Abstract
A subset of cells, tentatively called cancer stem cells (CSCs), in breast cancer have been associated with tumor initiation, drug resistance, and tumor persistence or aggressiveness. They are characterized by CD44 positivity, CD24 negativity, and/or ALDH1 positivity in flow cytometric studies. We hypothesized that the frequency or density of these cells may be associated with more aggressive tumor behavior. We borrowed these multiplexed, flow-based methods to develop an in situ method to define CSCs in formalin-fixed paraffin-embedded breast cancer tissue, with the goal of assessing the prognostic value of the presence of CSCs in breast cancer. Using a retrospective collection of 321 node-negative and 318 node-positive patients with a mean follow-up time of 12.6 years, we assessed TMAs using the AQUA method for quantitative immunofluorescence. Using a multiplexed assay for ALDH1, CD44, and cytokeratin to measure the coexpression of these proteins, putative CSCs appear in variable sized clusters and in 27 cases (of 490), which showed significantly worse outcome (log rank P = 0.0003). Multivariate analysis showed that this marker combination is independent of tumor size, histological grade, nodal status, ER-, PR,- and HER2-status. In this cohort, ALDH1 expression alone does not significantly predict outcome. We conclude that the multiplexed method of in situ identification of putative CSCs identifies high risk patients in breast cancer.
A series of studies over the last few years have defined a new model for tumorgenesis based on the observation that only a very small percentage of cells in solid tumors are capable of generating new tumors in mice. These cells have been observed to initiate new tumors that resemble the tumor of origin in structure, behavior, and molecular characteristics. Although not all scientists agree, these tumor initiating cells have been termed cancer stem cells (CSCs). They are defined through their ability to self-renew, differentiate, and create new tumors in nude mice.1,2,3,4,5,6,7 These cells were also found to show different properties than the rest of the cells in a given tumor.8 One of the properties that may be unique to these cells is their general drug resistance. CSCs have mechanisms for protection from chemotherapeutic agents that has been raised as a potential pathway for tumor recurrence.9,10,11 That is, if a therapeutic agent kills all of the non-CSC cells, the tumor may shrink or appear to disappear, but then later these cells may be responsible for regrowth.12
CSCs have presented a challenge in definition in that to prove their stem-like properties they must be removed, selected, and grown in animal models. Recent advances in the identification of cell surface markers for various tissue and tumor types have provided evidence for the presence of CSCs in those tumor types. In solid tumors, as breast, liver, lung, prostate cancer, an enriched population of 20 to 1000 CSCs is required to successfully generate tumors in animal models.5,7,13,14,15,16,17,18,19,20,21 For breast cancer it has been demonstrated that a CD44+/CD24−/lin− cell population is meeting the characteristics of CSCs. Furthermore, this cell population is sufficient for invasion but does not translate into real metastasis.22 Flow cytometry, followed by cell sorting, growing mammospheres, and transplanting these sorted cells into nude mice, allowed to identify various subsets of cells capable of self renewal, differentiation, and tumor initiation characterized through different cell surface markers, which seem to be specific for each tumor type.23,24 In breast cancer different cell surface markers that have been described include CD44+/CD24−/lin−, α 6 Integrin, CD133, β 1 Integrin/CD29, and ALDH1, which is not a cell surface marker.22,23,24,25,26
ALDH1 is an aldehyde dehydrogenase, responsible for oxidation of retinol to retinoic acid, important for normal development and homoeostasis in several organs and crucial during embryogenesis. It is an important detoxifying enzyme in the liver, also expressed in kidney, as well as hematopoeitic progenitor cells.27,28,29,30 ALDH1 is described to play a crucial role within normal differentiation of stem cells. It was demonstrated that ALDH1-positive cells can easily be identified by the flow cytometry-based Aldefluor assay that detects intracellular ALDH1 expression in viable cells.31 ALDH1 expressing breast epithelial cells isolated from reduction mammoplasties have been shown to be capable of growing mammospheres as well as growth and duct formation in the fat pad of NOD/scid mice. In contrast, Aldefluor-negative cells fail to grow mammospheres or to show differentiated outgrowth