Abstract
Regions of metazoan genomes replicate at defined times within S phase. This observation suggests that replication origins fire with a defined timing pattern that remains the same from cycle to cycle. However, an alterative model based on the stochastic firing of origins may also explain replication timing. This model assumes varying origin efficiency instead of a strict origin-timing programme. Here, we discuss the evidence for both models.
Models for the organization of eukaryotic DNA replication have to account for two diametrically opposed conceptions of replication. On one side of the spectrum is the replicon paradigm, exemplified by the mechanism of bacterial replication, in which origins are well-defined sites that fire once in each cell cycle, leading to uniform replication that is identical in every cell1. At the opposite end of the spectrum is the mechanism of replication in frog and fly embryos, in which replication initiates asynchronously throughout S phase at indiscriminate sites, presumably leading to a different, random pattern of replication in each cell2,3. It is clear that neither of these extreme mechanisms reflect the reality of replication in most eukaryotic somatic cells, but it is also clear that both models contain a kernel of truth. So how do we reconcile these apparently competing views of replication? Recent work suggests a way to bridge the gap and to reconcile stochastic origin firing with defined patterns of genome replication.
It is useful to first establish which parts of each model generally apply to eukaryotic replication. The replicon paradigm, which serves as the foundation for most textbook models of eukaryotic replication, describes eukaryotic replication reasonably well, with two major exceptions: first, most eukaryotes do not seem to have well-defined, sequence-specific origins4,5. Consequently, it has been difficult to identify metazoan replication origins, and in the few cases in which they have been identified, they seem to have no particular sequence specificity. However, too few metazoan origins have been defined to exclude the possibility of origin specific sequences. The question of what constitutes a eukaryotic replication origin remains an active field of research, and as knowledge of the exact location of origins is not necessary for the ideas presented here, we will not pursue it further. Second, eukaryotic origins are inefficient and fire in only a subset of S phases6 — only a fraction of the origins established in G1 are fired in S phase and it is a different set in each cell. In fission yeast, most origins fire about 30% of the time, whereas in mammals, the well-studied dihydrofolate reductase (DHFR) origins fire approximately 20% of the time7,8.
In terms of these two exceptions to the replicon paradigm, eukaryotic replication looks more like the example of frog and fly embryos. Frog embryos go to the extreme of establishing origins randomly across the genome, without regard to DNA sequence2,9. This is not the case for eukaryotes in general, which tend to have defined origins of replication4. Nevertheless, the redundant and inefficient nature of eukaryotic origins is more similar to the stochastic embryonic model than to the deterministic replicon model. In particular, eukaryotic replication shares an important similarity with the embryonic example: in both cases replication is heterogeneous. Because each cell uses a different combination of origins, each cell has a different pattern of replication.
It should be noted that neither of these exceptions apply entirely to budding yeast. Budding yeast has well-defined, site-specific origins, many of which are efficient and fire in as many as 90% of S phases10,11. Theses characteristics lead to fairly homogeneous replication kinetics12. The fact that budding yeast more closely fits the replicon model has made it much easier to understand replication in budding yeast, and has supported application of the replicon paradigm to eukaryotes in general. However, the observation that budding yeast’s strategy of well-spaced, efficient origins is not conserved in its distant cousin, fission yeast, let alone in metazoans, suggests that it does not serve as a general model for the global regulation of eukaryotic origins8,13,14. Thus, budding yeast seems to be the exception, rather than the rule, in the organization of eukaryotic replication.
