Abstract
Metabolic profiling has received increasing recognition as an indispensable complement to genomics and proteomics for probing biological systems and for clinical applications. 1H nuclear magnetic resonance (NMR) is widely used in the field but is challenged by spectral complexity and overlap. Improved and simple methods that quantitatively profile a large number of metabolites are sought to make further progress. Here, we demonstrate a simple isotope tagging strategy, in which metabolites with carboxyl groups are chemically tagged with 15N-ethanolamine and detected using a 2D heteronuclear correlation NMR experiment. This method is capable of detecting over one hundred metabolites at concentrations as low as a few micromolar in biological samples, both quantitatively and reproducibly. Carboxyl-containing compounds are found in almost all metabolic pathways, and thus this new approach should find a variety of applications.
Keywords: Metabolic Profiling, Metabolomics, Metabonomics, NMR, Heteronuclear correlation, 15N Isotope Tagging, Carboxylic Acid
The improved characterization of the metabolome promises immense benefits for better understanding cellular activities, linking genotypes and phenotypes, and providing new understanding of complex biological states, including those of health and disease1–11. The significant interest in developing improved methods for metabolic profiling (including metabolomics and metabonomics) stems from the high sensitivity of metabolite profiles to even subtle stimuli, which is potentially important for detecting the earliest onset of various adverse biological perturbations. However, while most eukaryotic organisms are now thought to possess 3–5,000 metabolites, and possibly many more5, current high throughput analytical technologies are only capable of detecting a small fraction of these metabolites owing to severe limitations of sensitivity, resolution and/or reproducibility. These drawbacks critically limit metabolic profiling applications since the information extracted from a small metabolite subset may be too non-specific to draw a meaningful conclusion.
The information-rich analytical techniques of mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy are the primary methods used in metabolite studies, and efforts are currently underway to improve their ability to access and measure a greater number of metabolites12–14. MS is extremely attractive in metabolite studies due to its exquisite sensitivity, experimental flexibility and ability to determine unknown molecules. While MS is more sensitive compared to NMR, the data from NMR are often more quantitative and reproducible. In particular, the same nuclei detected in an NMR experiment have the same sensitivity, independent of the properties of metabolite molecules. Therefore, the absolute quantities of different metabolites can be measured with a single internal standard. NMR does not require much sample preparation or separation, and it is nondestructive. However, the complexity and spectral overlap apparent in NMR spectra of biofluids remain significant obstacles to the identification and quantification of a large number of metabolites. Methodological innovations which alleviate the current bottlenecks of NMR will greatly advance metabolomics applications.
Targeted profiling has recently shown to be a powerful approach for better understanding the metabolome in complex biological systems. As every metabolite contains at least one chemical functional group, the use of chemoselective tags can reduce the molecular complexity of the samples and potentially improve the detection of low-concentration metabolites by reducing the contribution of less interesting chemical signals. For example, liquid chromatography-coupled mass spectrometry (LC/MS) has been used for targeted metabolite detection in combination with chemoselective tagging and enrichment12. This approach provided over 300 signals from metabolites with carboxyl, amino, mercapto, or aldehyde/ketone groups in cancer cells. Such strategy is analogous to the use of chemical probes in targeted proteomics15, 16. Efforts have also been made for relative quantification of metabolites with certain functional groups by using isotopic variant tags with LC/MS17 and two-dimensional gas chromatography/mass spectrometry experiments18. We recently reported a proof-of-principle approach for NMR-based targeted metabolic profiling in which amine-containing metabolites were tagged with 13C to achieve improved detection limits and resolution, and applied this approach to the study of inborn errors of metabolism19.
In the present work, we report a robust method for detecting a large number of low-concentration carboxyl-containing metabolites using a straightforward method to tag metabolites of interest with 15N. Carboxyl-containing compounds represent a large and important class of metabolites or exogenous compounds, and they appear in essentially all important metabolic pathways20. A number of these compounds are known biomarkers or potential biomarkers for various diseases21–23. Advantages of the NMR method detailed below include its quantitative accuracy, improved resolution that reduces spectral overlap, and a concomitant improvement in identification. The approach makes use of high resolution 1H-15N 2D NMR and results in the detection of nearly two hundred well-resolved signals corresponding to well over 100 carboxyl-containing metabolites that can be routinely and reproducibly detected in serum and urine samples.
