl-CoA dehydrogenases. A mechanistic overview. Eur J Biochem. 2004;271:494–508. doi: 10.1046/j.1432-1033.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 58.He XY, Yang SY. Histidine-450 is the catalytic residue of L-3-hydroxyacyl coenzyme A dehydrogenase associated with the large alpha-subunit of the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry. 1996;35:9625–9630. doi: 10.1021/bi960374y. [DOI] [PubMed] [Google Scholar]
- 59.Kispal G, Sumegi B, Alkonyi I. Isolation and characterization of 3-hydroxyacyl coenzyme A dehydrogenase-binding protein from pig heart inner mitochondrial membrane. J Biol Chem. 1986;261:14209–14213. [PubMed] [Google Scholar]
- 60.Sumegi B, Porpaczy Z, Alkonyi I. Kinetic advantage of the interaction between the fatty acid beta-oxidation enzymes and the complexes of the respiratory chain. Biochim Biophys Acta. 1991;1081:121–128. doi: 10.1016/0005-2760(91)90016-b. [DOI] [PubMed] [Google Scholar]
- 61.Sumegi B, Srere PA. Complex I binds several mitochondrial NAD-coupled dehydrogenases. J Biol Chem. 1984;259:15040–15045. [PubMed] [Google Scholar]
- 62.Sumegi B, Srere PA. Binding of the enzymes of fatty acid beta-oxidation and some related enzymes to pig heart inner mitochondrial membrane. J Biol Chem. 1984;259:8748–8752. [PubMed] [Google Scholar]
- 63.Fahien LA, Kmiotek E. Complexes between mitochondrial enzymes and either citrate synthase or glutamate dehydrogenase. Arch Biochem Biophys. 1983;220:386–397. doi: 10.1016/0003-9861(83)90428-9. [DOI] [PubMed] [Google Scholar]
- 64.Fahien LA, Kmiotek E, Smith L. Glutamate dehydrogenase–malate dehydrogenase complex. Arch Biochem Biophys. 1979;192:33–46. doi: 10.1016/0003-9861(79)90069-9. [DOI] [PubMed] [Google Scholar]
- 65.Fahien LA, MacDonald MJ, Teller JK, Fibich B, Fahien CM. Kinetic advantages of hetero-enzyme complexes with glutamate dehydrogenase and the alpha-ketoglutarate dehydrogenase complex. J Biol Chem. 1989;264:12303–12312. [PubMed] [Google Scholar]
- 66.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