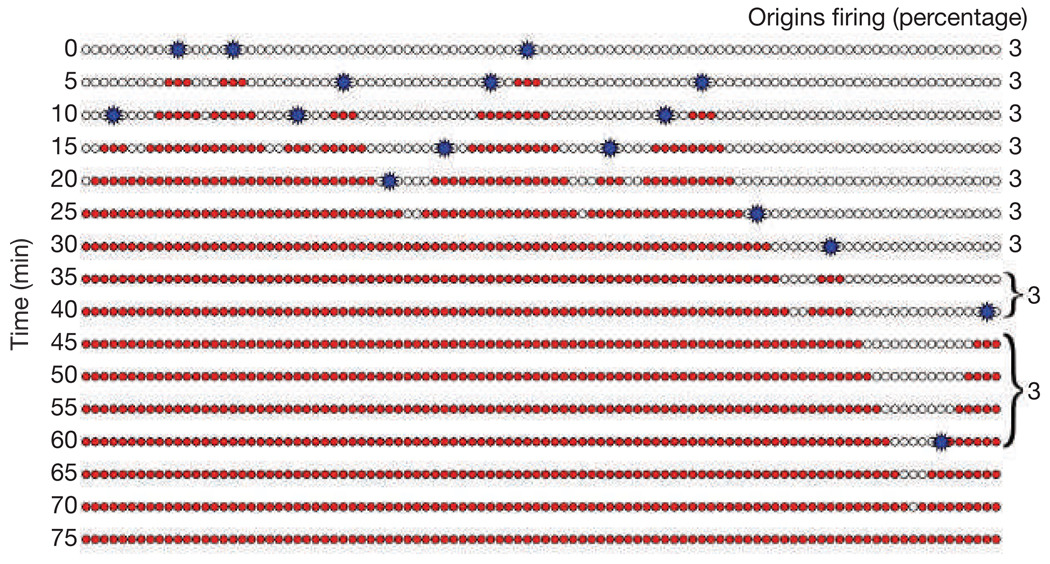
So why not accept the notion that replication in eukaryotes is less well organized than in bacteria? There are two considerations that have been difficult to reconcile with the hypothesis that the firing of eukaryotic replication origins is stochastic: first, metazoan genomes replicate with a defined timing15–20 and this observation has been interpreted to mean that eukaryotes must have a defined pattern of origin firing21. However, it is important to distinguish between the reproducible replication timing of broad regions of the genome, which has been well established, and the specific replication timing of individual origins, for which there is little evidence outside of budding yeast. We will return to this point later. Second, stochastic firing of origins leads to the so-called ‘random gap’ problem — randomly distributed origin firing will occasionally lead to large gaps between replication bubbles that would take an inordinately long time to replicate (Fig. 1)6,22. To avoid the random gap problem, it has been argued that there must be some mechanism to coordinate origin firing so as to avoid large random gaps22. Surprisingly, in two cases in which the distribution of origin firing has been carefully examined — frog embryo extracts and fission yeast — the distribution was in fact random and large random gaps were observed8,9,23,24.
Figure 1.
The random gap problem. A random distribution of origin firing will occasionally lead to large gaps in which no origins fire. This could greatly delay the completion of S phase. In this example, the chromosome is depicted as an array of 100 potential origins (white circles), all of which are licensed in G1 and each of which can fire (blue star) with a probability of 3% per 5 min step. Replication proceeds bidirectionally from the fired origins, turning the unreplicated loci (white circles) to replicated loci (red circles). At time 0, three origins fire (3% of the total). However, by 15 min, only 57 potential origins remain, so only two origins fire (~3% of the total). In this example, by random chance, no origins on the right side of the chromosome fire in early S phase. Because the chance of any one origin firing is fairly low, later firing origins do not help to replicate the gap.
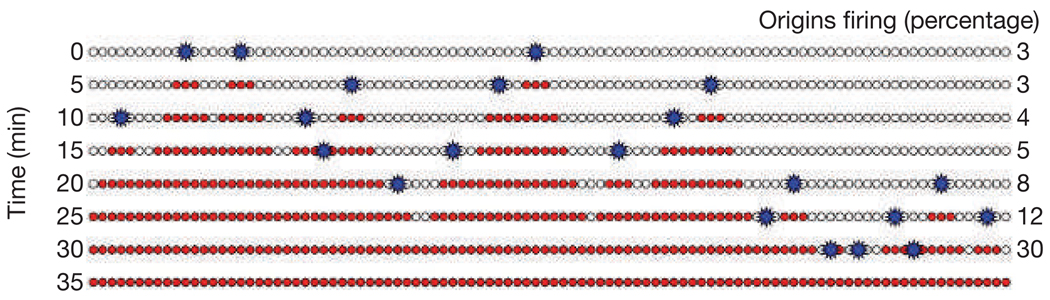
An elegant solution to the random gap problem has been proposed that reconciles the random distribution of origin firing with efficient replication6,25: the proposition is that origins fire stochastically, but the efficiency of origin firing increases as S phase progresses (Fig. 2). Therefore, the longer a large random gap persists, the more likely origins within it are to fire. This situation would ensure that no large gaps will persist beyond late S phase. This ‘increasing efficiency model’ is particularly powerful because it is applicable to any eukaryotic genome or cell cycle, and, as discussed below, can accommodate different types of replication organization, from fast and heterogeneous in embryos, to slow and ordered in somatic cells. Furthermore, it can be modified to incorporate realistic descriptions of chromosome dynamics26. It should be noted that this model only addresses the random gaps caused by stochastic firing during S phase of established origins; it does not address the separate issue of how and where origins are established in G1.
Figure 2.
A progressive increase in origin efficiency solves the random gap problem. In this example, replication begins as in Fig. 1, with a large random gap on the right side of the chromosome. However, the efficiency of origin firing increases from 3% at time 0 to 30% at 30 min. Therefore many more origins fire late in S phase in this model than in Fig. 1, and as most of the potential origins remaining in late S phase are in the gap on the right, origins fire there, closing the gap and efficiently finishing replication.
How would the increased efficiency of origin firing later in S phase in this model be achieved? One possible mechanism is polymerase recycling. Origin firing may be limited by the ability to recruit essential replication-fork proteins, such as polymerases. If so, once all the polymerases have been recruited, no more origins could fire. However, as forks merge, polymerases would be released and could be reenlisted to allow further origins to fire. This situation would lead to a roughly constant number of active replication forks throughout S phase, as polymerases are recycled from old replication forks to new ones. It would also maintain continual origin firing. However, as S phase progresses, there are fewer potential origins to fire. The reduction in potential origins is due to the fact that when an origin fires, or when it is passively replicated by a replication fork from a neighboring origin, the origin is stripped of its initiation proteins27. These proteins cannot be reloaded during S phase, preventing origins from firing more than once per cell cycle28. Thus, the number of origins firing would constitute an increasing percentage of the remaining potential origins and origin efficiency would increase (Fig. 2). This increase in origin efficiency would, in turn, ensure that random gaps would be efficiently closed by new origin firing. Both the constant number of replication forks and the increasing density of origin firing predicted by this model have been observed in budding yeast and frog embryo extracts3,9,12,25. Moreover, in the case of frog embryo extracts, the polymerase-loading and replication-fork protein Cdc45 seems to be limiting for replication, suggesting that, in this system, regulation may involve polymerase recycling29.
A constant number of active replication forks could also be maintained by a checkpoint feedback loop that monitors fork numbers and inhibits new origin firing until old replication forks merge30–32. The ATM and ATR DNA-damage checkpoint kinases, which regulate a number of initiation factors including Cdk2, Cdc7 and Cdc45, have been implicated in origin regulation during normal replication and have been proposed to regulate fork numbers in just such a manner33–36.
Alternatively, increasing origin efficiency could be caused by a diffusible, rate-limiting activator, such as Cdk2/cyclin E or Cdc7/Dbf4. Both of these replication kinases are required throughout S phase for origin firing27. If either was rate limiting, it would limit the efficiency of origins during S phase. At first, the number of firing origins would be a small faction of potential origins, but as S phase progresses and the number of potential origins declines, that number would become an ever larger fraction of potential origins and the efficiency of the remaining origins would increase. Other regulators of origin firing, such as Cdc45 or the GINS complex 27, could also function as rate-limiting activators.
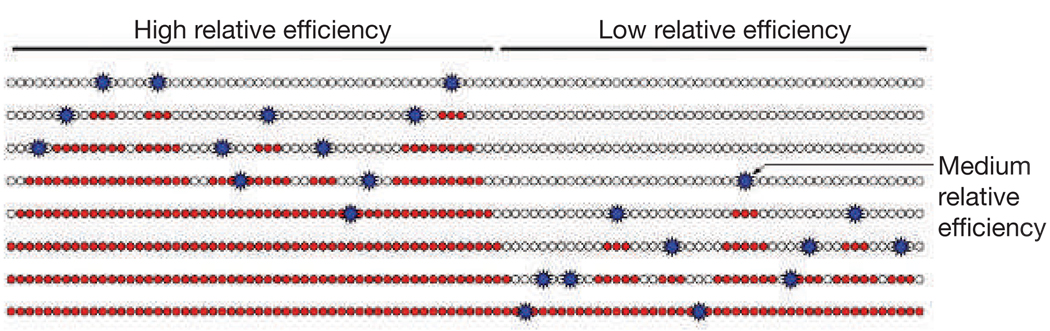
Although the increasing efficiency model would solve the random gap problem for frog embryos and fission yeast, it produces random patterns of replication, which seem to be inconsistent with the defined patterns of replication timing of metazoan genomes. However, by simply varying the relative efficiencies of individual origins, the model can explain differential timing (Fig. 3). For instance, if the origins on the left side of Fig. 3 are all more efficient that the origins on the right, the left origins are likely to fire first. Therefore, the left side will usually replicate early, and the right will usually replicate late. Furthermore, if one origin on the right is less efficient than all of the origins on the left, but more efficient than the others on the right (labelled as medium relative efficiency in Fig. 3), it will usually fire after the origins on the left, but before the others on the right, thus behaving as an efficient late origin. The existence of origins that fire efficiently late in S phase has been taken as evidence for a deterministic replication-timing programme, which would activate specific origins at specific times37. However, as shown in Fig. 3, the relative efficiency model incorporates both late efficient origins and stochastic origin firing.
Figure 3.
Differences in the relative efficiency of stochastic origins can produce defined replication-timing patterns. If all of the origins on the left are more efficient that any of the origins on the right, the origins on the left will usually fire first and the left side of the chromosome will replicate early. However, the increasing efficiency of all origins later in S phase (Fig. 2) ensures that once the left side of the chromosome is replicated the origins on the right will fire and the right side of the chromosome will replicate late. Furthermore, if one origin on the right is less efficient than all of the origins on the left, but more efficient than the others on the right (labelled as medium relative efficiency), it will usually fire late, but will usually be the first on the right to fire. Therefore, it will act as an efficient late origin.
This relative efficiency model fits well with what is known about eukaryotic replication timing. Early and late replicating sequences have not been mapped to specific early- or late-firing origins. Instead, they are associated with broad regions of early or late replicating DNA15,18–20. In the relative efficiency model, individual origins would fire stochastically, but regions that have many efficient origins would almost always replicate early because some subset (albeit a random subset) of those origins would fire early in each S phase. Thus, the relative efficiency model reconciles stochastic firing of individual origin with defined replication-timing patterns of genomic regions. This model has the added advantage that it does not require an independent mechanism to keep track of S-phase timing and firing of specific origins at defined times.
The relative efficiency model may also explain the observed correlation between active transcription and early replication38. The open chromatin structure associated with active transcription may make neighbouring origins more accessible to replication factors, and therefore more efficient and more likely to fire early39. Indeed, histone acetylation at an origin makes it more transcriptionally active and it also makes the origin replicate earlier40. This explanation suggests that replication timing is regulated over large chromosomal domains, consistent with the observation that the correlation between active transcription and early replication is strongest over fairly long distances (approximately 180 kb in flies18).
A test of the relative efficiency model awaits high resolution kinetic studies of replication that can identify specific origins and measure their efficiency at different points in S phase. Until that time, it provides one plausible explanation for how cells can accommodate the apparently disparate characteristics of having both organized replication timing and stochastic origin firing.
ACKNOWLEDGMENTS
I am grateful to J. Bechhoefer, D. Clarke, B. Kobertz and M. Munson for critical reading of the manuscript.
References
- 1.Jacob F, Brenner S. On the regulation of DNA synthesis in bacteria: the hypothesis of the replicon. C. R. Hebd. Seances. Acad. Sci. 1963;256:298–300. [PubMed] [Google Scholar]
- 2.Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21:761–771. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- 3.Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert DM. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DePamphilis ML. Replication origins in metazoan chromosomes: fact or fiction? Bioessays. 1999;21:5–16. doi: 10.1002/(SICI)1521-1878(199901)21:1<5::AID-BIES2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Hyrien O, Marheineke K, Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 2003;25:116–125. doi: 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- 7.Dijkwel PA, Hamlin JL. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell Biol. 1995;15:3023–3031. doi: 10.1128/mcb.15.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Mol. Biol. Cell. 2006;17:308–316. doi: 10.1091/mbc.E05-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrick J, Stanislawski P, Hyrien O, Bensimon A. Replication fork density increases during DNA synthesis in X. laevis egg extracts. J. Mol. Biol. 2000;300:1133–1142. doi: 10.1006/jmbi.2000.3930. [DOI] [PubMed] [Google Scholar]
- 10.Fangman WL, Brewer BJ. Activation of replication origins within yeast chromosomes. Annu. Rev. Cell Biol. 1991;7:375–402. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- 11.Newlon CS, et al. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics. 1991;129:343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghuraman MK, et al. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 13.Dubey DD, Zhu J, Carlson DL, Sharma K, Huberman JA. Three ARS elements contribute to the ura4 replication origin region in the fission yeast, Schizosaccharomyces pombe. EMBO J. 1994;13:3638–3647. doi: 10.1002/j.1460-2075.1994.tb06671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dijkwel PA, Wang S, Hamlin JL. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell Biol. 2002;22:3053–3065. doi: 10.1128/MCB.22.9.3053-3065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drouin R, Lemieux N, Richer CL. Analysis of DNA replication during S-phase by means of dynamic chromosome banding at high resolution. Chromosoma. 1990;99:273–280. doi: 10.1007/BF01731703. [DOI] [PubMed] [Google Scholar]
- 16.Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J. Cell Sci. 2004;117:5353–5365. doi: 10.1242/jcs.01412. [DOI] [PubMed] [Google Scholar]
- 17.Taljanidisz J, Popowski J, Sarkar N. Temporal order of gene replication in Chinese hamster ovary cells. Mol. Cell Biol. 1989;9:2881–2889. doi: 10.1128/mcb.9.7.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodfine K, et al. Replication timing of human chromosome 6. Cell Cycle. 2005;4:172–176. doi: 10.4161/cc.4.1.1350. [DOI] [PubMed] [Google Scholar]
- 20.Woodfine K, et al. Replication timing of the human genome. Hum. Mol. Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- 21.Donaldson AD. Shaping time: chromatin structure and the DNA replication programme. Trends Genet. 2005;21:444–449. doi: 10.1016/j.tig.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Laskey RA. Chromosome replication in early development of Xenopus laevis. J. Embryol. Exp. Morphol. 1985;89:285–296. [PubMed] [Google Scholar]
- 23.Herrick J, Jun S, Bechhoefer J, Bensimon A. Kinetic model of DNA replication in eukaryotic organisms. J. Mol. Biol. 2002;320:741–750. doi: 10.1016/s0022-2836(02)00522-3. [DOI] [PubMed] [Google Scholar]
- 24.Marheineke K, Hyrien O. Aphidicolin triggers a block to replication origin firing in Xenopus egg extracts. J. Biol. Chem. 2001;276:17092–17100. doi: 10.1074/jbc.M100271200. [DOI] [PubMed] [Google Scholar]
- 25.Lucas I, Chevrier-Miller M, Sogo JM, Hyrien O. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol. 2000;296:769–786. doi: 10.1006/jmbi.2000.3500. [DOI] [PubMed] [Google Scholar]
- 26.Jun S, Herrick J, Bensimon A, Bechhoefer J. Persistence length of chromatin determines origin spacing in Xenopus early-embryo DNA replication: quantitative comparisons between theory and experiment. Cell Cycle. 2004;3:223–229. [PubMed] [Google Scholar]
- 27.Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- 28.Diffley JF. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 29.Edwards MC, et al. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 30.Diaz-Martinez L, Clarke DJ. Self-regulating model for control of replication origin firing in budding yeast. Cell Cycle. 2003;2:576–578. [PubMed] [Google Scholar]
- 31.Shechter D, Gautier J. ATM and ATR Check in on origins: a dynamic model for origin selection and activation. Cell Cycle. 2005;4:235–238. [PubMed] [Google Scholar]
- 32.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 33.Marheineke K, Hyrien O. Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J. Biol. Chem. 2004;279:28071–28081. doi: 10.1074/jbc.M401574200. [DOI] [PubMed] [Google Scholar]
- 34.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nature Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 35.Miao H, Seiler J, Burhans WC. Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J. Biol. Chem. 2003;278:4295–4304. doi: 10.1074/jbc.M204264200. [DOI] [PubMed] [Google Scholar]
- 36.Sorensen CS, Syljuasen RG, Lukas J, Bartek J. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- 37.Ferguson BM, Brewer BJ, Reynolds AE, Fangman WL. A yeast origin of replication is activated late in S phase. Cell. 1991;65:507–515. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- 38.Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert N, et al. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]