EXPERIMENTAL SECTION
Chemicals and biological samples
Sixty-two carboxyl-containing metabolite standards (Table 1), 3-(trimethylsilyl) propionic-2, 2, 3, 3-d4 acid sodium salt (TSP) (all from Sigma - Aldrich), 4- (4, 6- dimethoxy [1. 3. 5] triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM) (Acros) and 15N-ethanolamine (Cambridge Isotope Laboratories) were used without further purification. Human serum and urine samples were obtained either from commercial sources or from healthy volunteers in accordance with the Internal Review Board at Purdue University. Sodium azide (0.1% wt/vol) was added to urine samples to prevent bacterial growth. The urine and serum samples were then filtered with a centrifugal filter device Centriprep YM-10 (Millipore, Billerica, MA), aliquoted, and frozen at −80 °C until used.
Table 1.
Chemical shifts for the 15N-tagged metabolite standard compounds [H2O:D2O (95:5); pH = 5.0; 298 K].
| Label | Name | 1H (ppm) | 15N (ppm) | Label | Name | 1H (ppm) | 15N (ppm) |
|---|---|---|---|---|---|---|---|
| 1 | Acetic Acid | 7.99 | 120.87 | 32 | 4-Hydroxy-L-proline | 8.25 | 116.72 |
| 2 | cis-Aconitic Acid | 8.19 | 121.80 | 8.42 | 115.63 | ||
| 8.26 | 120.49 | 33 | Isocitric Acid | 8.25 | 116.36 | ||
| 8.58 | 118.58 | 8.04 | 120.22 | ||||
| 3 | Adipic Acid | 8.01 | 120.40 | 8.29 | 121.78 | ||
| 4 | L-Alanine | 8.21 | 113.65 | 7.94 | 121.63 | ||
| 5 | β-Alanine | 8.10 | 119.80 | 34 | Isoleucine | 8.28 | 118.75 |
| 6 | L-2-Aminoadipic Acid | 8.20 | 116.41 | 35 | alpha-Ketoglutaric Acid | 8.58 | 116.45 |
| 8.27 | 115.70 | 8.40 | 118.20 | ||||
| 7 | L-2-Aminobutyric Acid | 8.28 | 115.90 | 36 | L-Lactic Acid | 8.40 | 113.69 |
| 8 | L-Arginine | 8.26 | 116.13 | 8.12 | 112.96 | ||
| 9 | L-Asparagine | 8.22 | 115.28 | 37 | L-Leucine | 8.24 | 115.19 |
| 10 | L-Aspartic Acid | 8.23 | 115.28 | 8.31 | 115.99 | ||
| 8.13 | 121.49 | 38 | L-Lysine | 8.26 | 115.84 | ||
| 11 | Benzoic Acid | 8.46 | 114.85 | 39 | Maleic Acid | 8.40 | 121.77 |
| 12 | Betaine | 8.56 | 124.23 | 40 | Malic Acid | 8.05 | 121.77 |
| 13 | Butyric Acid | 7.99 | 120.60 | 8.19 | 114.22 | ||
| 14 | Citraconic Acid | 8.00 | 120.08 | 41 | Malonic Acid | 8.23 | 122.01 |
| 8.26 | 118.55 | 42 | L-Mandelic Acid | 8.66 | 115.38 | ||
| 15 | Citric Acid | 8.02 | 122.59 | 43 | L-Methionine | 8.25 | 115.70 |
| 8.17 | 113.75 | 44 | Methylmalonic Acid | 8.09 | 118.90 | ||
| 8.16 | 121.70 | 45 | 3-Methyl-2-oxovaleric acid | 8.62 | 113.10 | ||
| 16 | L-Cysteine | 8.30 | 116.36 | 46 | Octanoic Acid | 7.95 | 120.47 |
| 17 | N,N-Dimethylglycine | 8.39 | 117.94 | 47 | Oxalic Acid | 8.73 | 113.89 |
| 18 | Formic Acid | 8.05 | 123.93 | 8.40 | 116.22 | ||
| 19 | Fumaric Acid | 8.53 | 124.40 | 48 | Phenylacetic Acid | 8.05 | 120.68 |
| 20 | Glucuronic Acid | 8.29 | 118.91 | 49 | L-Phenylalanine | 8.11 | 117.24 |
| 8.22 | 118.47 | 50 | L-3-Phenyllactic Acid | 8.26 | 116.43 | ||
| 21 | L-Glutamic Acid | 8.20 | 115.55 | 7.94 | 115.42 | ||
| 8.01 | 120.37 | 51 | L-Proline | 8.25 | 114.94 | ||
| 22 | L-Glutamine | 8.27 | 115.87 | 52 | Propionic Acid | 7.92 | 118.49 |
| 23 | Glycine | 8.11 | 114.70 | 53 | L-Pyroglutamic Acid | 8.20 | 115.55 |
| 24 | Glycolic Acid | 8.29 | 114.65 | 54 | Pyruvic Acid | 8.55 | 110.99 |
| 8.12 | 114.10 | 55 | L-Serine | 8.25 | 116.69 | ||
| 25 | Guanidineacetic Acid | 8.16 | 114.98 | 56 | Suberic Acid | 7.97 | 120.34 |
| 26 | Hippuric Acid | 8.09 | 114.79 | 57 | Succinic Acid | 8.00 | 119.30 |
| 27 | L-Histidine | 8.22 | 116.52 | 58 | L-Tartaric Acid | 8.34 | 116.51 |
| 28 | 4-Hydroxybenzoic Acid | 8.55 | 115.34 | 59 | L-Threonine | 8.27 | 117.49 |
| 29 | 3-Hydroxybutyric Acid | 8.04 | 121.94 | 60 | L-Tryptophan | 8.01 | 116.84 |
| 30 | 2-Hydroxyphenyl Acetic Acid | 7.91 | 119.78 | 61 | L-Tyrosine | 8.17 | 117.23 |
| 31 | 4-Hydroxyphenyl Acetic Acid | 8.06 | 120.83 | 62 | L-Valine | 8.29 | 118.40 |
General procedure of 15N-ethanolamine tagging
3 μL 15N-ethanolamine (50 μmol) was added to 400 μL of the sample in a dry glass vial, and the pH of the mixture was adjusted to 7.0 with 1 M HCl. DMT-MM (21 mg) was added to initiate the reaction24, 25. The mixture was continuously stirred at room temperature for 4 hrs to complete the reaction. In order to maintain 15N amide protonation, the pH was adjusted to 5.0 by adding 1 M HCl or 1 M NaOH, and then the solution volume was adjusted to 600 μL by adding water prior to NMR detection26. The mixture of metabolite standards was prepared by mixing 10μL of a 20 mM stock solution for each compound. Urine was used with no pretreatment. Serum was deproteinated by a procedure described in the Supporting Information prior to tagging.
NMR spectroscopy
The sample solutions (570 μl each) were mixed with 30 μl of D2O containing TSP (0.5% wt/vol) and placed in 5 mm NMR tubes. NMR experiments were carried out at 298 K on a Bruker Avance-III-800 spectrometer equipped with a room temperature 1H inverse detection Z-gradient probe unless otherwise noted. Additional spectra were acquired using a Bruker DRX-500 spectrometer equipped with a 1H inverse detection Z-gradient cryoprobe (Figure S3 & S5). 1H NMR spectra were obtained using the water Pre-SAT180 sequence27. A 1H-1H double quantum-filtered COSY spectrum was obtained with WATERGATE solvent suppression. The sensitivity-enhanced 1H-15N 2D HSQC experiments employed an INEPT transfer delay of 5.5 ms corresponding to a 1JNH of 90 Hz. Spectral widths of approximately 10 kHz for the 1H dimension and 5 kHz for 15N were used at 800 MHz and spectral widths of approximately 6 kHz for the 1H dimension and 3 kHz for 15N were used at 500 MHz. 128 or 256 free induction decays of 2,048 data points each were collected in the indirect (t1) dimension with either 4 or 8 transients per increment. 15N decoupling during the direct detection dimension (t2) was achieved with the GARP (Globally Optimized Alternating-Phase Rectangular Pulses) sequence. The resulting 2D data were zero-filled to 1,024 points in the t1 dimension after forward linear prediction to 512 points. A 45°-shifted sinebell window function was then applied to both dimensions before Fourier transformation. Chemical shifts were referenced to the 1H signal of TSP for the 1D spectra or the derivatized formic acid signal (1H: 8.05 ppm; 15N: 123.93 ppm) in the HSQC spectra. The signal-to-noise ratios of peaks in the 2D spectra were calculated by dividing the peak intensity by the standard deviation of 50 noise peaks randomly chosen from the nearby spectral region (i.e., within ± 0.1 ppm in 1H dimension and ± 1 ppm in 15N dimension of the peak itself). NMR data were processed using Bruker Topspin 2.0 spectrometer software on a Redhat Linux platform and Bruker XWINNMR 3.5 on a SGI/IRIX platform.
RESULTS
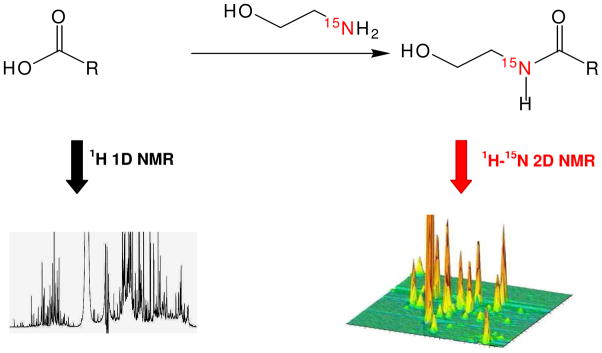
15N-ethanolamine was found to meet the criteria for a good isotope tag for NMR analysis of carboxyl-containing metabolites. Ethanolamine easily and selectively combines with carboxyl-containing metabolites under aqueous conditions through the formation of amide bonds (Figure 1). The use of 15N-enriched reagent ensures that other N-containing metabolites are invisible in the spectrum because of the low natural abundance of 15N (0.37%). Because the tagged metabolite contains at least one polar group (hydroxyl in ethanolamine), it retains its solubility in aqueous medium. The magnetic characteristics of the detected pairs of nuclei (15N and 1H) from ethanolamine, such as the J-couplings and relaxation times, are very similar for different tagged metabolites, which reduces detection bias. J-couplings of ~90 Hz between 1H and 15N were observed in all formed 15N-amides ensures high sensitivity detection in the 2D heteronuclear coherence single quantum (HSQC) NMR experiment. On the other hand, the varied chemical environment of individual metabolites leads to high 1H-15N peak dispersion in the 2D spectrum. As illustrated in Figure 1, this tagging approach leads to a well-resolved 2D spectrum that is much less complex than its 1D counterpart. The improved resolution results in part due to the fact that each reacted carboxyl group is represented by a single peak in the 2D spectrum.
Figure 1.
Schematic figure illustrating the approach used for metabolite 15N tagging. 15N-ethanolamine reacts with carboxyl-containing metabolites in the presence of DMT-MM to form 15N-amides. The 2D 1H-15N HSQC experiment detects the tagged metabolites with wide dispersion that greatly improves the resolution and limit of detection.
15N-ethanolamine tagging was evaluated using a mixture of 62 known carboxyl-containing compounds (Table 1) that were chosen because of their importance in a number of biological systems. In our experience, most normal carboxyl-containing compounds (having no strong electron-donating or withdrawing functional groups at α-carbons) are converted into 15N-amides quantitatively or with yields greater than 95% using an excess amount of 15N-ethanolamine. Some complications do arise: α-amino acids that undergo intramolecular side reactions and have lower tagging efficiencies (~30%), and compounds with α-hydroxyls (lactic acid) or conjugated carboxyl groups also show reduced yields, yet the reproducibility is still maintained in both cases (See Note in Supporting Information).
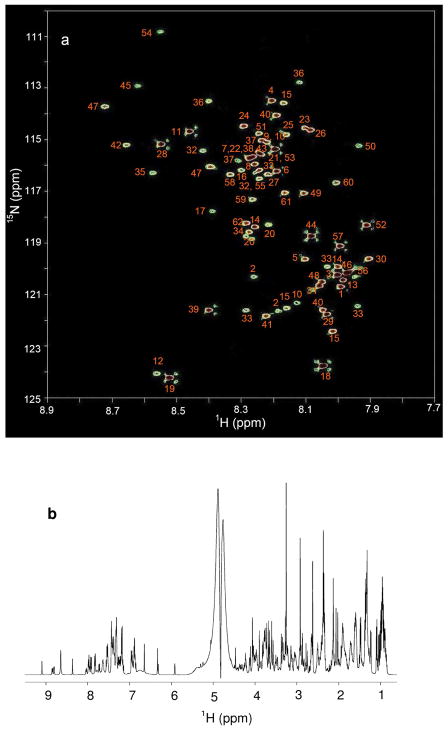
Figure 2a shows the 2D 1H -15N HSQC spectrum of the 62 compound mixture after tagging with 15N-ethanolamine. The peaks in the 2D spectrum are well dispersed and span a range of ~15 ppm in the 15N dimension and ~1.0 ppm in the 1H dimension. Each peak in the 2D HSQC spectrum represents one carboxyl-containing metabolite. However, metabolites that contain multiple carboxyl groups may give rise to additional peaks depending on the molecular structure. Metabolites having identical carboxyl groups such as adipic, maleic, and succinic acids (labeled 3, 57, and 39, respectively) show only one peak while those having two non-identical carboxyl groups such as aspartic and malic acids (labeled 10 and 40, respectively) show two peaks in the spectrum. All assigned peaks were identified using a library of chemical shifts constructed from 2D HSQC spectra of individual standard compounds reacted with 15N-ethanolamine (Table 1). In contrast to the well-resolved 2D HSQC spectrum shown in Figure 2, the 1D 1H NMR spectrum of the same mixture without tagging produces a complex and highly overlapped spectrum (Figure. 2b) from which it is difficult to identify many of the same metabolites without spiking individual compounds.
Figure 2.
(a) A 2D 1H-15N HSQC NMR spectrum (with annotations) of a mixture of 62 metabolite standards tagged with 15N-ethanolamine and acquired at 800 MHz within 40 min. The identified metabolites correspond to the numbered entries in Table 1. (b) 1D 1H NMR spectrum of a mixture of 62 metabolite standards acquired at 800 MHz with a pre-SAT180 pulse sequence before tagging.
The detection linearity was evaluated using a set of mixtures of 11 standard compounds that were reacted with 15N-ethanolamine and analyzed by HSQC. The integrated volumes of the 2D signals were plotted against the corresponding concentrations measured by 1D 1H NMR (See Figure S1). All metabolites exhibited good linearity with coefficients of regression (R2) greater than 0.99, including compounds with one and two carboxyl groups. The detection limit was evaluated using an 8 min 2D experiment at 800 MHz. 15N labeled metabolites could be detected with concentrations as low as 8 μM (SNR ~3).
Tagging metabolites in biological samples
15N-ethanolamine tagging of metabolites in urine samples resulted in a rich 2D 1H-15N HSQC spectrum (Figure 3). Nearly 200 well-dispersed peaks were detected that arise entirely from carboxyl-containing metabolites. This result demonstrates the facility with which 15N tagging can be utilized to detect a large number of such metabolites, well over 150, assuming that most of these metabolites do not have multiple unique carboxyl groups. Twenty-two metabolites were identified by comparing the chemical shifts of 2D peaks in Table 1 and compounds are labeled in Figure 4. This group includes metabolites such as oxalic acid that cannot be detected by 1D 1H NMR experiment. In contrast, approximately 30–40 metabolites (including less than 20 containing carboxyl groups) can be identified in a typical urine 1D 1H NMR spectrum28. For the urine sample used in this study, the 1D 1H NMR spectrum contained identifiable signals from only 13 carboxyl-containing metabolites because of the high degree of spectral overlap (See Figure S2 and Table S1). Spectral overlap further impedes reliable quantitative analysis, unless selective methods or spiking experiments are employed29, 30. Even the use of high resolution two-dimensional experiments, such as 1H-1H correlation spectroscopy (COSY), did not appreciably increase the number of detected metabolites (Figure S3).
Figure 3.
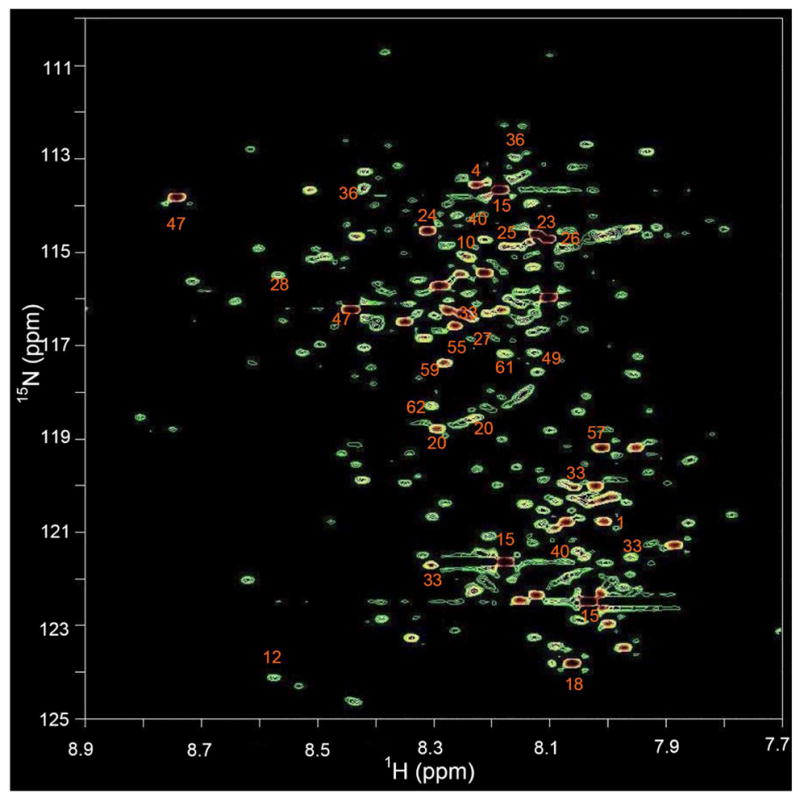
A 2D 1H-15N HSQC spectrum of human urine obtained after tagging carboxyl-containing metabolites with 15N-ethanolamine acquired at 298K within 40 min. Nearly 200 metabolites are detected and 23 of these are identified and annotated by comparing the chemical shifts with those of metabolites in Table 1.
Figure 4.
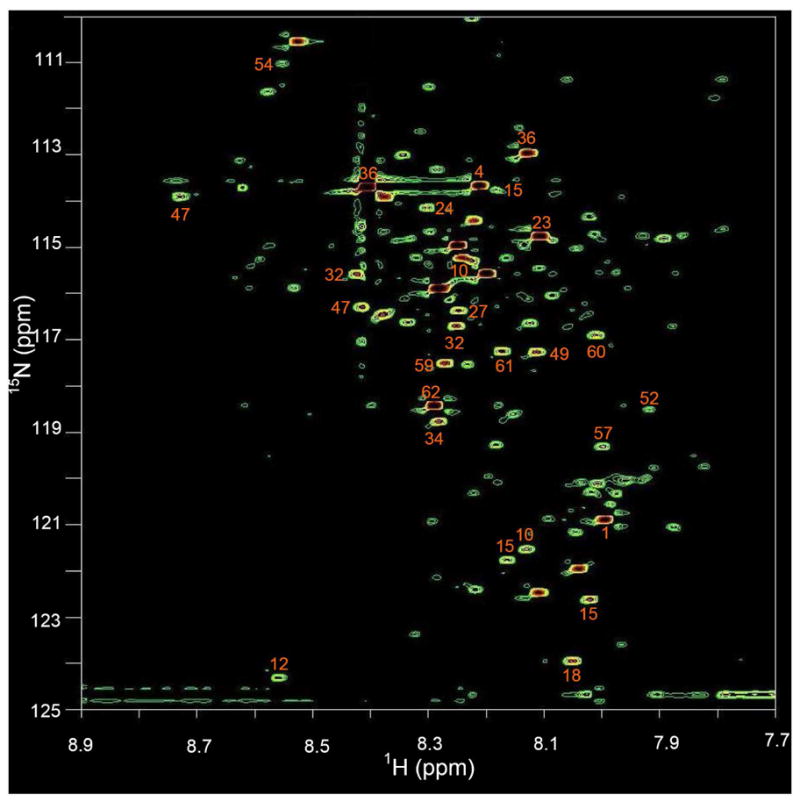
A 2D 1H-15N HSQC spectrum of human serum obtained after tagging carboxyl-containing metabolites with 15N-ethanolamine. Nearly 180 metabolite peaks are detected and 21 of these stronger peaks are identified as annotated by comparing the chemical shifts with those of metabolites in Table 1.
Figure 4 shows the 1H-15N 2D spectrum of a serum sample after treating with 15N-ethanolamine. Nearly 180 well-dispersed peaks corresponding to a similar number of carboxyl-containing metabolites can be seen in the spectrum. Twenty-one metabolites identified by comparison with the chemical shift library of Table 1 are labeled in the figure. Typically, 30–50 metabolites can be identified from the 1D 1H NMR spectrum of a serum sample, and less than 20 of these metabolites contain carboxyl groups. With the combination of JRES, COSY and HMQC NMR experiments, up to 40 carboxyl-containing metabolites have been identified from human serum samples31, 32. From the 1D proton NMR spectrum of the serum sample used here, spectral features for only 15 carboxyl- containing metabolites could be found (Figure S4 and Table S2). Not surprisingly, peaks from almost all these metabolites are overlapped or are obscured by the varying baseline, which thus significantly complicates reliable quantitative analysis.
The reproducibility was tested by performing triplicate reactions and analyses on a split serum sample. Identical 2D spectra were observed for the three samples, with all the peaks matching on a one-to-one basis (Figure S5 & S6), which indicates the excellent reproducibility of this tagging method. Automated peak picking and integration using Bruker Topspin gave an average coefficient of variation (CV) of 7% for the 101 most intense peaks and an average CV of 5% for the 47 most intense peaks detected using a higher peak-picking threshold.
DISCUSSION
Selection of the 15N-labeled ethanolamine ‘tag molecule’ was based on the following criteria. First, the 15N isotope imparts a large dispersion to the individual metabolite signals because of the broad chemical shift dispersion of 15N nuclei (~900 ppm for 15N in varied chemical environments, and ~15 ppm for 15N-labeled ethanolamides investigated here). To utilize such dispersion efficiently, the isotopically enriched nucleus was chosen to be as close as possible (one bond) to the metabolite upon tagging. Second, the sensitivity of metabolite detection utilizing the heteronuclear correlation NMR experiment benefits from having the 15N of the tagging molecule coupled relatively strongly to a more sensitive nucleus such as 1H. To retain quantitation, both the magnitude of the coupling and the relaxation properties of the nuclear pair (15N and 1H) do not appreciably vary across the metabolites of interest. Third, the 15N tagging process yields a simple NMR spectrum (one peak per carboxyl group) to ensure high resolution in the NMR spectrum. Fourth, the tagging reaction should have a high yield to ensure good detection sensitivity and quantitation. Aqueous reaction conditions are preferred to avoid an additional change of solvent that could reduce reproducibility; for the same reason the metabolites should continue to be soluble after tagging. The use of 15N-ethanolamine meets these criteria and offers a simple method to tag the carboxyl-containing metabolites selectively and directly in the complex biofluid. 1H-15N 2D HSQC NMR detection provides access to several hundred low concentrated metabolites (as low as a few μM) in a single experiment within one hour.
The 15N tag shows excellent reproducibility for a complex biological sample such as serum. These characteristics strongly reinforce the advantages of NMR for quantitative metabolic profiling. The resolution improvement is imparted from the combined dispersion of the chemical shifts of the tagging nuclei (15N and 1H) and the high sensitivity is derived from the combination of factors such as isotope labeling and the strong J-coupling between the observed nuclei. In addition, a single peak (devoid of multiplicity) for each tagged metabolite and effective filtering of non-tagged metabolites significantly add to the sensitivity and background suppression. These characteristics are important for advanced metabolic profiling as well as for identifying unknown potential metabolite biomarkers.
In contrast, one-dimensional (1D) 1H NMR experiments, commonly used in metabolic profiling, primarily emphasize signals arising from highly concentrated (mM) metabolites. These metabolites (often referred to as the “usual suspects”) are often found to be too non-specific to be used reliably as biomarkers. Most low-concentration metabolites in crowded 1D 1H NMR spectra are buried beneath the dominant signals of high concentration metabolites. This situation leads to low numbers of detected metabolites and challenges in direct quantitation without spiking. Considering such limitations in the 1D and 2D NMR experiments commonly used for detecting relatively low concentrated metabolites in metabolomics studies, the high number of quantifiable metabolites available for detection using the 15N tagging approach shown here is a significant improvement.
The growth of metabolic profiling applications critically depends on the ability to measure the onset of the biological perturbations. Therefore, technological advancements in both detecting and identifying larger numbers of low concentrated metabolites in the spectra will be a key driver for future progress and applications in metabolite profiling. Further improvements in detection limits may be obtained by the use of preconcentration and microcoil NMR33, or potentially by new methods in dynamic nuclear polarization34. Although the 2D experiment used here may require more time than a simple 1D 1H NMR, incorporation of latest technological advancements for data collection and processing will potentially make this approach suitable for high throughput applications35–37 Alternatively, this approach could be used for biomarker discovery, while selective 1D NMR approaches could then be applied for quantitative and faster analysis29, 30. Regarding the identification of unknown metabolites, we anticipate that the structures of individual metabolites can be established from NMR experiments such as 2D HSQC-TOCSY which could provide crucial molecular connectivity for the tagged metabolites38. We are also pursuing LC/microcoil NMR based methods for structural analysis. These approaches will be particularly important for identifying any new metabolites that do not exist or are not well confirmed in existing databases and for constructing larger chemical shift libraries of tagged metabolites.
In conclusion, we have demonstrated a simple isotope tagging approach for conducting a multidimensional NMR based targeted profiling with the aim of detecting a large number of low concentrated metabolites. Carboxyl-containing metabolites represent a major part of the metabolome, and thus profiling these metabolites is vital, particularly since they are believed to be present in virtually all the important metabolic pathways. 15N-ethanolamine is straightforwardly coupled to carboxyl groups and allows the reproducible and quantitative 2D NMR detection of over one hundred metabolites in commonly studied biofluid samples such as serum and urine. The inherent characteristics of this tagging approach add significantly to the number of metabolites that can be detected and quantified by NMR. Simultaneous detection of a class of metabolites of this large number, we believe, will be of high utility towards the advancement of metabolic profiling.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grant 1 R01GM085291-01. Daniel Raftery is a member of the Purdue Cancer and Oncological Sciences Centers.
Footnotes
SUPPORTING INFORMATION AVAILABLE
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nicholson JK, Lindon JC, Holmes E. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 2.Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Nature Biotechnology. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 3.Fiehn O. Plant Molecular Biology. 2002;48:155–171. [PubMed] [Google Scholar]
- 4.Kell DB. Current Opinion in Microbiology. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Nature Reviews Molecular Cell Biology. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 6.van der Greef J, Smilde AK. Journal of Chemometrics. 2005;19:376–386. [Google Scholar]
- 7.Van Dien S, Schilling CH. Molecular Systems Biology. 2006;2:1–2. doi: 10.1038/msb4100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serkova NJ, Niemann CU. Expert Review of Molecular Diagnostics. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- 9.Pan ZZ, Raftery D. Analytical and Bioanalytical Chemistry. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 10.Gowda GAN, Zhang SC, Gu HW, Asiago V, Shanaiah N, Raftery D. Expert Review of Molecular Diagnostics. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang SC, Gowda GAN, Asiago V, Shanaiah N, Barbas C, Raftery D. Analytical Biochemistry. 2008;383:76–84. doi: 10.1016/j.ab.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson EE, Cravatt BF. Nature Methods. 2007;4:429–435. doi: 10.1038/nmeth1038. [DOI] [PubMed] [Google Scholar]
- 13.Welthagen W, Robert SA, Joachim S, Michael R, Ralf Z, Oliver F. Metabolomics. 2005;1:65–73. [Google Scholar]
- 14.Dunn WB, Bailey NJC, Johnson HE. Analyst. 2005;130:606–625. doi: 10.1039/b418288j. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Li XJ, Martin DB, Aebersold R. Nature Biotechnology. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 16.Brittain SM, Ficarro SB, Brock A, Peters EC. Nature Biotechnology. 2005;23:463–468. doi: 10.1038/nbt1076. [DOI] [PubMed] [Google Scholar]
- 17.Lamos SM, Shortreed MR, Frey BL, Belshaw PJ, Smith LM. Analytical Chemistry. 2007;79:5143–5149. doi: 10.1021/ac062416m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang XD, Regnier FE. Analytical Chemistry. 2008;80:107–114. doi: 10.1021/ac071263f. [DOI] [PubMed] [Google Scholar]
- 19.Shanaiah N, Desilva MA, Gowda GAN, Raftery MA, Hainline BE, Raftery D. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galli V, Garcia A, Saavedra L, Barbas C. Electrophoresis. 2003;24:1951–1981. doi: 10.1002/elps.200305473. [DOI] [PubMed] [Google Scholar]
- 21.Dionisi-Vici C, Deodato F, Roschinger W, Rhead W, Wilcken B. Journal of Inherited Metabolic Disease. 2006;29:383–389. doi: 10.1007/s10545-006-0278-z. [DOI] [PubMed] [Google Scholar]
- 22.Wolfsdorf J, Glaser N, Sperling MA. Diabetes Care. 2006;29:1150–1159. doi: 10.2337/diacare.2951150. [DOI] [PubMed] [Google Scholar]
- 23.Hiraoka A, Akai J, Tominaga I, Hattori M, Sasaki H, Arato T. Journal of Chromatography A. 1994;680:243–246. doi: 10.1016/0021-9673(94)80073-1. [DOI] [PubMed] [Google Scholar]
- 24.Kunishima M, Kawachi C, Morita J, Terao K, Iwasaki F, Tani S. Tetrahedron. 1999;55:13159–13170. [Google Scholar]
- 25.Kunishima M, Kawachi C, Hioki K, Terao K, Tani S. Tetrahedron. 2001;57:1551–1558. [Google Scholar]
- 26.Zhang YZ, Paterson Y, Roder H. Protein Science. 1995;4:804–814. doi: 10.1002/pro.5560040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mo HP, Raftery D. Journal of Magnetic Resonance. 2008;190:1–6. doi: 10.1016/j.jmr.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foxall PJD, Parkinson JA, Sadler IH, Lindon JC, Nicholson JK. Journal of pharmaceutical and biomedical analysis. 1993;11:21–31. doi: 10.1016/0731-7085(93)80145-q. [DOI] [PubMed] [Google Scholar]
- 29.Sandusky P, Raftery D. Analytical Chemistry. 2005;77:7717–7723. doi: 10.1021/ac0510890. [DOI] [PubMed] [Google Scholar]
- 30.Sandusky P, Raftery D. Analytical Chemistry. 2005;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson JK, Foxall PJD, Spraul M, Farrant RD, Lindon JC. Analytical Chemistry. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 32.Lindon JC, Nicholson JK, Everett JR. Annual Reports on NMR Spectroscopy. 1999;38:1–88. [Google Scholar]
- 33.Djukovic D, Liu S, Henry I, Tobias B, Raftery D. Analytical Chemistry. 2006;78:7154–7160. doi: 10.1021/ac0605748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang FL, Brüschweiler R. Journal of the American Chemical Society. 2004;126:13180–13181. doi: 10.1021/ja047241h. [DOI] [PubMed] [Google Scholar]
- 36.Frydman L, Blazina D. Nature Physics. 2007;3:415–419. [Google Scholar]
- 37.Blinov KA, Larin NI, Williams AJ, Mills KA, Martin GE. Journal of Heterocyclic Chemistry. 2006;43:163–166. [Google Scholar]
- 38.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Nature Biotechnology. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.