in mice. ALDH1-positive tumor cells were able to generate tumors in nude mice with as low as 500 cells. When ALDH1 positivity was combined with the marker set of CD44+/CD24−/lin− the enriched population of cells was able to successfully generate tumors with as few as 20 cells in nude mice when transplanted orthotopically into fat pad. The population of cells that were ALDH1− but CD44+/CD24−/lin− were not able to generate tumors when transplanted into fat pad of nude mice even with as many as 50,000 cells.23 Thus, tumor cells expressing ALDH1 and CD44+/CD24−/lin− are highly tumorigenic and were considered to be cancer stem cells. Thus ALDH1 presents a potential method to define CSCs without removing the cells from their tissue environment. Wicha and colleagues attempted to visualize and assess CSCs using an immunohistochemical assay for ALDH1. Two breast cancer cohorts, consisting of 341 and 136 patients, were analyzed for ALDH1 expression and ALDH1 positivity was seen in 20% to 30% of the patients, and a strong correlation was seen between ALDH1 positivity and poor prognosis.23
Given the properties described above, we hypothesized that multiplexed antibody assessment may provide a mechanism for identification of stem cells in situ. Furthermore, given their ability to evade chemotherapy and their ability to form new tumors, we hypothesized that the presence and density of CSCs may be associated with tumor aggressiveness. Toward that goal, we have designed a multiplexed assay for CSCs and tested these in a large cohort of breast cancer patients.
Materials and Methods
Yale Breast Cancer Cohort
Formalin-fixed, paraffin-embedded (FFPE) tumors of 642 patients, who underwent surgery at the Yale University New Haven Hospital between 1962 and 1983, were obtained from the archives of the Pathology Department of Yale University (New Haven, CT). The follow-up time for these patients ranges between 4 months and 41 years with a median follow-up of 12.6 years. Patient age at diagnosis ranged from 28 to 88 years with a mean age of 58 years. 321 patients were node-negative, and 318 patients were diagnosed as node-positive (Table 1). Postsurgical treatment information was not available for all of the patients; however, most patients were treated with local irradiation, and none of the node-negative patients received adjuvant systemic therapy. Approximately 15% of the node-positive patients received adjuvant chemotherapy. Postmenopausal patients with ER-positive tumors, diagnosed after 1978, received tamoxifen (5.6% of the patients). By standard immunohistochemistry, 52% of the tumors showed positivity for ER, 46% for PR, and 14% for HER2. Nuclear grade 3—on a scale 1 to 3—was seen in 28% of the specimens, tumor size ranged between 2 and 5 cm in 44% of the cases and more than 5 cm in 15% of the cases. 81% were diagnosed as infiltrating ductal carcinoma, 2% as lobular carcinoma, and 17% as mixed types or other histology.
Table 1.
Characteristics of the Yale Breast Cancer Cohort, Showing Information on Age, Histology, Tumor Size, Nodal Status, ER-, PR-, and HER2 Assessment, All of These Factors within the Expected Range for a Breast Cancer Cohort of 642 Patients
| Variable | n (%) | Median (range) |
|---|---|---|
| Number of patients | 642 (100) | |
| Follow-up, y | 8.9 (0.19–41) | |
| Age, y | 58 (24–88) | |
| <50 | 170 (26) | |
| >50 | 465 (73) | |
| Not specified | 7 (1) | |
| Histology | ||
| Infiltrating duct | 520 (81) | |
| Infiltrating lobular | 14 (2) | |
| Carcinoma (not other spec.) | 83 (13) | |
| Other | 25 (4) | 2.5 (0.13–14.5) |
| Tumor size, cm | ||
| <2 | 212 (33) | |
| 2 to 5 | 279 (44) | |
| >5 | 99 (15) | |
| Not specified | 52 (8) | |
| Nodal status | ||
| Negative | 321 (50) | |
| Positive | 318 (49) | |
| Not specified | 3 (1) | |
| ER | ||
| Negative | 287 (45) | |
| Positive | 320 (50) | |
| Not available | 35 (5) | |
| PR | ||
| Negative | 290 (46) | |
| Positive | 298 (47) | |
| Not available | 49 (7) | |
| HER2 | ||
| Negative | 487 (77) | |
| Positive | 109 (16) | |
| Not specified | 46 (7) |
Tissue Microarray Construction
The tissue microarrays (TMAs) for this study were constructed as previously described.32 Representative tumor areas from FFPE of primary breast tumors were placed in a recipient block using 0.6 mm core size. Liver tissue and cell lines were used as internal controls. The cell lines MB231, A431, BT549, SKBR3, SW480, BAF3, HT29, MB435, T47D, MCF7, MB436, MB468, BT474, NCIH28, and HST2A were purchased from ATCC (Manassas, VA) and cultured in the lab. Culture conditions and construction of cell line arrays have been previously described.33 The cell lines were represented in duplicate as internal controls.
Multiplexed Immunohistochemical Staining for Single Target, Multiple Targets, and Antibodies
Slides were deparaffinized with xylene, rehydrated with alcohol, and antigen retrieval was achieved by pressure cooking with citrate buffer (pH = 6) for 10 minutes. After blocking of endogenous peroxidase with methanol and hydroxyl peroxide, slides were preincubated with 0.3% bovine serum albumin in 0.1 mol/L of Tris-buffered saline (triethanolamine-buffered saline, pH = 8) for 30 minutes at room temperature. For multiplexed immunohistochemistry targeting a single target slides were then incubated with a cocktail of the primary antibody ALDH1 (BD Biosciences, immunogen human ALDH1, Clone 44, dilution 1:1000) and a wide-spectrum rabbit anti-cow cytokeratin antibody (Z0622, DAKO, dilution 1:100) in bovine serum albumin/Tris-buffered saline at 4°C overnight. To target multiple targets slides were incubated overnight at 4°C with the following antibodies: ALDH1– BD Biosciences at 1:1000, CD44 rabbit monoclonal antibody – Abcam, clone EPR1013Y, at 1:1000 and guinea pig Anti-Keratin, Sigma, at 1:200. As secondary antibodies we used mouse EnVision reagent (DAKO, neat) and Alexa 546 conjugated goat anti-rabbit secondary antibody (Molecular Probes, Eugene, OR; 1:100) for targeting ALDH1 as a single target. For multiple targets we used mouse EnVision and rabbit EnVison (DAKO, neat), after blocking horseradish peroxidase as previously described,34 and Alexa Fluor 546 goat anti-guinea pig secondary antibody (Molecular Probes) for 1 hour at room temperature. Cy5-tyramide (Perker Elmer, Life Science, MA) and biotinylated tyramide (Perker Elmer) followed by streptavidin-conjugated Alexa Fluor 750 (Invitrogen, Molecular Probes) were used to visualize the targets. DAPI staining containing 4′6-diamidino-2phynilindol was used to identify tissue nuclei.
Automated Quantitative Analysis
Automated quantitative analysis (AQUA®) is a method that allows exact and objective measurement of protein concentration within a defined tumor area, as well as within subcellular compartments, as described elsewhere.33 Briefly, a series of monochromatic high-resolution images were captured using an Olympus AX-51 epifluorescent microscope using a previously described algorithm for image collection.33 For each histospot an in- and out-of-focus image were obtained using the signals from the 4′6-diamidino-2phynilindol, from the cytokeratin – Alexa 546, from ALDH1 – Cy5 channel and CD44 – Alexa 750 channel. A tumor mask was created by binarizing the cytokeratin signal, which allowed us to exactly measure the ALDH1-, respectively the CD44 protein expression within the epithelial compartment of the tumor and to distinguish between stromal, lymphocytic, and tumor protein expression. Only expression within the tumor mask was accounted for a positive score. To measure coexpression of ALDH1 and CD44, we binarized the CD44 signal within the tumor mask and created a CD44 compartment, allowing us to measure the ALDH1 pixel intensity within the CD44 compartment. AQUA scores of a given target within the tumor mask, the CD44 compartment, or different subcellular compartments were calculated by dividing the signal intensity by the area of the tumor mask, CD44 compartment, or the subcellular compartment within the histospot. Patient sample histospots showing 5% tumor or less, as determined by the percentage of area of the spot that was positive for cytokeratin, were excluded from the analysis.
Western Blot
The cell lines MCF7, BT474, MB453, HT29, A431, 1650, CaCo, and Mell888 were purchased from ATCC (Manassas, VA). Western blotting of protein extracts was performed using standard methods. ALDH1 expression was detected by overnight incubation with the mouse monoclonal anti-ALDH1 antibody (BD Biosciences, Clone 44) at 1:250. Protein loading was assessed using rabbit anti–β-tubulin at 1:4000 overnight.
Statistical Analysis
Pearson correlation coefficient (R) was used to assess the correlation of AQUA scores between corresponding histospots on boutique arrays and on the Yale Breast Cancer Cohorts. Survival curves were calculated using the Kaplan–Meier analysis, accounting for breast cancer specific mortality with a follow-up time of 120 months. Patients were also stratified for nodal status and Kaplan–Meier analysis for node-positive and node-negative patients was performed separately. Multivariate analysis using the Cox proportional hazards model was performed to assess the independent prognostic significance of ALDH1 expression within the tumor mask and within the CD44 compartment on nonnodal relapse and overall survival. Bivariate χ2 tests were used to determine the association between ALDH1 and clinicopathologic parameters.
All P values were based on two-sided testing, and P < 0.05 was considered as statistically significant.
All statistical analysis was done using Statview software.
Results
Antibody Validation and Cell Line Expression
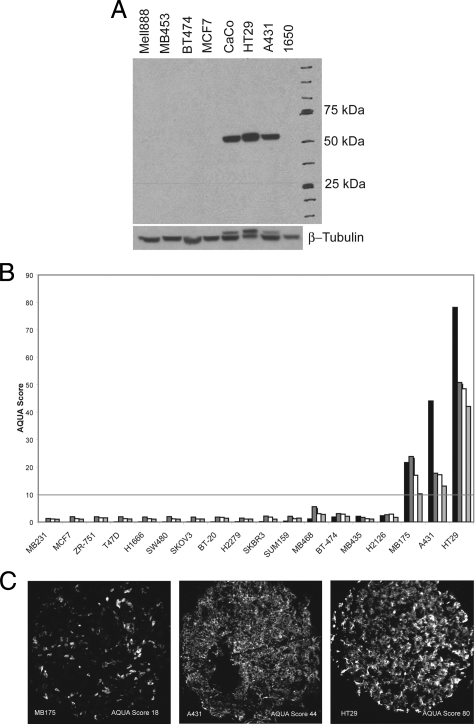
To assess the expression of ALDH1 and to validate the antibody for use in situ, we prepared cell lysates from a series of immortalized cell lines and fractionated them by Western blot. The Western blot showed bands that migrate at 55 kDa consistent with ALDH1 expression in CaCo2, HT 29, and A431 cell lines (Figure 1A). The same antibody was used in immunofluorescent assays to show detection of HT29, A431, and MB175 when prepared as FFPE specimens as a control tissue microarray containing 15 other cell lines that did not express ALDH1. Examples of these cell lines are shown in Figure 1C and quantified in fourfold redundancy as shown in Figure 1B. The cell line with the lowest expression of ALDH1 was MB175, which corresponded with an AQUA score of 10. The AQUA score of 10 is defined as the threshold of detection using this assay system (Figure 1). The CD44 antibody was validated in a similar manner, and an AQUA score of 8 was used as a cutoff for CD44 positivity (data not shown).
Figure 1.
Antibody validation and cell line expression of ALDH1 through Western blot and immunofluorescent staining followed by AQUA analysis on a series of control cell lines. A: The Western blot shows positive protein expression for ALDH1 in the cell lines CaCO, A431, and HT29 with a band at 55 kDA. No ALDH1 expression was detected in MB453, BT474, MCF7, 1650, and Mell888. β Tubulin serves as a control. B: Representative images of the three cell lines, MB175, A431, and HT29, with positive ALDH1 expression. The cell lines are represented on a tissue microarray; immunohistochemical staining was performed and the images were captured on the Cy5 channel with the PM1000 microscopy platform. Subsequent AQUA analysis resulted in an AQUA Score, which was used to define positive ALDH1 staining for breast cancer tissue. C: Immunohistochemical staining and AQUA analysis of ALDH1 were performed in fourfold redundancy on a control TMA consisting of cell lines, breast cancer samples, liver, and normal breast tissue. AQUA scores for each cell line are shown in fourfold redundancy. Cell lines showing positive expression for ALDH1 with an AQUA score above the threshold of 10 are MB175, A431, and HT29, thus confirming the Western blot results for ALDH1 positivity in A431 and HT29 cell lysates. The threshold of 10 was determined as the lowest AQUA Score for MB175 on these experiments performed in fourfold redundancy (no Western blot was performed for MB175 lysate).
Assessment of ALDH1 and CD44 Expression in the Yale Breast Cancer Cohort
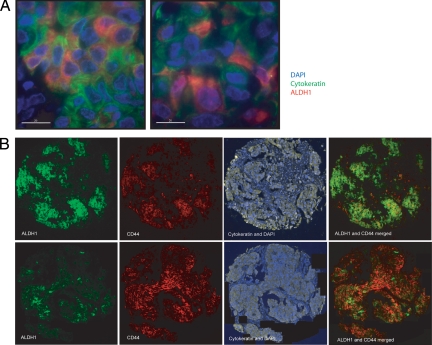
ALDH1 expression was measured by AQUA analysis in fourfold redundancy on the Yale Breast Cancer Cohort. In most cases, no ALDH1 expression was seen anywhere in the histospots. Some cases showed patchy epithelial and/or stromal expression patterns. Most commonly in ALDH1-positive histospots, it appeared as small clusters of epitheliod tumor cells (Figure 2). Less often it was expressed in small islands of cells that were morphologically indistinct from surrounding cells. Very rarely, it was expressed in nearly every cell in a given histospot.
Figure 2.
Representative examples for ALDH1 and ALDH1/CD44 staining patterns in our breast cancer cohort. A: ×60 magnification images captured with a convolution/deconvolution imaging system microscope (Deltavision) showing ALDH1 expression in groups or in single cells within the epithelial tumor. The epithelial tumor is defined through cytokeratin staining, shown in green; ALDH1 is shown in red, and DAPI staining in blue. B: Breast cancer sample 1 and 2 show different staining patterns for ALDH1 and CD44 coexpression as found on our TMAs. Sample 1 shows high expression for ALDH1, where CD44-positive cells can be found in clusters. Sample 2 shows high expression for CD44, where ALDH1-positive cells can be found in clusters. In both tissue samples cells showing coexpression of ALDH1, CD44, and cytokeratin illustrate the common clustering pattern.
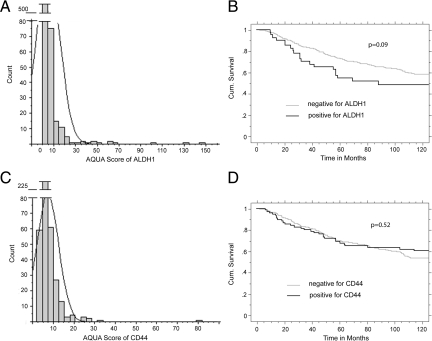
As per the validation above, AQUA scores above 10 were considered above the threshold of background nonspecific staining. Because most histospots were negative, the highest AQUA score from all of the TMA spots for each patient was used to represent that patient. We were able to assess ALDH1 expression for 626 patients within this cohort, 45 (7%) of them showing positive AQUA scores for ALDH1. Because we were attempting to define epithelial CSCs, only protein expression colocalizing within the cytokeratin mask was scored. There was some staining within the stroma as well as in the lymphocytic and histocytic areas of the tumor tissue, but this staining was excluded from the analysis. Only 4 of the 45 positive histospots express ALDH1 at high levels homogeneously within the cytokeratin stain, whereas the majority of the positive cases are represented by groups and clusters of ALDH1-positive cells or low protein expression just above the threshold (Figure 2A). A frequency distribution of cases is shown in Figure 3A. Kaplan–Meier analysis of ALDH1 expression showed that those patients expressing ALDH1 above the threshold showed worse breast cancer specific survival compared with those without ALDH1 expression (P = 0.09; Figure 3B). Stratifying by nodal status did not show significance in either subset. ALDH1 expression did not show correlation with expression of ER, PR, HER-2, CK5, or CA IX (data not shown).
Figure 3.
A: Distribution of AQUA scores for ALDH1 Expression on our Yale Breast Cancer Cohort. The majority of the patients show an AQUA score below the threshold of 10, thus they scored negative for ALDH1 expression within the epithelial tumor. 7% of the breast cancer cases show positive scores for ALDH1. B: Kaplan–Meier analysis for ALDH1 as single target on the Yale Breast Cancer Cohort. Assessment of ALDH1 expression was performed in fourfold redundancy, and maximal scores were counted. The P value shows a trend for poorer survival for ALDH1 expressing patients without reaching statistical significance. Stratification for nodal status does not change this trend (data not shown). C: Distribution of AQUA scores for CD44 expression on the Yale Breast Cancer Cohort, the threshold for CD44 is 8, as defined through cell line positivity on the control TMA (data not shown). Every patient with an AQUA Score for CD44 of 8 and above was counted as positive. D: Kaplan–Meier analysis for CD44 as single target on the Yale Breast Cancer Cohort, which does not show any significance looking at breast cancer specific mortality in patients with low or high CD44 expression.
We found a heterogeneous, largely membranous, and cytoplasmic staining pattern for CD44 consistent with the previously described expression pattern in breast cancer.35,36,37,38,39,40,41 CD44 expression was seen in 13 of 20 cell lines. The threshold for expression was an AQUA score of 8. The distribution of scores for CD44 is shown in Figure 3C. Survival analysis for CD44 in a Kaplan–Meier analysis shows CD44 expression is not associated with outcome (Figure 3D). Stratification for nodal status shows that node-negative breast cancer patients with high expression of CD44 trend toward better prognosis (P = 0.09) as described previously for the CD44 splice form CD44s.42 In node-positive patients CD44 expression does not show any association with outcome (supplemental Figure S1 at http://ajp.amjpathol.org).
Multiplexed Immunofluorescent Staining for ALDH1 and CD44 for in Situ Identification of CSCs
In efforts to mimic the method for identification of CSCs used in dispersed cells, but without the capability to use negative selection for in situ identification, we developed a protocol to assess ALDH1 co-expression with CD44 in the tissue compartment defined by cytokeratin expression as described in the Materials and Methods section. Several tumors expressed ALDH1 but not CD44. Other tumors expressed CD44 but not ALDH1. In some cases with high levels of ALDH1, CD44 was seen only in clusters or islands of cells within the tumor spot. The most common pattern was a heterogeneous pattern of CD44 staining with islands representing a few cells that expressed ALDH1 (Figure 2B) There was also occasional ALDH1 expression colocalized with CD44 within the stroma, on lymphocytes or histocytes. Because they were not in regions positive for cytokeratin, these cases had AQUA scores below the threshold of 10. To exclude the possibility that multiplexing these markers resulted in variable or decreased expression, we compared the multiplexed cases analysis with the assessment of each marker in a single marker assay. In each case the single assay showed close correlation with the multiplexed assay (supplemental Figure S2 at http://ajp.amjpathol.org).
Although we do not know the average or expected density of CSCs in tumors we decided to use the standard approach of assessment of the assay in twofold redundancy. The highest AQUA score of the two spots was selected as the score for the case, on the basis of the fact that TMAs may undersample for rare events like CSCs. Of the 620 cases on the original build of the array, only 490 were scoreable in 2× redundancy with greater than 5% cytokeratin-positive tumor area. We were able to identify 27 patients of 490 (5.5%) with an AQUA score above threshold for ALDH1 in the CD44 compartment, thus showing coexpression for ALDH1, CD44, and Cytokeratin.
Statistical Analysis
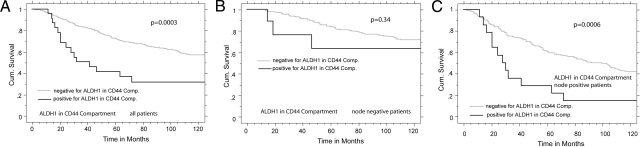
Statistical analysis including log rank testing and Kaplan–Meier analysis showed that patients coexpressing ALDH1 and CD44 within the epithelial compartment have a high breast cancer specific mortality, with a P value of 0.0003 (Figure 4A). Furthermore, assessment by the Cox proportional Hazard Model shows a hazard ratio of 2.15 (95% CI; 1.198 to 3.846; P = 0.0103) for breast cancer specific death when the CSCs are found by this assay, independent of ER-, PR-, HER2-status, nodal status, nuclear grade, and tumor size (Table 2). Even though the assay is independent of the patient’s nodal status, stratification for nodal status was done to determine whether CSCs are seen more frequently in patients with known metastasis to lymph nodes. The CSC assay was highly significant within the node-positive patient group but not significant in the node-negative group (Figure 4, B and C). Although this finding is consistent with the biological hypothesis, the observation may be attributable to the small subset of patients coexpressing ALDH1 and CD44, especially in the node-negative group.
Figure 4.
Kaplan–Meier analysis for ALDH1 in a CD44 compartment assessed through AQUA analysis after immunohistochemical staining for multiple targets on our cohort. A: Assessment of ALDH1 and CD44 coexpression was performed in twofold redundancy, an AQUA score of 10 and above for ALDH1 in the CD44 compartment was defined as positive, and maximal scores were counted. The P value shows significance with patients coexpressing both markers showing poor prognosis. B: Kaplan–Meier analysis for node-negative patients coexpressing ALDH1 and CD44. Eight patients score positive for this marker set, two events only. C: Kaplan–Meier analysis for node-positive patients coexpressing ALDH1 and CD44. The P value is highly significant, thus this marker set is identifying a subset of node-positive patients who are at high risk.
Table 2.
The Multivariate Analysis for ALDH1 in the CD44 Compartment Shows that the Prognostic Value of this Marker Set Is Independent of ER-, PR-, HER2-Status, of Tumor Size, Nuclear Grade, and Nodal Status
| Parameter | HR | 95% Cl | P value |
|---|---|---|---|
| Nuclear grade | |||
| Low | 1.00 | ||
| High | 1.200 | 0.870–1.655 | 0.2664 |
| Tumor size | |||
| <2 cm | 1.00 | ||
| 2 to 5 cm | 2.117 | 1.453–3.083 | <0.0001 |
| >5 cm | 3.187 | 2.000–5.076 | <0.0001 |
| Nodal status | |||
| Negative | 1.00 | ||
| Positive | 2.37 | 1.582–3.165 | <0.0001 |
| ER | |||
| Low | 1.00 | ||
| High | 0.786 | 0.558–1.107 | 0.1680 |
| PR | |||
| Low | 1.00 | ||
| High | 0.797 | 0.573–1.110 | 0.1795 |
| HER2 | |||
| Low | 1.00 | ||
| High | 1.523 | 1.017–2.226 | 0.0411 |
| ALDH1 in CD44 compartment | |||
| Negative | 1.00 | ||
| Positive | 2.150 | 1.198–3.846 | 0.0103 |
Discussion
The potential existence of cancer stem cells and their influence on tumor initiation, growth, and metastasis is an old concept.43 But the concept is hard to prove, in part because the method of proving the stem-like qualities of the cells requires assays that remove the cells from their native environment. One set of identifying proteins used to define possible cancer stem cells in breast cancer are the combination of CD44+/CD24−/lin−,22 though it has been shown that this marker set is not relevant in terms of prognostic and predictive value after analyzing breast cancer tissue for these proteins.44 ALDH1 is of special interest, because it is an enzyme crucial for embryogenic development but also necessary for normal differentiation of stem cells. ALDH1 has been shown to identify cells with characteristics of stem cells and/or early progenitor cells for normal breast epithelium as well as breast cancer. Furthermore, breast cancer cells sorted for ALDH1 positivity and CD44 positivity are highly tumorigenic, with only 20 cells capable of initiating tumor growth in NOD/scid mice.23 Thus in the present study we combined the best markers identified so far to identify CSCs in situ.
In the present study we are able to develop an assay to reproducibly measure the coexpression of ALDH1 and CD44 within the epithelial (cytokeratin) compartment of breast tumor tissue. This method allowed us to identify a subset of patients with poor prognosis, independent of conventional risk factors in breast cancer. We believe these cell clusters may represent CSCs as the frequency of their appearance and their morphological characteristics are consistent with descriptions of stem cells in mammospheres.45 However, to date, the validation that cells are indeed cancer stem cells has been their growth in a mouse model. This is obviously not possible in this system. Thus, although this work provides a new assay and an interesting result, we are unable to conclude that we have truly identified cancer stem cells.
Some of the data in this study are inconsistent with previous work on CSCs. In one key study using ALDH1 as a marker for stem cells, expression was seen in 20% to 30% of breast cancer patients on two different cohorts.23 In our Yale Breast Cancer Cohort we showed positivity in only 7% of the cases. This discrepancy may be attributable to the increased stringency we used for assessment of CSCs (multiplexing), or it may be attributable to the fact that we were strictly measuring ALDH1 expression within the epithelial cells, where other studies counted any staining by ALDH1 including the stroma and infiltrating lymphocytes. Also, other studies have shown significant correlations between ALDH1 expression and HER2 status and CK5/6 expression,23 as well as a regulatory effect of HER2 on ALDH1 expression in breast cancer cell lines.46 Our work shows no correlation between HER2 expression and the cells identified as CSCs by our assay. However, our assay does not assess colocalization of HER2 expression within the CSC cells, but rather compared the HER2 status of the case to CSC status. It is possible that the rare CSC cell islands express some HER2 but were below the threshold for definition of HER2 positivity for the entire case.47
Another limitation of this study is the fact that it was performed on TMAs. The amount of tumor sampled by two TMA spots is less than 0.0001 percentage of the tumor, yet there have been hundreds of studies using TMAs48 and many studies have shown concordance between TMA studies and whole section analysis.49 We are concerned that the area of a TMA spot may be insufficient for this assay, given the presumed low frequency of CSCs and the heterogeneity of tumors. Thus, although we report a frequency of 7% of CSCs in our population, this number may be proven to be a low estimate once studies are done with larger samples, either on whole slides on at higher-fold redundancy by TMA. However, the association of poor outcome in the 7% with CSCs supports our hypothesis that tumors with higher frequency of CSCs are associated with more aggressive behavior.
Finally, some of the results of this work are highly supportive of previous efforts related to CSCs. Perhaps most convincing is that CSCs are thought to allow tumors to grow independently and aggressively. That is consistent with the poor disease-specific survival seen in CSC-positive patients identified by our assay. CSCs are also thought to mediate metastasis because stem cells are known to be able to both self-replicate and differentiate. This study shows our assay identifies CSCs with much greater frequency in cases with nodal metastasis at presentation. However, despite these promising findings, further assays need to be done to validate these observations. Specifically, we are working toward assessment of the density of CSCs on conventional sections of breast tissue to assess the heterogeneity and density of CSCs. Finally, we are also working on confirmation of this observation in an independent cohort. If these findings are consistent with this first report, this multiplexed CSC assay may be valuable in the future in both prognostic and predictive settings.
Footnotes
Address reprint requests to David L Rimm, M.D., Ph.D., Professor of Pathology, Department of Pathology, BML 116, Yale University School of Medicine, 310 Cedar St., PO Box 208023, New Haven, CT 06520-8023. E-mail: david.rimm@yale.edu.
Supported by R33 CA 110511 Predicting Metastasis to Lymph Nodes (in breast cancer) to D.L.R.
D.L.R. declares a duality of interest, as a founder, consultant, and stockholder of a biotech company called HistoRx. This company was founded by Yale and is the exclusive Yale licensee of the AQUA® technology developed in Dr. Rimm’s lab. R.L.C. is also a stockholder in this company and serves as a consultant.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Gil J, Stembalska A, Pesz KA, Sasiadek MM. Cancer stem cells: the theory and perspectives in cancer therapy. J Appl Genet. 2008;49:193–199. doi: 10.1007/BF03195612. [DOI] [PubMed] [Google Scholar]
- Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- Kakarala M, Wicha MS. Cancer stem cells: implications for cancer treatment and prevention. Cancer J. 2007;13:271–275. doi: 10.1097/PPO.0b013e318156da4e. [DOI] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Wiestler OD. Mechanisms of disease: cancer stem cells–targeting the evil twin. Nat Clin Pract Oncol. 2008;5:337–347. doi: 10.1038/ncponc1110. [DOI] [PubMed] [Google Scholar]
- