Abstract
Luminal-like breast tumor cells express estrogen receptor α (ERα), a member of the nuclear receptor family of ligand-activated transcription factors that controls their proliferation, survival, and functional status. To identify the molecular determinants of this hormone-responsive tumor phenotype, a comprehensive genome-wide analysis was performed in estrogen stimulated MCF-7 and ZR-75.1 cells by integrating time-course mRNA expression profiling with global mapping of genomic ERα binding sites by chromatin immunoprecipitation coupled to massively parallel sequencing, microRNA expression profiling, and in silico analysis of transcription units and receptor binding regions identified. All 1270 genes that were found to respond to 17β-estradiol in both cell lines cluster in 33 highly concordant groups, each of which showed defined kinetics of RNA changes. This hormone-responsive gene set includes several direct targets of ERα and is organized in a gene regulation cascade, stemming from ligand-activated receptor and reaching a large number of downstream targets via AP-2γ, B-cell activating transcription factor, E2F1 and 2, E74-like factor 3, GTF2IRD1, hairy and enhancer of split homologue-1, MYB, SMAD3, RARα, and RXRα transcription factors. MicroRNAs are also integral components of this gene regulation network because miR-107, miR-424, miR-570, miR-618, and miR-760 are regulated by 17β-estradiol along with other microRNAs that can target a significant number of transcripts belonging to one or more estrogen-responsive gene clusters.
The cellular responses to hormones and other homeostatic stimuli are characterized by timed and coordinated transcriptional regulation of specific gene subsets. In the case of mitogens, for example, the stimulus induces ordered gene activation and repression events occurring at specific times after exposure, coinciding with cell cycle progression. The overall response represents a transcriptional cascade that stems from the primary genomic targets of the mitogen into a gene network that ultimately comprises a large number of intracellular effectors. Through analysis of gene expression profiles, we attempt to reconstruct these gene regulatory circuits and to identify genes that represent focal nodes for exchanges between multiple pathways, provided the possibility to define relationships between regulatory inputs, changes in gene activity, and cellular functions.1,2
Estrogens control multiple functions of hormone-responsive breast cancer (BC) cells, where they act as potent mitogens for a subset of tumor cells fostering cell cycle progression and cell proliferation, and contribute to the maintenance of epithelial luminal A or B phenotypes.3,4 Estrogen actions are mediated primarily by the estrogen receptors (ERs), ligand-inducible transcription factors of the nuclear receptor family of homeostatic regulators. ERs drive gene cascades comprising primary genes, whose transcription is directly controlled by the hormone through physical interaction of ERs with regulatory sites in the genome (genomic pathway of ER action) and/or with signal transduction effectors (nongenomic pathway), as well as downstream genes whose activity depends on the functions encoded by the primary responders.5,6 The identification of primary estrogen target genes, and the downstream gene cascade that stems from them, is being actively pursued because it will provide essential information on the molecular and genomic pathways of the hormone-responsive BC phenotype. This is an important asset in clinical oncology because effective endocrine treatments of BC with ERα antagonists or aromatase inhibitors would be greatly improved by reliable predictors of tumor sensitivity to these drugs in the adjuvant or metastatic settings, as well as by the availability of markers of pharmacological resistance and new therapeutic targets to overcome it.7 Estrogen responsive BC cell models have proven to be very useful in this case because ERα-expressing breast tumors and cell lines share significant similarities in their transcriptomes8 and the expression profiles of estrogen-responsive gene sets identified in vitro in luminal-like BC cell models have been found to be an intrinsic genetic signature of ERα-expressing breast tumors.9 These gene signatures have been useful to define BC prognosis and response to endocrine therapy.10 Similarly, the expression pattern of antiestrogen-responsive genes identified in BC cell models in vitro correlated significantly with disease recurrence after Tamoxifen treatment in women with ERα-positive cancers.11 A detailed definition of the estrogen-responsive gene network in BC cell models is thus a prerequisite not only to be able to identify defined breast tumor subtypes according to their estrogen-dependent intrinsic molecular signatures, but also to understand ERα contribution to the establishment and maintenance of specific BC clinical phenotypes, characterized by diverse disease prognosis and responsiveness to therapy.12
For these reasons, a number of studies have focused on identification of gene sets regulated by estrogen in ERα-expressing luminal-like BC cell models, including MCF-7,13,14,15 T47D,8 and ZR-75.116,17,18 cells. Although these studies led to the identification of a significant number of hormone-regulated genes, an integrated view of the estrogen-responsive genetic pathway of luminal-like BC cells is still missing, due in part to biological and technical variables among experiments.19 Furthermore, data available do not permit accurate identification of functional hierarchies and interactions among estrogen-responsive genes, nor do they reveal novel regulatory components of the estrogenic pathway.
To overcome these limitations, we performed a comprehensive analysis of ERα-based estrogen signaling in 17β-estradiol (E2) stimulated MCF-7 and ZR-75.1 cells by using high-density bead-arrays for high resolution time-course mRNA expression profiling designed to map hormone-induced transcriptional changes. The results obtained in the two cell lines were combined to define common patterns of gene regulation by building a comprehensive map of the dynamic gene expression changes that characterize the cell response to estrogen. To this end, genes showing identical kinetics of response to the hormone in both cell lines were clustered and the resulting gene sets, highly homogeneous in kinetics terms, were analyzed to identify common sequence signatures, within transcription units (TUs) belonging to the same kinetic cluster, including transcription factor binding sites in promoter regions. In parallel, global mapping of ERα binding to MCF-7 cells genome in vivo by chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-Seq) was performed under these experimental conditions to identify primary response genes.
MicroRNAs (miRNAs) are a class of small RNAs that regulate protein-coding gene expression mainly at the level of mRNA stability and translation efficiency. miRNAs can target simultaneously a relatively large number of mature transcripts within the same cells and have been shown to act on cellular signaling pathways by preferentially targeting downstream components of the cascade, rather than ligands and receptors, suggesting that they may contribute to fine-tuning of signal-dependent regulatory networks.20 Recent evidence indicates that the cellular concentration of miRNAs can be modulated by extracellular stimuli, particularly nuclear receptor ligands.21,22,23,24,25,26,27 To further study this, we took advantage of the highly concordant changes in mRNA levels that characterize the kinetic clusters identified in this study and searched for enrichment in miRNA target transcripts within each gene cluster identified and performed miRNA expression profiling in MCF-7 cells after estrogen stimulation. The results obtained identify several miRNAs involved in hormonal control of gene activity in luminal-like BC cells.
Materials and Methods
Cell Culture and RNA Purification
Human breast cancer MCF-7 and ZR-75.1 cells were propagated as monolayer cultures in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (both from Invitrogen Spa, San Giuliano Milanese, Italy), 100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml Amfotericin-B. Cells were routinely tested for mycoplasma infection by using MycoAlert mycoplasma detection kit (Cambrex Bio Science, Rockland, ME). For G0–G1 synchronization cells were plated at 20% to 40% confluence in steroid-free medium (phenol red-free Dulbecco’s modified Eagle’s medium with 5% fetal bovine serum pretreated with dextran-coated charcoal and antibiotics) and maintained for 4 days with replacement of same fresh medium before stimulation with 10−8 M E2.
Total RNA was extracted from control (+ ethanol vehicle, −E2) or hormone stimulated (+E2) cell cultures by RNA extraction with Trizol (Invitrogen), as described previously.17,28
For mRNA expression profiling, RNA was extracted from MCF-7 and ZR-75.1 cells before (0) or after 1, 2, 4, 6, 8, 12, 16, 20, 24, 28, and 32 hours of stimulation with 17β-estradiol 10−8 M (+E2) or ethanol vehicle. For miRNAs expression profiling, RNA was extracted from MCF-7 cells before (0) or after 2, 12, 24, and 72 hours of stimulation with 17β-estradiol 10−8 M (+E2) or ethanol vehicle.
In each case RNA derived from three independent experiments was used. Before use, RNA concentration in each sample was determined with a ND-1000 spectrophotometer (NanoDrop, Wilmington, DE) and quality assessed with the Agilent 2100 Bioanalyzer with Agilent RNA 6000 nanokit (Agilent Technologies, Santa Clara, CA).
Microarray Analyses
For mRNA expression profiling, 500 ng total RNA were reverse transcribed, as described previously,29 and used for synthesis of cDNA and biotinylated cRNA according to the Illumina TotalPrep RNA amplification kit (Ambion, Austin, TX; category number IL1791) sample preparation kit protocol. For each sample, 700 ng of cRNA were hybridized for 18 hours at 55°C on Illumina HumanWG-6 version 2.0 BeadChips containing 48701 probes (Illumina Inc., San Diego, CA), according to the manufacturer’s protocol, and subsequently scanned with the BeadArray Reader 500 according to the manufacturer’s standard methods.
For miRNA expression profiling, instead, 600 ng of total RNA were fluorescently labeled and hybridized in triplicate to Illumina Human version 2 MicroRNA Expression BeadChips, according to the protocols provided by the array manufacturer. The human microRNA panel used comprises 1145 probes designed on miRNA sequences present in miRBase database (release 12.0) and on additional novel content derived using Illumina sequencing technology.
mRNA Expression Profiling Data Analysis
Hybridization reactions were performed in duplicate for each sample, except for the reference sample (before stimulation, 0 hours), which was at least in quadruplicate. For mRNA expression profiling, the microarray analysis protocol previously described18 was implemented as follows: data were preprocessed by using BeadStudio Software version 3.2 (Illumina Inc., San Diego, CA) with the quantile30 normalization; values below a detection score of 0.05 were filtered out; missing values were imputed. Probes with more than two out of four (MCF-7) or three out of five (ZR-75.1) missing values for the reference sample arrays (0 hours) were excluded from further analysis. Data were log2-transformed, and ratios of each signal against the average reference signal were calculated. Probes with more than 20% missing values of the resulting time series were filtered out.
The probes showing a signal profile that was significant over time were identified with Bayesian Analysis of Time Series software.31,32 For this analysis, two different models (normal and double-exponential) were used for the noise estimation, and three different values of the lambda parameter, which influences the degree of the estimated polynomials describing the probe expression profiles, were applied. Using the different combination of parameters, six partially overlapping probe lists were obtained in each cell line. We considered as a more robust selection the one made up of probes present in all six lists (intersection) and as less robust selection the one made up of probes present in at least one list (union).
For gene identification, probe annotations of the array’s manifest were controlled by remapping all of the probe sequences onto the human genome hg18 and checking their concordance to Refseq database entries (National Center for Biotechnology Information Build 36.1). An “in house” method was developed and applied to obtain the alignments of each 50 oligonucleotides probe sequence along each chromosome. Only complete alignments (100% identity) were considered. Probes having a single match were called “one match probes,” whereas probes with more than one match on the same or different chromosomes (“multiple match probes”) were discarded from further analysis. The “one match probes” were mapped onto the UCSC Known Genes track of the UCSC Genome Browser.33 All multiple transcripts referring to the same gene name were merged in a single TU. A limited number of probes mapping to the same TU were checked for signal coherence and median analyzed at each time-point to obtain a single signal for each gene. All of the probes regulated in both cell lines mapping to a position where no UCSC Known Genes record was present were assigned (in order) to a RefSeq gene, an Ensembl gene, a National Center for Biotechnology Information transcript, or, if none of the above was available, labeled with the corresponding Illumina PROBE_ID.
Cluster analysis on the gene profiles significantly affected by estrogen stimulation and on the shorter list of putative primary targets was performed by using SplineCluster,34 a Bayesian functional based software. Data were prepared by imputing missing data points with the Bioconductor package ‘impute,” implementing the “K nearest neighbor” method,35 and by averaging the replicates at each time point for each selected gene. All of the clustered gene profiles are shown as heatmaps, generated with the MeV application.36
To identify statistically overrepresented “biological process” gene ontology terms in our regulated gene lists, we used the Database for Annotation, Visualization, and Integrated Discovery functional annotation tool.37 We analyzed the overrepresentation of biological process terms in the differentially expressed genes regulated in both cell lines against the whole expressed (detected) gene list as a background, with a P value threshold of 0.01.
miRNA Expression Profiling Data Analysis
For miRNA expression profiling raw data analysis quality control and the fluorescence intensity files were loaded into the Illumina BeadStudio Gene Expression module version 3.1.3.0. A quantile normalization algorithm was applied on the raw datasets. For differential expression analysis, technical replicates of each sample were grouped together, and miRNAs with a detection P value below 0.01, corresponding to a false-positive rate of 1%, were considered as expressed. Differently expressed miRNAs were selected with a DiffScore cut-off set at ±20, corresponding to a P value of 0.01. Probe sequences were aligned both on the genome, the pre-miR sequences from the UCSC genome browser,33 and the sno/miRNA (wgRna) track,38,39,40 using the Biostrings (Pages H, Aboyoun P, Gentleman R, DebRoy S. Biostrings: string objects representing biological sequences, and matching algorithms. R package version 2.12.1), BSgenome (Pages H. BSgenome: infrastructure for Biostrings-based genome data packages. R package version 1.12.0), and BSgenome.Hsapiens.ucsc.hg18 (Pages H. BSgenome.Hsapiens.UCSC.hg18: homo sapiens full genome: UCSC version hg18. R package version 1.3.11) Bioconductor packages; no mismatches were allowed. Probes matching repeat elements, having multiple matches in the genome outside the expected targets, and probes not matching a known or putative miRNA gene were discarded.
Using the software SigTerms,41 we calculated microRNA target enrichment in the clusters of estrogen-responsive genes (Figure 1, A–C). To this aim, we generated a custom annotation file (the reference background for the analysis) according to SigTerms’ instruction. We downloaded the TargetScanS 5.0 miRNA family-mRNA target pair predictions42 and removed from the lists both mRNAs and miRNAs or miRNA families not expressed in the BC cell lines. All detected mRNA genes that were not predicted to be the target of any microRNA, but were present in the list of 17,839 genes against which the original prediction analysis was run, were added and assigned to a virtual “none” microRNA category. This was done considering that for target enrichment analyses, using all predicted mRNA targets as background, independently from their effective expression in the cell lines under study, would introduce a bias in the enrichment calculation. Furthermore, genes without the assignment of an Entrez Gene ID,43 such as those recognized by “orphan probes,” were removed from the clusters before the analysis.
Figure 1.
High resolution time-course of gene expression profiling in estrogen-stimulated MCF-7 and ZR-75.1 BC cells and transcription factor binding site overrepresentation analysis in the promoter region of genes showing identical kinetics of response to hormonal stimulation. A: Changes in expression (log2 of the fold-change) of 1270 genes occurring in both cell lines after exposure to 10−8 M 17β-estradiol; black triangles indicate the time of cell exposure to the hormone (left to right: from 1 to 32 hours), and vertical bars point to the 33 gene clusters identified in MCF-7 cells and shown in detail in B, where the number of genes within each gene cluster are reported in brackets. C: TFBS matrix enrichment (overrepresentation) analysis of the promoter region (−2000/+800 from the transcription startsite) of genes belonging to each estrogen-responsive gene cluster reported in B is shown. Z score cut-off was 3.0 and only TFBSs showing an overrepresentation score ≥2.5 in at least one of the gene clusters are reported; gray cells indicate a Z score <3.0. Only clusters where at least one matrix scored above these thresholds are shown. The V$ER.01, V$ER.02, and V$ER.03 matrices represent different variants of the ERα binding site (ERE) and are shown for comparison; in this case a cut-off for the Z score was not applied and the overrepresentation threshold was 1.5.
Enrichment analysis was performed for all clusters with more than 10 genes by using the classical one-sided Fisher’s exact test provided by SigTerms, with a P value threshold of 0.01. The threshold was set after running several Monte-Carlo simulation tests with simulated clusters having a varying size, in the range of the actual cluster sizes. Furthermore, only miRNA families showing at least 15% predicted target in a given cluster were considered to avoid the possibility that an excessively low number of targets might artificially generate significant P values.
Chromatin Immunoprecipitation
Human breast cancer MCF-7 cells were hormone deprived for 4 days (as described before). Chromatin was extracted as replicates before (−E2) and after stimulation for 45 minutes with 10−8M 17β-estradiol (+E2). Chromatin was prepared with the Millipore/Upstate Chromatin Immunoprecipitation (ChIP) assay kit (Millipore, Vimodrone, Italy) according to the manufacturer’s instruction, using a variation of the protocol described at the Upstate website. For each assay, a total of 5 × 106 cells were fixed with 1% of formaldehyde for 10 minutes at room temperature, and the reaction was stopped by adding glycine at final concentration of 0.125 M. Fixed cells were washed twice with ice-cold PBS, harvested by scraping, centrifuged, and the cell pellets were resuspended in SDS lysis buffer. Samples were sonicated with a Diagenode Bioruptor (Diagenode SA, Belgium) for 12 cycles of 30 seconds at high power, centrifuged at 13,200 rpm for 15 minutes, and diluted eightfold in ChIP dilution buffer. After removing an aliquot (whole-cell extract input), the chromatin samples were incubated at 4°C overnight with antibodies against the C-terminus (HC-20, from Santa Cruz Biotechnology Europe, Heildelberg, Germany) or the N-terminus (anti-estrogen receptor 18-32, from Sigma-Aldrich, Milano, Italy) of human ERα (+E2 and −E2). As control, aliquots of the same chromatins were processed in the same way, but the Abs were omitted from the reaction (+E2/−Abs). The samples were then precipitated by binding to protein A-Agarose/Salmon Sperm DNA beads (Millipore, Billerica, MA), and beads were washed sequentially with “low-salt immune complex wash buffer,” “high salt immune complex wash buffer,” “LiCl immune complex wash buffer,” and Tris-EDTA (TE) buffer, before elution in Elution buffer by over-night (ON) incubation at 65°C and treatment with proteinase K. DNA was purified from immunoprecipitated chromatin by extraction with phenol/chloroform/isoamyl alcohol (25:24:1) and ethanol precipitation according to standard procedures. DNA pellets were dissolved in nuclease-free water and kept frozen before further use.
Samples Preparation for ChIP-Seq
About 20 ng of ChIP DNA was purified by using the MinElute PCR Purification Kit (Qiagen, Milano, Italy) with a recovery of 55% to 70%, as assessed with the Quant-IT DNA Assay Kit-High Sensitivity and a Qubit Fluorometer (Invitrogen). Preparation of immunoprecipitated DNA libraries for massively parallel sequencing was performed from 10 ng purified DNA according to the Illumina ChIP-Seq sample preparation kit protocol (Illumina catalog IP-102-1001). Libraries were sequenced with the Illumina Cluster Station and Genome Analyzer according to the manufacturer’s instructions.
ChIP-Seq Data Analysis
The sequence tags generated by massively parallel sequencing were aligned on the genome with the software ELAND, allowing up to two mismatches. Enriched regions from MCF-7 cell stimulated with 17β-estradiol for 45 minutes (+E2) were compared with the same from MCF-7 cultured maintained in steroid-free medium (−E2). The enriched ChIP-Seq peaks were identified by using Site Identification from Short Sequence Reads44 with a window width of 30 bp and a P value threshold of 0.001. Sites comprising a number of tags with a site/length ratio in the 25% lower part of the range were discarded. Because the DNA samples were sequenced twice, the analysis was performed independently on the two replicates. The resulting site lists were merged.
Computational Searches for Estrogen Response Element Sequences
The estrogen response element (ERE) binding motif was searched in the ERα binding sites with the MatInspector application,45 a part of the GenomatixSuite software (Genomatix Software GmbH, Munich, Germany). The matrices ER.01, ER.02, and ER.03 (Genomatix Matrix Library 8.0) were used with a core similarity threshold of 0.75 and a matrix similarity threshold of optimal −0.02. We classified the sequences bearing a match of any of the three matrices as ERE+ sequences. On the remaining sequences, the hemi-palindromic ERE motif was searched with the same application by defining a custom half ERE matrix PuGGTCA, because the half-ERE was not present in the database, with the similarity threshold of 1.00 (“perfect matches” only). The remaining sequences were classified as ERE− and were scanned for other transcription factor binding sites motifs contained in the Genomatix Matrix Library.
Transcription Factor Binding Site Overrepresentation Analysis
Transcription factor binding sites (TFBSs) overrepresentation analysis was performed with the RegionMiner application of the Genomatix software suite.46 Enrichment was calculated with respect to the whole human genome or to Genomatix annotated promoters expressed as Z scores.47 Overrepresentation values with a Z score less than 3.0 were not considered further. A filter of 1.5, 2.5, 5, or 10 was applied on overrepresentation values, depending on the range of values set for each analysis (see Results), to highlight only the stronger associations. Results are shown as heatmaps representing overrepresentation ratios, generated with MeV software.36
Real-Time Quantitative PCR Analysis of ChIP DNA Samples
The PCR primers used are listed in Table 1 and were designed to amplify 60 to 120 bp fragments from selected genomic regions. Real-time PCR reactions were performed by using Power SYBR green PCR master mix (Applied Biosystems, Monza, Italy) in an MJ Research PTC-200 Opticon Instrument. Each PCR reaction generated only the expected specific amplicon, as shown by the melting-temperature profiles of final products (dissociation curve). Fold-enrichment was determined by using 2 μl ChIP DNA (input) as template. Each enrichment ratio was determined as described48 from multiple, independently performed ChIP assays, each evaluated in duplicate by real-time PCR.
Table 1.
List of Primers Used for qPCR Validation of ERα Binding Sites Identified by ChIP-Seq
| Site | Chromosome | Forward primers | Coordinates* | Reverse primers | Coordinates* |
|---|---|---|---|---|---|
| ERαC291 | 1 | 5′-TGTTTAGTGCTTGCTCCATCTC-3′ | 200348918-39 | 5′-TGGAAAGCTGCCACAGTTC-3′ | 200348960-78 |
| ERαC370 | 2 | 5′-GAGCTGACCTTGTGGTAGGC-3′ | 11556327-46 | 5′-GCTGACAGAGGAGACAAAACG-3′ | 11556371-91 |
| ERαC375 | 2 | 5′-CAGCTGACTGTCTTCCACCA-3′ | 11597092-111 | 5′-CAGTACGCTGTGCAAAGAACA-3′ | 11597140-60 |
| ERαC393 | 2 | 5′-ACCAGTTCTCAGGGTTGTATCTG-3′ | 20538276-98 | 5′-AGTACTCCCCCAGCTGCAC-3′ | 20538334-52 |
| ERαC449 | 2 | 5′-CTGGGACATAATCCCCCTTT-3′ | 74926006-25 | 5′-TCCTCCTGTCCTCAGACATTC-3′ | 74926051-71 |
| ERαC466-467 | 2 | 5′-CTCTGCTGGTGCTGGTTG-3′ | 100274256-73 | 5′-GAGGAGCGGAGTGCCTTT-3′ | 100274299-316 |
| ERαC523 | 2 | 5′-GATCTGATGTGGTGGCCTGT-3′ | 160786599-618 | 5′-CTGAGGCTCACCCTTCACC-3′ | 160786650-68 |
| ERαC797 | 4 | 5′-CCCTGCGAGCTATTTTGAA-3′ | 3434825-43 | 5′-CGGTCATCTTCTGTCATGGTT-3′ | 3434887-907 |
| ERαC850 | 4 | 5′-ATCAGTTGGGCTCTGCAAGT-3′ | 140051385-404 | 5′-GGTGACCCAGAACACTCAAAA-3′ | 140051424-44 |
| ERαC1125 | 6 | 5′-TGAAGCTCAAAAGGGCATAGA-3′ | 151979252-72 | 5′-CTGCTTGACACATGAACCAAA-3′ | 151979322-42 |
| ERαC1199 | 7 | 5′-CTGTTCCAAGCCTGAGTGG-3′ | 56000280-98 | 5′-TGCTTTACTCTTTTATTCCCTACTCC-3′ | 56000344-69 |
| ERαC1229 | 7 | 5′-GGGCCGCACATAATTTCA-3′ | 92076038-55 | 5′-TGGCAGGAGAGAGGCAGT-3′ | 92076103-20 |
| ERαC1417 | 8 | 5′-ACTCTGCACTGCCAGACAAA-3′ | 128749887-906 | 5′-TGGAAACCACATTTTGGTCA-3′ | 128750005-24 |
| ERαC1531 | 9 | 5′-TGAGCAGCTGGGTCAACTC-3′ | 96441308-26 | 5′-CGACACGGACGTCAACAC-3′ | 96441376-93 |
| ERαC1544 | 9 | 5′-GGAGCTATCTTGACCGTGAGA-3′ | 109343882-902 | 5′-AGAAACTGCCCGTGAGGTC-3′ | 109343923-41 |
| ERαC1573 | 9 | 5′-CAGCTGTCCAACTGCCAAC-3′ | 130437654-72 | 5′-CCCTGTGTCCCAGGTCAG-3′ | 130437753-70 |
| ERαC1591 | 9 | 5′-GGGGAGGGGTGTGACCTA-3′ | 136391265-82 | 5′-GGGAGGGAGGGAAAACAAT-3′ | 136391306-24 |
| ERαC1592 | 9 | 5′-CATCTCAGGTTGGGCAATG-3′ | 136444210-28 | 5′-AATGCTGCGGAACGAGTC-3′ | 136444260-77 |
| ERαC1609 | 9 | 5′-ACGACCAGACAATCTGAGAGG-3′ | 139528141-61 | 5′-GTCACCGGCAAGCTCATT-3′ | 139528195-212 |
| ERαC1750 | 10 | 5′-AGGCCCTCAAAGATCCTAGC-3′ | 76173221-40 | 5′-ACACCTCAGGGTGACAGGAC-3′ | 76173307-26 |
| ERαC1910 | 11 | 5′-CAGACCACAGCATGGCTCTA-3′ | 68961082-101 | 5′-CACCTGTCTGCCCCTGTC-3′ | 68961135-52 |
| ERαC2027 | 12 | 5′-GGGCGACTGAATGGAAAG-3′ | 38123182-99 | 5′-GAGCCGTGAATCACCTCCT-3′ | 38123228-46 |
| ERαC2117 | 12 | 5′-CTACCCTGTCTGTGGGGTTG-3′ | 130911657-76 | 5′-ACTCCCCTGTTGCATCCAC-3′ | 130911721-39 |
| ERαC2123 | 12 | 5′-GAACCTTCTGGCCTCGTTC-3′ | 131709045-63 | 5′-GGCCTGACATCCCATATTGA-3′ | 131709089-108 |
| ERαC2268 | 14 | 5′-ATCCCAAACCACCTCTGATG-3′ | 98439511-30 | 5′-GTGTATTCCAGGCAGAAACCTT-3′ | 98439573-94 |
| ERαC2790 | 17 | 5′-GTGCATGGCTCCCATACC-3′ | 63800004-21 | 5′-GGGGCACGGACTGTCTAA-3′ | 63800054-71 |
| ERαC2800 | 17 | 5′-CGCCTGTGGCTTAGAAAGAC-3′ | 68148502-21 | 5′-GGACAAATCTGCGATGTGAA-3′ | 68148577-96 |
| ERαC3084 | 20 | 5′-ATGATGCCCTGAGGAAAGG-3′ | 29724208-26 | 5′-CAGGATGGGAACTAAAAGATCAG-3′ | 29724280-302 |
| ERαC3389 | 20 | 5′-CATCAGAGGCTCGTGTCAGA-3′ | 61171612-31 | 5′-CTCCCTGTGCTTCCCAAGT-3′ | 61171656-74 |
| ERαC3450 | 21 | 5′-CTAGACGGAATGGGCTTCAT-3′ | 42660143-62 | 5′-GCTTGGCCTGACAACAGTG-3′ | 42660075-94 |
| ERαC1615 | X | 5′-GGCAGCCTGAACTTTCTGAG-3′ | 12957559-78 | 5′-GGGTCACCCTAGTCCTCCAG-3′ | 12957613-32 |
| ERαC1628 | X | 5′-CAGTGCCAGCAAAATACCTG-3′ | 109081916-35 | 5′-TGTGTTGCCTTTATTAAGGATCTTC-3′ | 109081963-87 |
Coordinates from the hg18 human genome sequence release. Information relative to the reverse primers used for PCR are reported in italic.
Time-Series Network Identification Analysis
To identify primary target genes downstream of the estrogenic stimulus, we used Time-Series Network Identification (TSNI).49 The TSNI algorithm was applied both on the genes regulated in MCF-7 and ZR-75, separately. Because TSNI needs equally spaced series of points, Bayesian Analysis of Time Series32 estimation was used of each selected gene profile to generate 1-hour spaced series from 1 to 32 hours. As baseline, a constant perturbation P = 1 or the fluorescence signal relative to ERα mRNA was used. Genes with a TSNI score associated to a P value lower than 0.005 were selected.
Raw Data Availability
Complete microarray and ChIP-Seq datasets relative to all experiments reported are available in the ArrayExpress database (http://www.ebi.ac.uk/microarray-as/ae/) with the following accession numbers: E-TABM-742 and E-MTAB-131.
Results
Genome-Wide Analysis of the Dynamic Gene Expression Changes Induced by Estrogen in Hormone-Responsive BC Cells
ERα-positive, hormone-responsive MCF-7 and ZR-75.1 cells represent a well established experimental model to investigate the molecular and genomic mechanisms mediating hormonal control of cell functions in vitro.8,50 We performed a high resolution time-course analysis of the estrogen-regulated transcriptome in both of these cell lines with a whole-genome microarray platform with the aim of identifying all known genes showing identical kinetics and type of response to the hormone in this luminal-like BC cell models. For this, total RNA was extracted from hormone-starved, growth-arrested cells at time t = 0 and at 11 time-points, for up to 32 hours, into stimulation with 10−8 M E2, a timing that corresponds to completion of a cell cycle under these conditions.28,50 The RNA samples, obtained in replicas, were then labeled and hybridized to Illumina HumanWG-6 BeadChips.
To identify the genes whose expression profile was significantly modified by estrogen treatment, the datasets of both cell lines were analyzed separately with Bayesian Analysis of Time Series software.32 Using stringent statistical criteria for estrogen-regulated probe selection, 2225 and 1699 probes were first identified in MCF-7 and ZR-75.1 cells, respectively. To increase the number of significant probes in common between the two cell lines, we applied a slightly less stringent selection threshold to MCF-7 cell-derived expression data. This was applied to include also “borderline” probes from the first analysis, yielding 3214 regulated probes from MCF-7 cells. Intersection of this dataset with that from ZR-75.1 cells generated a common set of 1399 probes revealing identical changes in the corresponding RNAs in both cell lines (Figure 1). It should be mentioned here that the probe-gene association provided by the array manufacturer was verified by remapping all probes present in the arrays to the human genome sequence (hg18) to discard unreliable or multiple matching probes.51,52 In this way we were able to map 40,931 of 48,701 probes present on the array, and 38,650 (79.4%) matched a unique sequence and 2281 (4.7%) showed multiple “hits” in the genome, whereas the remaining 7770 probes did not match any genome sequence. The last two groups were discarded from further analyses. After retrieving the genomic coordinates for the 38,650 useful probes, each of them was assigned to the corresponding gene by matching it against transcripts from UCSC Known Genes, RefSeq Genes, Ensembl Genes, and/or National Center for Biotechnology Information mRNAs databases (correct strand only), and when no match was found the probe was labeled “orphan.” We then merged all transcripts deriving from the same gene locus in a unique TU to avoid possible redundancies in the dataset due to genes with multiple transcripts and to simplify distance calculations (see below). For this reason, we use the terms “gene” and “TU” as synonyms. When multiple probes were associated to the same TU, coherence in their expression profile was checked. If coherence was confirmed, we calculated for each experimental point the average probe signal and considered it as gene-specific signal. Through this procedure we unambiguously identified 10,128 expressed TUs in MCF-7 cells, including 2873 estrogen-regulated ones, of which 1270 were also regulated in ZR-75.1 cells (Supplemental Table S1, see http://ajp.amjpathol.org).
The common gene list was clustered by using SplineCluster34 and 33 highly homogeneous gene clusters were observed, each comprising genes showing identical kinetics of induction or repression in hormone stimulated cells (Figure 1, A and B). Only two genes (GREB1 and CALB2; Figure 1B) were not clustered with other estrogen-regulated genes because of their very strong response, which largely exceeded that of all other genes. The expression profiles appeared highly similar in the two cell lines (Figure 1A), and the only significant difference observed was represented by a slight delay (2 to 4 hours) in hormonal response to the hormone observed in MCF-7 cells, a difference that has already been described for cyclin genes and reflects a more rapid cell cycle start and progression in ZR-75.1 cells after estrogen stimulation.53
Each identified gene cluster contained genes that respond to estrogen in a coordinated fashion and are therefore most likely to share a common transcriptional and/or posttranscriptional control. Therefore, we searched for common regulatory sequence elements within promoters of genes belonging to each of the kinetic clusters shown in Figure 1A, excluding clusters 1, 10, and 11 because these did not include a sufficient number of genes for this analysis. We defined as promoter region the sequence located between positions −2000 and +800 with respect to the transcription startsite of the gene, considering the upstream-most transcription startsite when multiple transcripts were reported for a given TU. We chose this approach to avoid redundancies that might have attributed a promoter region to a gene that is poorly characterized and thus is not matched to an entry in the promoter databases. In Figure 1C we show the results obtained for transcription factor (TF) binding matrices enriched in one or more clusters and recognized by at least one factor expressed in MCF-7 and ZR-75.1 cells. The V$ER.01, V$ER.02, and V$ER.03 matrices represent different variants of the ERα binding site, ERE. They were not significantly enriched in any gene cluster (Z score <3.0 and overrepresentation value <2.5) and have been reported in the figure for comparison. This first result indicates that genes containing an ERE sequence in their promoter are not abundant in the estrogen-regulated gene set identified. This suggests that either the primary ERα targets represent a small fraction of all hormone regulated genes or, alternatively, that the canonical ERE is not the prevailing docking site for the receptor in target genes promoters and/or that transcriptional regulation by ERE-bound ER complexes occurs predominantly from promoter-distal sites in the genome. On the other hand, genes with binding sites for E2F4/DP-1 and/or RB/E2F1/DP-1 complexes were clearly overrepresented within clusters 12 to 19, which contain a large number of cell cycle-related genes. Because the E2F family of TFs plays a key role in cell cycle progression, this result is consistent with the known functions of the genes comprised in the three enriched clusters and with the mitogenic action of estrogen in BC cells. Interestingly, expression of E2F1, 2, 4, and DP-1 genes is enhanced by estrogen in BC cells, and E2F1 and 2 are primary targets of ERα in MCF-7 cells (see below). The other most overrepresented TF binding matrices were as follows: CHOP.02 (clusters 12 and 16), recognized by the C/EBP homologous CHOP/DDIT3 factor; GAGA.01 (cluster 21), whose binding factor(s) has not been identified yet; GSC.01 (clusters 29 and 33), which binds bicoid-type homeodomain TFs; GTF3R4.01 (cluster 19), which binds GTF2IRD1 whose gene is down-regulated by estrogen (see below); LXRE.01 (clusters 6, 12, 18, and 25) that, together with RAR_RXR.02 (clusters 8, 17, 18, 27, and 33), heterodimerizes with nuclear receptor like RXRα-γ and retinoid, thyroid, vitamin D3, and liver X receptors; MYEF2 and MEF2.07 (cluster 21), recognized by the myocyte-specific enhancer factors MEF2—that repress target gene transcription by binding to class II histone deacetylases; TFII-IR4.01 (cluster 6), binding the general transcription factor TFII-I, which has been shown to mediate recruitment of ERα to target gene promoters by tethering54; and USF1.01 (cluster 16), recognized by the E-box binding factors USF and very similar to binding sites for other members of this TF family such as ATF6, MYC, MAX, and SREBP.
A functional analysis of the entire estrogen-regulated gene set based on gene ontology55 and performed with the Database for Annotation, Visualization, and Integrated Discovery37 revealed significantly enriched biological process terms related to cell proliferation and characterizing all cell cycle phases, from G1 to mitosis, confirming a significant effect of the hormone on cell cycle regulation and cell cycle clock processes. Other biological processes significantly affected by estrogen included responses to stimuli (including stress and phosphoinositide-mediated signaling), DNA recombination and repair, nucleosome and chromatin assembly, cell motility and adhesion, ribosome biogenesis and assembly, intracellular transport, metabolic processes including aromatic compound, amino acid metabolism, and ion homeostasis, and cell differentiation (Supplemental Table S2, see http://ajp.amjpathol.org). The majority of these processes reflect known effects of estrogen on BC cell phenotype and confirm our previous results, obtained with a more limited estrogen-responsive gene dataset from ZR-75.1 cells.28 When the analysis was performed on the individual timed gene clusters, the results provided a more detailed view of the dynamics of all these biological processes after cell exposure to the hormone and in particular during cell cycle progression (Figure 2; Supplemental Table S3, see http://ajp.amjpathol.org). Indeed, all terms associated with DNA replication (regulation of DNA replication, DNA replication initiation, DNA repair, etc) are enriched in clusters 12 to 18, which comprise genes showing maximal response to the hormone (activation) after 16 to 28 hours, the time of cell transition through the S-phase under these conditions in both lines.28,53 Terms associated with M-phase (mitotic spindle organization and biogenesis, sister chromatid segregation, chromosome segregation, etc) are enriched in clusters 19 and 20, where genes show a maximal response 28 to 32 hours into stimulation, coincident with S phase completion and cell division. This result confirms that the gene set obtained reflects timing and nature of known cellular responses to estrogen and is thus a useful tool to extract novel information concerning key cellular and molecular pathways controlled by the hormone via ERα in BC cells.
Figure 2.
Functional significance of the dynamic gene expression changes induced by estrogen in hormone-responsive BC cells. The genes of estrogen-responsive timed cluster were analyzed by using the Database for Annotation, Visualization and Integrated Discovery to identify enriched gene ontology biological process (GO-BP) categories in each cluster. The heatmap shows only clusters and terms above significance threshold (Fisher’s exact test P value <0.01), where gray tonality is proportional to significant P value and white boxes indicate that the GO-BP term did not reach the P value cut-off.
Identification of Primary Estrogen-Responsive Genes by Global Mapping of ERα Binding to MCF-7 Cell Genome in Vivo by ChIP-Seq
ERα is known to control transcription by binding in and around TUs as well as via long range mechanisms in chromatin.56,57 For this reason, we decided to experimentally map all ERα binding sites in MCF-7 genome by ChIP-Seq58,59,60 under the same experimental conditions applied for gene expression profiling. ChIP-Seq analysis was performed after 45 minutes of hormone stimulation because preliminary tests on six genomic ERα binding sites indicated that the ChIP results obtained at this time-point were more pronounced and reproducible than those measured after longer (>3 hours) hormonal treatment times, in line with previous results from MCF-7 cells.61 This is possibly due to receptor cycling and/or epitope masking by associated cofactor protein complexes. Also, this timing allowed us to compare the results obtained in this same cell line by ChIP-on-chip62 and ChIP-PET.63 Of 10 million to 13 million 32-mer sequence tags obtained by sequencing immunoprecipitated DNA, 45% of which mapped uniquely (with a maximum of two mismatches allowed) to the human genome (hg18), whereas 38% did not align. The remaining tags matched either ribosomal and adapter sequences, or showed multiple matches in the genome. These results are comparable with what has been reported in the literature.58,59,60
To identify the actual ERα binding sites, we defined sequence tag enrichment (peaks) in the +E2 versus −E2 and +E2 versus +E2/−Ab samples by using the Site Identification from Short Sequence Reads software,44 and we mapped 3561 specific ERα binding sites (Supplemental Table S4, see http://ajp.amjpathol.org). Interestingly, no constitutive ER sites were found by comparing tag distribution in −E2 versus +E2/−Ab samples, suggesting that ER-chromatin interaction in the absence of hormone might not be stable enough to be detected by this approach or, alternatively, that the peak density in −E2 cells was below the filter set, due to the low concentration of ERα present in cell nuclei in the absence of estrogen. Indeed, the number of binding sites identified in +E2 chromatin is compatible with that of ERα molecules present in the nuclei of estrogen-stimulated MCF-7 cells, measured by enzyme-linked immunosorbent assay and by direct labeling with 3H-E2 (5500 to 7000 molecules cell; data not shown).
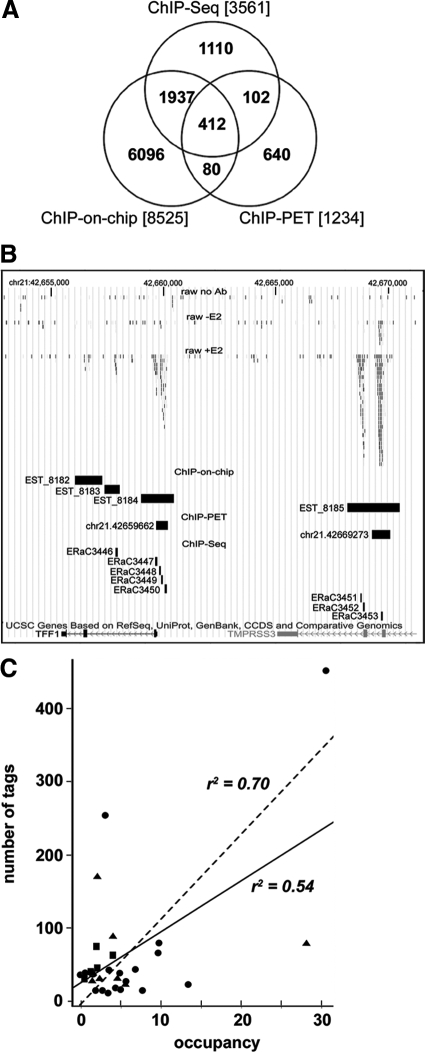
About 70% of the binding sites identified by ChIP-Seq were independently detected also by two different genome-wide approaches (ChIP-on-chip62 and ChIP-PET63; Figure 3A), and paired comparisons showed a stronger overlap with ChIP-on-chip (66%) than with ChIP-PET (46%) data. A similar result was recently reported by Welboren et al,64 despite the fact that a larger number of ER binding sites were mapped in that study. Figure 3B shows a genome browser view of a representative result in the genomic context of the ERα target gene pS2/TFF1, where binding sites were detected by all techniques around the TU. All three approaches revealed multiple binding sites in this locus, but ChIP-Seq shows much better peak resolution, allowing us to distinguish multiple, clearly separated sites where the other techniques detect only one, broader binding region. ERα ChIP followed by real-time quantitative PCR of 30 different sites identified in this study (Figure 3C) confirmed in 29 cases the ChIP-Seq results, demonstrating also a good correlation (solid line, R2 = 0.54) between the number of tags detected by sequencing with the relative strength (percent occupancy) of the binding interaction measured by quantitative PCR. ERE+ binding regions show an even higher correlation (dashed line, R2 = 0.70).
Figure 3.
Global mapping of ERα binding to MCF-7 cells genome by ChIP-Seq. The Venn diagram in A shows a comparison of ERα binding measured under comparable conditions (45 minutes stimulation of estrogen-deprived MCF-7 cell cultures with E2) by ChIP-Seq (this study), ChIP-on-chip,65 and ChIP-PET66; B shows, as an example, the position and extension of the ERα binding sites identified upstream and within the well-characterized ERα-responsive TFF1/pS2 gene in chromosome 21 by the same genome-wide mapping approaches. C shows the correlation between ChIP-Seq signal (number of tags) and quantitative PCR ChIP (occupancy) for 30 binding regions selected for validation. The regions selected contain an ERE (circles), a half-ERE (triangles), or no canonical ERE sequences (squares).
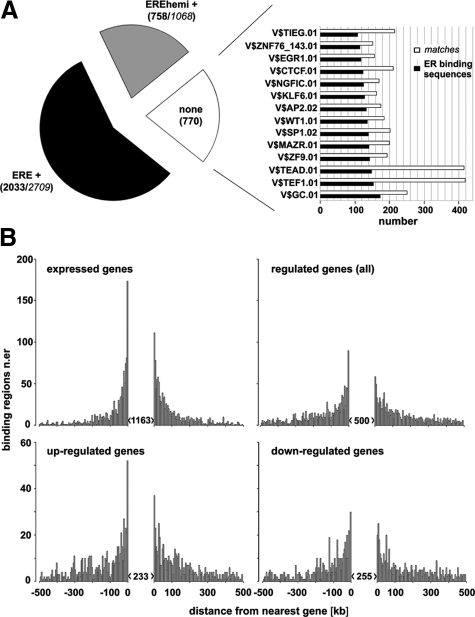
The identified binding sites were classified with respect to the nature of the sequence elements they contain and their distance from expressed and regulated genes (Figure 4). Ligand-activated ERα dimerizes and binds directly the perfectly or imperfectly palindromic ERE sequences (consensus: GGTCAnnnTGACC) and, with lower affinity, to the half-palindromic EREhemi (PuGGTCA), which is generally present in multiple copies in enhancers or proximal to binding sites for other TFs, which in this case are believed to help stabilize ER interaction with DNA.65 Moreover, ERα can bind to chromatin by physically interacting with DNA-bound TFs (tethering), as demonstrated for example for Sp166,67 and the AP-1 complex.68 ERE search was performed with the Genomatix MatInspector software (Figure 4A), whose TF binding matrix library comprises an ERE family (V$EREF) with five individual matrices, where V$ER.01, V$ER.02, and V$ER.03 are slightly different definitions of the canonical ERE sequence. For this search, the binding peaks were extended 5′ and 3′, where necessary, to reach a uniform size (240 bp) and to include also cases where the Site Identification from Short Sequence Reads-generated peak might not exactly overlap the actual receptor binding site and is located, instead, in very close proximity. The matrix similarity threshold was kept slightly lower than the optimized value to relax the stringency for analysis of these experimentally identified binding sites. In this way, 2033 binding regions (57%) were found to contain a canonical ERE, whereas 758 of the remaining binding sites (21% of total) showed one or more EREhemi sequences (Figure 4A). To identify sequence elements mediating ER binding in the remaining 770 sites, we first considered the possibility that these might comprise a novel sequence element recognized directly by the receptor. Direct sequence alignment failed, however, to identify a similar sequence in a significant number of these sites (data not shown), and for this reason we considered the possibility that receptor targeting to these genomic sites might be mediated by tethering to one or more TFs. We thus scanned these binding regions for TF matrix overrepresentation against the genome background. This led to identification of 14 significantly overrepresented TF binding matrices (right panel in Figure 4A). The sequences identified bind members of the Sp-1 and Krueppel-like factor families (V$GC.01, V$KLF6.01, V$SP1.02, and V$TIEG.01), the zinc finger proteins ZNF148, 202, 219, and 281 (V$ZNF9.01), ZNF76 and ZNF143 (V$ZNF76_143.01), the CCCTC-binding factor CTCF (V$CTCF.01), a transcriptional repressor of c-myc,69 the MYC-associated zinc finger protein-related MAZ and PATZ1 (V$MAZR.01), members of the EGR/NGFI and WT (V$EGR1.01, V$NGFIC.01, and V$WT1.01) or AP-2 (V$AP-2.02) families, and TEA domain-containing TEAD factors (V$TEAD.01 and V$TEF.01). The majority of these TFs bind GC-rich sequences and are structurally related to each other, suggesting the possibility that a significant portion of the sequence elements listed above, with the notable exception of V$AP-2.02, might indeed bind Sp-1 factors. On the other hand, binding motifs for forkhead box factors, found enriched within ERα binding regions detected in MCF-7 cells by ChIP-on-chip56 where they have been shown to bind FOXA1, suggesting that this factor can be intimately associated with ERα-mediated transcriptional regulation,70 are only slightly enriched in our ERα ChIP-Seq data, a result that is supported by data from other laboratories.13,64,71
Figure 4.
Sequence and positional analysis of ERα binding regions mapped by ChIP-Seq in estrogen-stimulated MCF-7 cells. A shows the result of TFBS matrix search within all ERα binding regions identified. At first, the binding regions were classified according to the presence of one or more palindromic (ERE+) or half-palindromic (EREhemi±) estrogen response element or of none of the above (none; left diagram). Numbers between parentheses indicate in each case the number of binding regions (in bold) and, where present, the total number of sequence matches they comprise (in italic). Sequences not bearing a recognizable ERE element were then searched for the presence of other TFBSs, and the 14 most abundant are shown in the diagram to the left. B shows the relative position of the ERα binding regions identified with respect to the nearest TU expressed in MCF-7 cells (upper left) or regulated by estrogen (upper right), these last subdivided in up-regulated (lower left) and down-regulated (lower right). The number of binding site located up-stream (negative distance), within (numbers in brackets), or down-stream (positive distance) of TUs are shown, for a distance up to 500 kbp, grouped in bins of 5 kbp each.
To establish functional correlations between estrogen-regulated gene expression profiles (Figure 1, A and B) and ERα binding data, we determined the distance between all binding regions identified by ChIP-Seq and the nearest mRNA-encoding genes found regulated by E2 under the same conditions (Figure 4B). This analysis revealed that 40% receptor binding sites map at >10 kb from known genes and a similar fraction is located inside TUs (mainly in introns; upper left panel in Figure 4B). The distance in the genome between each binding site and the nearest estrogen-regulated gene was then calculated. This showed a clear bias for regulated genes, as in these cases the majority of the binding sites is found inside (14%) or ≤100 kb upstream or downstream of the gene (31%, among which 7% within 10 kb) with a Gaussian distribution centered on the TU. Furthermore, a significant fraction (14%) of ERα binding sites are located at significant distances (>100 kb) from any known mRNA genes, suggesting that some of these sites could be proximal to still unknown or noncoding RNA genes and/or that the recently described long-range effects of ERα in the genome57,72 might occur more frequently than previously considered.
To link ERα binding to its transcriptional effects on estrogen-responsive genes, we assigned any given receptor binding site to the nearest regulated TU only if the distance between the two was ≤10 kb, considering instead “intergenic” the sites located ≥100 kb away from any expressed gene. In this way, we identified 218 genes associated to one or more ERα binding sites (Supplemental Table S5, see http://ajp.amjpathol.org), which we termed “primary” to indicate that they are most likely to respond directly to the signal conveyed by E2-activated receptor. This definition is based on the well demonstrated assumption that changes in transcription rate of a primary gene are mediated by direct physical interaction of the receptor to the corresponding chromosomal locus.
To verify the possibility that intergenic receptor binding sites might represent sequence features distinctive from those associated with regulated genes and that differences might exist between up- and down-regulated genes, TF matrix overrepresentation analysis was performed separately in all three classes of binding sites. ERE motifs were clearly enriched in all binding sites categories, including the intergenic ones where no other TFBS was found overrepresented more than fivefold. This is less pronounced in sites associated to genes showing down-regulation by estrogen (Supplemental Figure 1A, see http://ajp.amjpathol.org), suggesting that these last are more often ERE-negative. By overrepresentation analysis, several binding matrices could be identified in ERE− binding regions (Supplemental Figure 1B, see http://ajp.amjpathol.org), including in particular V$CTCF.01, V$EGR.01, 0.02, and 0.03; V$SP1.01 and 0.02; V$ZF5.01; V$E2F.01; binding E2F-factor family members; V$CDE.01, a cell cycle-dependent element recognized by CDE-1; and V$NRF1.01, binding the bZIP transcription factor NRF1 (nuclear respiratory factor 1), which acts on nuclear genes encoding mitochondrial proteins, is estrogen-regulated, and has been proposed to mediate estrogen stimulation of mitochondrial biogenesis73 and to correlate with BC progression.74
Identification of an Estrogen-Responsive Gene Regulation Cascade Triggered by ERα Binding to the Genome
The response kinetics to estrogen of the 218 primary estrogen-responsive genes are reported in Figure 5, A and B, where individual expression profiles are shown to cluster in 10 homogeneous groups. Interestingly, when compared with the whole estrogen-responsive transcriptome clustering (Figure 1, A and B), these data show that most of these genes respond readily to E2 with changes in mRNA level often detectable within 2 to 4 hours (Figure 5B). TFBS overrepresentation analysis was performed for all clusters except the smaller 4, 5, and 7 on the respective gene-associated ER binding sites. As shown in Figure 5C, ERE matrices are overrepresented in all cases ≥10-fold above the genome background, except for cluster 8 where the score was, however, only slightly below this value. All remaining matrices were essentially the same identified in previous analyses (Figures 1C and 4A), including in particular those for E2F and AP-2, CTCF, EGR1, NRF1, SP1, and ZF5 factors.
Figure 5.
Identification of primary estrogen target genes in MCF-7 cells by timed expression profiling and ChIP-Seq. A shows the heatmap of the expression values relative to the estrogen-regulated genes from Figure 1 showing at least one ERα binding site inside or within 10 kb of the transcription unit. Black triangles (top) indicate the time of cell exposure to the hormone (left to right: from 1 to 32 hours). Genes also selected by the TSNI algorithm for identification of primary response genes in gene networks are highlighted in red. The expression profiles relative to these primary genes underwent cluster analysis, generating 10 gene kinetic clusters (vertical bars), whose profiles are depicted in B. C shows the results of TFBS matrix enrichment (overrepresentation) analysis performed on ERα binding regions mapped by ChIP-Seq within or ±10 kb from genes belonging to each cluster shown in Figure 1, A and B. Z score cut-off was 3.0 and only TFBSs showing an overrepresentation score ≥10 in at least one of the gene clusters are reported; gray cells indicate a Z score <3.0 and/or an overrepresentation score <10. Only clusters where at least one matrix scored above these thresholds are shown.
Early responding genes were evaluated also by means of the network reconstruction algorithm TSNI.49 Merging MCF-7 and ZR-75.1 gene lists, TSNI identified 179 gene entries (P ≤ 0.005) common to both cell lines (Supplemental Table S6, see http://ajp.amjpathol.org). Of these, 59 were present in the 218-gene ChIP-Seq list (ie, did show evidence of proximal ERα binding sites; highlighted in red in Figure 5A). This may be due also to the fact that the majority of our ER-binding TUs excluded by TSNI, including the well known primary hormonal targets pS2/TFF1 and GREB2, showed a relative low change in activity at the earliest time points and were thus missed by TSNI analysis. Considering, instead, the early responder gene list identified by TSNI that do not appear to have a proximal ER binding site, these are likely to include genes regulated by the hormone via nongenomic signaling pathways, which have been shown to affect gene expression in BC cells without involvement of nuclear ERs.14 In addition, some of them are, instead, regulated via long-range chromosomal effects of ER, including, for example, the well known primary-response genes MYC and FOS, where the receptor binds at 50 kb from the TU (Hurtado et al62 and our results).
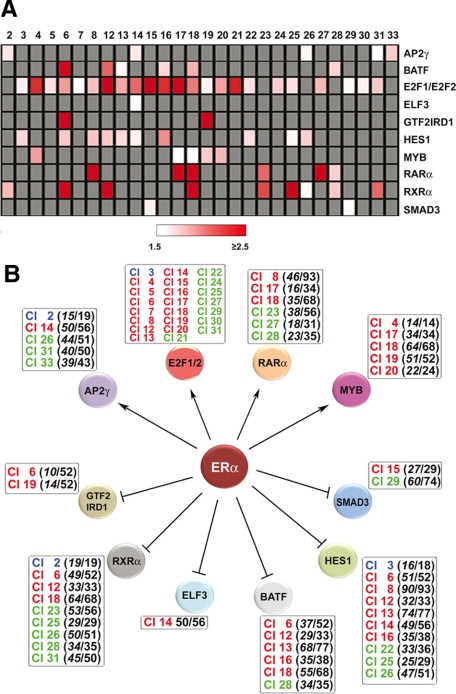
From the primary gene list reported in Figure 5, an estrogen-regulated gene network was built, stemming from ERα and reaching a large number of downstream genes via TFs encoded by primary genes. This was based on the rationale, derived by a large body of experimental observations, which the transcriptional response of a TF factor gene to a signal most often reflects a regulatory role of the stimulus on the activity of that TF in the cell. For this analysis we used Bibliosphere75 to identify among the primary genes binding ERα those encoding TFs associated to binding matrices present in available databases. This lead to 16 TF genes, including six up-regulated by estrogen (E2F1, E2F2, TGIF2, and RARα in cluster 3; TFAP2C/AP-2γ in cluster 4; and MYB in cluster 5) and 10 down-regulated (JARID1B, CREB3L4, and ARID5B in cluster 6; E74-like factor 3 (ELF3) and SMAD3 in cluster 8; B-cell activating transcription factor (BATF) and hairy and enhancer of split homologue-1 (HES1) in cluster 9; and GTF2IRD1, RXRα, and SOX13 in cluster 10). All promoter sequences (−2000 to +800) of the estrogen-regulated genes belonging to the estrogen-responsive gene clusters shown in Figure 1 were scanned by using the matrices associated to these transcription factors. A filter was applied to the TFBS overrepresentation analysis to exclude all cases with an associated Z score <3 and <1.5 overrepresentation in the cluster, and the results are reported in Figure 6A. E2F binding sites were again found to be overrepresented in promoter regions in the majority of the clusters, including in particular clusters 12 and 15 to 18, which, as already mentioned, comprise a significant number of G1- and S-phase genes (compare data in Figures 2 and 6A). In line with this result, the binding motif for GTF2IRD1, a protein that interacts in vitro and in vivo with pRb1 (retinoblastoma protein 176) and, like E2Fs, is under control of this oncosuppressor protein, is highly overrepresented in the cell cycle gene cluster 19. Marked overrepresentation values were found for binding sites of BATF (cluster 6), a basic leucine-zipper protein that belongs to the AP-1/ATF superfamily of TFs and dimerizes with JUN proteins to act as AP-1 complex inhibitor, and the nuclear receptors RARα (clusters 8, 17, 18, and 27) and RXRα (clusters 6, 12, 18, and 25), whereas those for AP-2γ, HES1, MYB, and SMAD3 are only moderately enriched in several clusters.
Figure 6.
An ERα-dependent gene regulation network in estrogen-stimulated breast cancer cells. In A the results of TFBS matrix enrichment (overrepresentation) analysis of the promoter region (−2000/+800 from the transcription startsite) of genes belonging to each estrogen-responsive gene cluster from Figure 1, A and B, are reported. The analysis was limited in this case only to TFs encoded by primary estrogen target genes from Figure 5, A and B. Z score cut-off was 3.0 and TFBSs showing an overrepresentation score ≥1.5 in at least one of the gene clusters are reported. Only clusters where at least one matrix scored above these thresholds are shown, and gray cells indicate a Z score <3.0. B shows the gene regulation network drawn by combining data relative to the estrogen-responsive transcriptome, TFBS matrix overrepresentation analysis of estrogen-responsive gene promoters, and ERα binding data in MCF-7 cells. Expression of the genes encoding transcription factors AP-2γ, E2F1 and 2, MYB, and RARα is enhanced by estrogen, whereas that of BATF, ELF3, GTF2IRD1, HES1, SMAD3, and RXRα is inhibited. The gene clusters where TFBS matrices associated with these TFs are enriched are listed in the boxes in red when up-regulated by estrogen, green when down-regulated, and blue when biphasic. For E2F1 and E2F2 TFs, the number of target genes/clusters were omitted from the graphic outlay of the network for clarity.
The matrix overrepresentation results obtained are represented in Figure 6B combined in a schematic model depicting an estrogen-responsive transcriptional regulation cascade in luminal-like BC cells, which stems from ERα and reaches about half the estrogen-regulated genes identified in this study (Figure 1) through 11 TFs encoded by primary response genes. In Supplemental Table S7 (see http://ajp.amjpathol.org), the list of genes carrying primary-response TF binding sites in their promoters is reported for each time cluster.
miRNA Involvement in Estrogen Control of Target Gene Expression in BC Cells
Considering the possibility that miRNAs77 might play a role in estrogen control of gene activity in BC cells, we performed a miRNA target enrichment analysis by identifying potential miRNA homing sites (short nucleotide stretches complementary to miRNA “seed” sequences) within philogenetically conserved sequences at the 3′ untranscribed regions (UTRs) of all estrogen-regulated transcripts identified in this study. We then searched for statistically significant enrichment of these miRNA target transcripts in each estrogen-responsive gene cluster from Figure 1, against a background represented by all expressed mRNAs, focusing only on miRNAs expressed in MCF-7 cells and using as a source of potential miRNA target transcripts those from TargetScanS 5.0, identified by miRNA family-mRNA target pair predictions.42 Enrichment analysis was performed for all clusters comprising more than 10 genes, considering as background all detectable (expressed) transcripts matching an entry in TargetScanS. The results are shown in Figure 7A, where on the x axis are reported the gene clusters showing at least 15% of the transcripts pairing to a given miRNA family (P ≤ 0.01), and on the y axis the corresponding miRNAs are reported (grouped for family). For comparison, the expression levels (log2 of the fluorescence intensity) of these miRNA in hormone-stimulated cells are reported in Figure 7B.
Figure 7.
Analysis of miRNAs involvement in estrogen regulation of target gene expression. In A the results of a search for potential miRNA target transcripts within the estrogen-regulated gene clusters from Figure 1, A and B, starting from miRNAs expressed at detectable levels in MCF-7 cells are summarized. The gene clusters shown comprise a significant number (≥15%; P value ≤0.01) of genes encoding an mRNA predicted by TargetScanS 5.0 as target of the indicated miRNA family. Statistical analysis (Fisher’s exact test) was performed considering as background all transcripts detectable (expressed) in MCF-7 cells. Only clusters where at least one miRNA family scored above the numerical and statistical thresholds are shown. The heatmap in B summarizes the relative level of expression in MCF-7 cells (log2 of fluorescence intensity) of individual members of the miRNA families shown in A; only expressed miRNAs are displayed. In C the early (6 to 24 hours) and late (3 days) effects of estrogen stimulation on the cellular level of the same miRNAs are shown, expressed as fold-change respect to the values measured in hormone-starved cells. In A and B, estrogen-regulated miRNAs are reported in red or green, according to their response to the hormone.
For the majority of miRNA families identified (13 of 16) potential mRNA targets were significantly enriched in only one gene cluster, whereas 5 of 9 clusters comprise a significant number of targets for more than one miRNA. Furthermore, in many instances a transcript showed multiple target sequences for the matched miRNA (10 for each miR-506 putative targets in cluster 22, for example) and in some cases target sequences for more than one miRNA family are found in the same transcript, like for example in FOS (miR-101 and -221/222), PHLDA1 (miR-101, -194, and -324-3p), RARA (miR-194 and -513a-5p), SLC25A25 (miR-513a-5p and -618), THBS1 (miR-101, -194, -221/222, and -618), and TLN2 (miR-194 and -324-3p) mRNAs in cluster 4 (Supplemental Table S8, see http://ajp.amjpathol.org).
To test the possibility that the intracellular concentration of some of these miRNAs might be regulated by E2, hormone-deprived MCF-7 cells were stimulated with E2, and miRNA expression profiling was performed after 6, 12, 24, or 72 hours of treatment (Figure 7C). Results showed progressive accumulation of miR-760 and -424 and decrease of miR-618, -570, and -107 in hormone-stimulated cells. This result was confirmed by opposite changes in the expression of the same miRNAs in MCF-7 cells maintained in complete medium after estrogen-deprivation (data not shown). This test showed also a less significant but reproducible effect of hormone deprivation on miR-101, -30e, and -340 levels, which were down-regulated by hormone removal from the medium, and miR-103, -125b, -222, -30d, and -513a-5p, which accumulated instead on estrogen withdrawal. Because treatment of the serum with dextran-coated charcoal eliminates not only estrogen but also many other compounds, these differences in miRNA expression cannot be ascribed directly to estrogen deprivation and for this reason were not considered further.
Taken together, the results reported in Figure 7 demonstrate the involvement of miRNAs in estrogen control of target gene activity in luminal-like BC cells. Furthermore, this demonstrates that these steroid hormones can modulate the intracellular concentration of specific miRNAs, which could effect the posttranscriptional activity of a larger number of genes than might be expected from their ability to regulate gene transcription directly, which is in agreement with what has been reported previously in this cell line.21,23,27,78 Interestingly, none of the genes encoding the E2-regulated miRNAs identified in this analysis showed a proximal ERα binding site, suggesting that regulation of these genes might occur via distal ERα binding to the genome, the nongenomic pathway of estrogen action, or posttranscriptional mechanisms acting at the level of primary transcript (pri-miR) processing.79
Discussion
The clinical management of hormone-responsive BC, as well as development of new drugs, needs accurate comprehension of the estrogen-regulated gene network active in tumor cells. In this study we obtained a comprehensive genome-wide identification of the entire E2-responsive transcriptome by using two BC lines. This was complemented by molecular characterization of regulated genes in terms of kinetics of response to the stimulus, role of ERα and other TFs in transcriptional control of gene activity, and potential involvement of miRNAs in posttranscriptional regulation events controlled by the hormone. Results yielded a detailed kinetic representation of the gene expression changes induced by estrogen through the identification of 33 homogenous gene clusters each characterized by well defined timing, intensity, and type of response to hormonal stimulation. On the basis of the well demonstrated postulate that genes behaving similarly are generally under control of the same regulatory mechanism, we exploited these clusters to search for common binding signatures for TFs in promoters and target sequences for miRNA in the transcribed regions. We could observe in this way that individual estrogen-responsive clusters are indeed enriched in TUs comprising such sequence elements in their regulatory regions. Through global mapping of ERα binding sites across the genome, we identified a large number of primary genes that appear to be directly targeted by the estrogen-receptor complex. By integrating the data obtained experimentally with computational analyses of TF binding elements in promoters, we identified a gene regulation network stemming from ERα binding to 11 genes encoding TFs and reaching a large proportion of the estrogen regulated TUs identified. To our knowledge, this result represents a significant advancement relative to other published work because in this way we provide a comprehensive view of primary, secondary, and downstream genes involved in this signaling pathway, in particular the primary estrogen-responsive TF genes that are central nodes in the network shown in Figure 6.
Transcription Factors Downstream of ERα in Estrogen Signaling
AP-2α and AP-2γ are the principal epithelial isoforms of the AP-2 TF family and are involved in estrogenic trans-repression of ERBB.80 Although the action of these TFs can be opposite in different nonepithelial cell lines, in ER-positive breast cells recent work indicated that AP-2γ, opposite to AP-2α, regulates mainly genes involved in cell cycle progression and developmental signaling, actually promoting cell proliferation.81 Because estrogen induces transcription of AP-2γ while down-regulating AP-2α, the ratio of these TFs could be an important part of estrogen control of the growth and cell differentiation status of hormone-responsive tumors.
E2F1 and E2F2, which show functional redundancies with each other but not with other members of this TF family, interact only with RB1 and, together with E2F3a, constitute one subfamily often referred as “activator E2Fs” because they were first proposed to function mainly in activating gene expression, although this has not yet been fully confirmed.82 In estrogen-responsive BC cell lines, E2F factors promote G1-S transition as a consequence of cyclin D1/cdk-4/6 activation by the hormone,53,83 whereas E2F1 overexpression causes hormone-independent proliferation and antiestrogen-resistance.84 Interesting results concerning E2F involvement in the mitogenic response of BC cells to estrogen came from a study in which expression profiles of E2-stimulated MCF-7 and MDA-MB-231ER+ (MDA) cells were compared.85 These authors observed that a set of cell cycle-related genes showing E2F binding site overrepresentation was activated in MCF-7 and inhibited in MDA cells and that RNA interference-mediated knockdown of E2F1 gene prevented hormonal regulation of these E2F-responsive genes and cell proliferation in MCF-7 cells.
The MYB gene is expressed in a high proportion of human breast tumors, and its expression correlates strongly with ER positivity.86 Proliferation of ER-positive, but not ER-negative, BC cell lines was inhibited when MYB was suppressed by antisense oligonucleotides or RNA interference,87 confirming that this gene product is an effector of ERα-mediated estrogen signaling in hormone-responsive BC cells.
The retinoic acid receptor subtype RARα is highly expressed in ER-positive BC cells, which are normally responsive to retinoids, whereas less differentiated, ER-negative cells are characterized by low RARα expression and resistance to these compounds. RARα protein has been shown to accumulate in BC cells after ER-mediated trans-activation of the RARA-1 gene promoter,88 and impairment of ERα DNA binding in vivo by expression of constitutively active AKT interferes with E2F- and retinoid-dependent pathways in MCF-7 cells.89 Interestingly, it was recently reported that RAR binding is highly coincident with ERα binding throughout the genome, resulting in a widespread cross talk of retinoid and estrogen signaling to antagonistically regulate cancer-associated genes.90
Concerning RXRα, a direct confirmation of the result reported here for the transcript encoding this nuclear receptor has been reported also in another study, where it was observed that RXRs, assessed by their DNA binding activity, are significantly reduced in nuclear extracts from MCF-7 and T47D cells after estrogen treatment.91 The biological significance of RXRα down-regulation by E2 in BC cells might be related to the anti-apoptotic actions of estrogen in this cell type.28
BATF is a nuclear basic leucine zipper protein of the AP-1/ATF superfamily of TFs, expressed most highly in hematopoietic tissues but also in many cancers, including BC. The leucine zipper of this protein mediates dimerization with members of the Jun family proteins, and BATF is thought to be a negative regulator of AP-1/ATF transcriptional events.92 Down-regulation of this gene expression is thus likely to contribute to potentiation of AP-1 activity in estrogen-stimulated BC cells, where this TF complex is known to mediate some of the effects of these hormones on gene regulation.16,68
The ELF3 is an ETS-domain transcriptional activator that binds ETS sequences containing the consensus nucleotide core sequence GGA[AT] and is expressed at higher levels in BC than normal mammary epithelia. ELF3 plays an important role in epithelial cell differentiation and tumorigenesis because its overexpression induces neoplastic transformation of mammary cells. The gene encoding this protein has been found up-regulated by estrogen during growth-inhibition of MDA-MB-231ER+ cells93 while being trans-repressed by the hormone in MCF-7 cells, a response prevented by AKT-induced inhibition of ERα DNA binding.89 This result is in line with what we observed for RARα, E2F1, and two genes in the same experimental model, providing further confirmation of a direct regulation of these TF genes by ERα binding to their promoters.
The protein encoded by the HES1 gene belongs to the basic helix-loop-helix family of transcription factors and acts as a repressor of genes that require a bHLH protein for their transcription. HES1 protein levels were found down-regulated by E2 in T47D and MCF-7 cells, where induction of the corresponding gene by all-trans retinoic acid or overexpression of HES1 protein by transfection of an expression vector, prevent the mitogenic effects of E2 and up-regulation of PCNA and E2F1 genes.94
GTF2IRD1 is a member of the transcription factor II-I family that contains five GTF2I-like repeats, each with a potential helix-loop-helix motif. It has been suggested that this TF may interact with other helix-loop-helix-proteins and function as transcriptional modulator, under the control of RB1.76 Interestingly, in MCF-7 and ZR-75.1 cells the pRb1 protein is phosphorylated by cyclin-dependent kinases in response to estrogen,53,83 suggesting a functional link between GTF2IRD1, RB1, and cell cycle regulation in BC cells.
SMAD3 belongs to a family of proteins, similar to the gene products of the Drosophila melanogaster gene “mothers against decapentaplegic” (Mad) and the Caenorhabditis elegans gene Sma, which act as signal transducers and transcriptional modulators downstream of multiple signal transduction cascades, including in particular the activin and transforming growth factor β pathways. Activin treatment, which enhances SMAD3 activity, has been shown to inhibit proliferation of MCF-7 but not ER-negative MDA-MB 231 cells.95 E2 treatment results in reduced phosphorylation of SMAD2 and 3 proteins, diminished SMAD2/3 gene reporter activity, and inhibition of cell migration in response to transforming growth factor β.96 Interestingly, SMAD proteins have been proposed to contribute to BC bone metastasis via transforming growth factor β and interleukin-11, suggesting that ER-mediated down-regulation of SMAD3 gene may affect this aggressive clinical phenotype in luminal-like, ER-positive breast tumors.
MicroRNA Circuitry in Estrogen-Stimulated Cells
Estrogens have long been known to affect stability of mRNAs by posttranscriptional mechanisms, and this has been postulated to impact the expression of a large population of genes in cells involved in reproductive functions.97,98 Among the cellular molecules able to influence mRNA concentration and activity by posttranscriptional mechanisms, miRNAs have emerged as key players. MicroRNAs are predicted to control the activity of approximately 30% of all protein-coding genes and have been shown to participate in the regulation of almost every cellular process investigated so far, both in normal and cancer tissues.99 We and others21,22,23,27,78,79,100 have recently observed that the intracellular concentration of certain miRNAs can be modulated by E2 via ERα in MCF-7 cells. Hence, the possibility that mRNA changes detected over time in E2-stimulated cells might also reflect the action and/or modulation of miRNAs was considered. A search for miRNA binding signatures within the 3′-UTR of mRNAs belonging to estrogen-responsive time clusters indicates significant enrichment for targets of 29 miRNAs expressed at detectable levels in MCF-7 cells, among which miRNA-424, -760 and, possibly, -101 were found up-regulated and miR-107, 570, and -618 down-regulated by estrogen treatment (Figure 7). These results, demonstrating an effect of estrogen on the intracellular levels of miRNA, suggest how these small RNAs may play a role in modulating the final gene responses to these hormones in BC cells. This could include a static control of the concentration and/or activity of hormone-regulated mRNAs by constitutively expressed miRNAs, finalized to fine-tuning gene regulation by estrogen, preventing for example excessive accumulation of specific gene products, as well as a dynamic posttranscriptional regulation of expressed mRNA levels and translation efficiency exerted by E-regulated miRNAs. The latter regulatory event could occur, for instance, during critical cell cycle phase transitions. These hypotheses can now be effectively addressed by evaluating the effects of miRNA antagonists and mimics on estrogen responsive genes and cellular processes, such as cell cycle progression, to define the actual relevance of these small RNAs in estrogen signaling and in the establishment and maintenance of the hormone-responsive BC phenotype.
Acknowledgments
We are grateful to Drs. Diego di Bernardo and Alessandra Vigilante for help with TSNI analysis, Michele Caselle and Davide Corà for miRNA target transcript searches, and Rosario Casale for technical assistance.
Footnotes
Address reprint requests to Alessandro Weisz, M.D., Dipartimento di Patologia generale, Seconda Università degli Studi di Napoli, Vico L. De Crecchio, 7, 80138 Napoli, Italy. E-mail: alessandro.weisz@unina2.it.
Supported by UE (CRESCENDO IP, contract n.er LSHM-CT2005-018652); Italian Association for Cancer Research, (grant IG-8586) Region Campania (L. 5 and POR); Ministry of Education, University and Research (grant 2008 CJ4 SYW_004, to A.W.); DISSPAPA (to M.L.C.); Region Piedmont (PRF Health 2006, 2007, and Applied Science 2004); Ministry of Health (PRF 2005, to M.D.B.); and the Ph.D. programs: Pathology of Cell Signal Transduction (O.P., O.M.V.G., and R.T.), Computational Biology (A.T.) of the Second University of Naples, and Toxicology, Oncology, and Molecular Pathology of the University of Cagliari (M.R.). M.M. is a recipient of a postdoctoral fellowship of the Second University of Naples, and O.P. is a recipient of a Doctoral Fellowship from the AIRC Naples Oncogenomics Center.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Wang E, Lenferink A, O'Connor-McCourt M. Cancer systems biology: exploring cancer-associated genes on cellular networks. Cell Mol Life Sci. 2007;64:1752–1762. doi: 10.1007/s00018-007-7054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlebach G, Shamir R. Modelling and analysis of gene regulatory networks. Nat Rev Mol Cell Biol. 2008;9:770–780. doi: 10.1038/nrm2503. [DOI] [PubMed] [Google Scholar]
- Conzen SD. Minireview: nuclear receptors and breast cancer. Mol Endocrinol. 2008;22:2215–2228. doi: 10.1210/me.2007-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC. A century of deciphering the control mechanisms of sex steroid action in breast and prostate cancer: the origins of targeted therapy and chemoprevention. Cancer Res. 2009;69:1243–1254. doi: 10.1158/0008-5472.CAN-09-0029. [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Ström A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Ordóñez-Morán P, Muñoz A. Nuclear receptors: genomic and non-genomic effects converge. Cell Cycle. 2009;8:1675–1680. doi: 10.4161/cc.8.11.8579. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Yamaguchi Y. Estrogen signaling pathway and hormonal therapy. Breast Cancer. 2008;15:256–261. doi: 10.1007/s12282-008-0070-z. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang A, Liu MC, Zwart A, Lee RY, Gallagher A, Wang Y, Miller WR, Dixon JM, Clarke R. Estrogen receptor alpha positive breast tumors and breast cancer cell lines share similarities in their transcriptome data structures. Int J Oncol. 2006;29:1581–1589. [PubMed] [Google Scholar]
- Weisz A, Basile W, Scafoglio C, Altucci L, Bresciani F, Facchiano A, Sismondi P, Cicatiello L, De Bortoli M. Molecular identification of ERalpha-positive breast cancer cells by the expression profile of an intrinsic set of estrogen regulated genes. J Cell Physiol. 2004;200:440–450. doi: 10.1002/jcp.20039. [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Sergio CM, Loi S, Inman CK, Anderson LR, Alles MC, Pinese M, Caldon CE, Schütte J, Gardiner-Garden M, Ormandy CJ, McArthur G, Butt AJ, Sutherland RL. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS ONE. 2008;3:e2987. doi: 10.1371/journal.pone.0002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasor J, Chang EC, Komm B, Lin CY, Vega VB, Liu ET, Miller LD, Smeds J, Bergh J, Katzenellenbogen BS. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–7340. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- Miller LD, Liu ET. Expression genomics in breast cancer research: microarrays at the crossroads of biology and medicine. Breast Cancer Res. 2007;9:206. doi: 10.1186/bcr1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdeau V, Deschênes J, Laperrière D, Aid M, White JH, Mader S. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2008;36:76–93. doi: 10.1093/nar/gkm945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madak-Erdogan Z, Kieser KJ, Kim SH, Komm B, Katzenellenbogen JA, Katzenellenbogen BS. Nuclear and extranuclear pathway inputs in the regulation of global gene expression by estrogen receptors. Mol Endocrinol. 2008;22:2116–2127. doi: 10.1210/me.2008-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner SA, Steffen DL, Hilsenbeck SG, Chen ES, Watkins C, McKenna NJ. GEMS (Gene Expression MetaSignatures), a Web resource for querying meta-analysis of expression microarray datasets: 17beta-estradiol in MCF-7 cells. Cancer Res. 2009;69:23–26. doi: 10.1158/0008-5472.CAN-08-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, Cancemi M, Caporali S, Caristi S, Scafoglio C, Teti D, Bresciani F, Perillo B, Weisz A. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24:7260–7432. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafoglio C, Ambrosino C, Cicatiello L, Altucci L, Ardovino M, Bontempo P, Medici N, Molinari AM, Nebbioso A, Facchiano A, Calogero RA, Elkon R, Menini N, Ponzone R, Biglia N, Sismondi P, De Bortoli M, Weisz A. Comparative gene expression profiling reveals partially overlapping but distinct genomic actions of different antiestrogens in human breast cancer cells. J Cell Biochem. 2006;98:1163–1184. doi: 10.1002/jcb.20820. [DOI] [PubMed] [Google Scholar]
- Mutarelli M, Cicatiello L, Ferraro L, Grober OMV, Ravo M, Facchiano AM, Angelini C, Weisz A. Time-course analysis of genome-wide gene expression data from hormone-responsive human breast cancer cells. BMC Bioinformatics. 2008;9(Suppl 2):12. doi: 10.1186/1471-2105-9-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Kraus WL. A global view of transcriptional regulation by nuclear receptors: gene expression, factor localization, and DNA sequence analysis. Nucl Recept Signal. 2008;6:e005. doi: 10.1621/nrs.06005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro L, Cicatiello L, Mutarelli M, Ravo M, Grober OMV, Sacconi G, Weisz A. MicroRNA expression profiling of hormone-responsive human breast cancer cells lines. Am J Pathol. 2008;173(Suppl):S16. [Google Scholar]
- Song G, Wang L. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res. 2008;36:5727–5735. doi: 10.1093/nar/gkn567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, Brown M, Hammond S, Srour EF, Liu Y, Nakshatri H. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchis ML, Ballarino M, Salvatori B, Puzzolo MC, Bozzoni I, Fatica A. A new molecular network comprising PU. 1, interferon regulatory factor proteins and miR-342 stimulates ATRA-mediated granulocytic differentiation of acute promyelocytic leukemia cells. Leukemia. 2009;23:856–862. doi: 10.1038/leu.2008.372. [DOI] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainer J, Ploner C, Jesacher S, Ploner A, Eduardoff M, Mansha M, Wasim M, Panzer-Grümayer R, Trajanoski Z, Niederegger H, Kofler R. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia. 2009;23:746–752. doi: 10.1038/leu.2008.370. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicatiello L, Scafoglio C, Altucci L, Cancemi M, Natoli G, Facchiano A, Iazzetti G, Calogero R, Biglia N, De Bortoli M, Sfiligoi C, Sismondi P, Bresciani F, Weisz A. A genomic view of estrogen actions in human breast cancer cells by expression profiling of the hormone-responsive transcriptome. J Mol Endocrinol. 2004;32:719–775. doi: 10.1677/jme.0.0320719. [DOI] [PubMed] [Google Scholar]
- Ravo M, Mutarelli M, Ferraro L, Grober OM, Paris O, Tarallo R, Vigilante A, Cimino D, De Bortoli M, Nola E, Cicatiello L, Weisz A. Quantitative expression profiling of highly degraded RNA from formalin-fixed, paraffin-embedded breast tumor biopsies by oligonucleotide microarrays. Lab Invest. 2008;88:430–440. doi: 10.1038/labinvest.2008.11. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Angelini C, De Canditiis D, Mutarelli M, Pensky M. A Bayesian approach to estimation and testing in time-course microarray experiments. Stat Appl Genet Mol Biol. 2007;6:24. doi: 10.2202/1544-6115.1299. [DOI] [PubMed] [Google Scholar]
- Angelini C, Cutillo L, De Canditiis D, Mutarelli M, Pensky M. BATS: a Bayesian user-friendly software for analyzing time series microarray experiments. BMC Bioinformatics. 2008;9:415. doi: 10.1186/1471-2105-9-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, Kober KM, Miller W, Pedersen JS, Pohl A, Raney BJ, Rhead B, Rosenbloom KR, Smith KE, Stanke M, Thakkapallayil A, Trumbower H, Wang T, Zweig AS, Haussler D, Kent WJ. The UCSC genome browser database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard NA, Holmes CC, Stephen DA. A quantitative study of gene regulation involved in the immune response of Anopheline mosquitoes: an application of Bayesian hierarchical clustering of curves. J Am Stat Soc. 2006;101:18–29. [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14:2290–2296. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E, Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Weigelt K, Moehle C, Stempfl T, Weber B, Langmann T. An integrated workflow for analysis of ChIP-chip data. Biotechniques. 2008;45:131–132. doi: 10.2144/000112819. [DOI] [PubMed] [Google Scholar]
- Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. OPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2080. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M, di Bernardo D. Inference of gene networks from temporal gene expression profiles. IET Syst Biol. 2007;1:306–312. doi: 10.1049/iet-syb:20060079. [DOI] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249–289. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Mecham BH, Kohane IS, Szallasi Z. Redefinition of Affymetrix probe sets by sequence overlap with cDNA microarray probes reduces cross-platform inconsistencies in cancer-associated gene expression measurements. BMC Bioinformatics. 2005;6:107. doi: 10.1186/1471-2105-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Wang F, Tu K, Xie L, Li YY, Li YX. Transcript-level annotation of Affymetrix probesets improves the interpretation of gene expression data. BMC Bioinformatics. 2007;8:194. doi: 10.1186/1471-2105-8-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicatiello L, Addeo R, Altucci L, Belsito Petrizzi V, Boccia V, Cancemi M, Germano D, Pacilio C, Salzano S, Bresciani F, Weisz A. The antiestrogen ICI 182,780 inhibits proliferation of human breast cancer cells by interfering with multiple, sequential estrogen-regulated processes required for cell cycle completion. Mol Cell Endocrinol. 2000;165:199–209. doi: 10.1016/s0303-7207(00)00243-4. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Azuma M, Tsuboi Y, Kabe Y, Yamaguchi Y, Wada T, Watanabe H, Handa H. TFII-I down-regulates a subset of estrogen-responsive genes through its interaction with an initiator element and estrogen receptor alpha. Genes Cells. 2006;11:373–381. doi: 10.1111/j.1365-2443.2006.00952.x. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology; the gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008;283:32977–32988. doi: 10.1074/jbc.M802024200. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, Davuluri RV, Huang TH. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, Jiang J, Howat WJ, Ali S, Carroll JS. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456:663–666. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei CL, Liu ET. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Kim KJ. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. Mol Endocrinol. 2008;41:263–275. doi: 10.1677/JME-08-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyhlidal C, Samudio I, Kladde MP, Safe S. Transcriptional activation of transforming growth factor alpha by estradiol: requirement for both a GC-rich site and an estrogen response element half-site. J Mol Endocrinol. 2000;24:329–338. doi: 10.1677/jme.0.0240329. [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Valentine C, Lopez GN, Kwok GR, McInerney E, Katzenellenbogen BS, Enmark E, Gustafsson JA, Nilsson S, Kushner PJ. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13:1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- Aulmann S, Bläker H, Penzel R, Rieker RJ, Otto HF, Sinn HP. CTCF gene mutations in invasive ductal breast cancer. Breast Cancer Res Treat. 2003;80:347–352. doi: 10.1023/A:1024930404629. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci USA. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–622. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida A, Smith AD, Imoto S, Tsutsumi S, Aburatani H, Zhang MQ, Akiyama T. Integrative bioinformatics analysis of transcriptional regulatory programs in breast cancer cells. BMC Bioinformatics. 2008;9:404. doi: 10.1186/1471-2105-9-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf M, Epple A, Werner T. The next generation of literature analysis: integration of genomic analysis into text mining. Brief Bioinform. 2005;6:287–297. doi: 10.1093/bib/6.3.287. [DOI] [PubMed] [Google Scholar]
- Yan X, Zhao X, Qian M, Guo N, Gong X, Zhu X. Characterization and gene structure of a novel retinoblastoma-protein-associated protein similar to the transcription regulator TFII-I. Biochem J. 2000;345:749–757. [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, Hannon GJ, Stebbing J. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K-I, Minami Y, O'Malley BW, Kato S. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Perissi V, Menini N, Cottone E, Capello D, Sacco M, Montaldo F, De Bortoli M. AP-2 transcription factors in the regulation of ERBB2 gene transcription by oestrogen. Oncogene. 2000;19:280–288. doi: 10.1038/sj.onc.1203303. [DOI] [PubMed] [Google Scholar]
- Williams CM, Scibetta AG, Friedrich JK, Canosa M, Berlato C, Moss CH, Hurst HC. AP-2gamma promotes proliferation in breast tumour cells by direct repression of the CDKN1A gene. EMBO J. 2009;28:3591–3601. doi: 10.1038/emboj.2009.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. E2F: at the crossroads of life and death. Trends Cell Biol. 2008;18:528–535. doi: 10.1016/j.tcb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Altucci L, Addeo R, Cicatiello L, Dauvois S, Parker MG, Truss M, Beato M, Sica V, Bresciani F, Weisz A. 17beta-Estradiol induces cyclin D1 gene transcription, p36D1–p34cdk4 complex activation and p105Rb phosphorylation during mitogenic stimulation of G(1)-arrested human breast cancer cells. Oncogene. 1996;12:2315–2324. [PubMed] [Google Scholar]
- Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Frasor J, Komm B, Chang KC, Kraus WL, Katzenellenbogen BS. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol Endocrinol. 2007;21:2112–2123. doi: 10.1210/me.2006-0474. [DOI] [PubMed] [Google Scholar]
- Guérin M, Sheng ZM, Andrieu N, Riou G. Strong association between c-myb and oestrogen-receptor expression in human breast cancer. Oncogene. 1990;5:131–135. [PubMed] [Google Scholar]
- Drabsch Y, Hugo H, Zhang R, Dowhan DH, Miao YR, Gewirtz AM, Barry SC, Ramsay RG, Gonda TJ. Mechanism of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc Natl Acad Sci USA. 2007;104:13762–13767. doi: 10.1073/pnas.0700104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgort MG, Zou A, Marschke KB, Allegretto EA. Estrogen and estrogen receptor antagonists stimulate transcription from the human retinoic acid receptor-alpha 1 promoter via a novel sequence. Mol Endocrinol. 1996;10:477–487. doi: 10.1210/mend.10.5.8732679. [DOI] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Wang G, Appaiah H, Luktuke N, Carroll JS, Geistlinger TR, Brown M, Badve S, Liu Y, Nakshatri H. AKT alters genome-wide estrogen receptor alpha binding and impacts estrogen signaling in breast cancer. Mol Cell Biol. 2008;28:7487–7503. doi: 10.1128/MCB.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Roth L, Lai C, Li X. Profiling activities of transcription factors in breast cancer cell lines. Assay Drug Dev Technol. 2006;4:293–305. doi: 10.1089/adt.2006.4.293. [DOI] [PubMed] [Google Scholar]
- Echlin DR, Tae HJ, Mitin N, Taparowsky EJ. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene. 2000;19:1752–1763. doi: 10.1038/sj.onc.1203491. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Murphy TC, Lim FL, Moore DJ, Stuckey R, Antrobus K, Kimber I, Orphanides G. Anti-proliferative effect of estrogen in breast cancer cells that re-express ER 127 1113088 is mediated by aberrant regulation of cell cycle genes. J Mol Endocrinol. 2005;34:535–551. doi: 10.1677/jme.1.01677. [DOI] [PubMed] [Google Scholar]
- Hartman J, Müller P, Foster JS, Wimalasena J, Gustafsson JA, Ström A. HES-1 inhibits 17beta-estradiol and heregulin-beta1-mediated upregulation of E2F-1. Oncogene. 2004;23:8826–8833. doi: 10.1038/sj.onc.1208139. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Fitzgerald K, Bryce KS, Jeruss JS. Effect of activin on smad 3 signal transduction on breast cancer cell lines. J Surg Res. 2008;144:379. [Google Scholar]
- Malek D, Gus R, Kleuser B. 17-Beta-estradiol inhibits transforming-growth-factor-beta-induced MCF-7 cell migration by Smad3-repression. Eur J Pharmacol. 2006;534:39–47. doi: 10.1016/j.ejphar.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Nielsen DA, Shapiro DJ. Insights into hormonal control of messenger RNA stability. Mol Endocrinol. 1990;4:953–957. doi: 10.1210/mend-4-7-953. [DOI] [PubMed] [Google Scholar]
- Ing NH. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of messenger RNAs. Biol Reprod. 2005;72:1290–1296. doi: 10.1095/biolreprod.105.040014. [DOI] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Maillot G, Lacroix-Triki M, Pierredon S, Gratadou L, Schmidt S, Bénès V, Roché H, Dalenc F, Auboeuf D, Millevoi S, Vagner S. Widespread estrogen-dependent repression of microRNAs involved in breast tumor cell growth. Cancer Res. 2009;69:8332–8340. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]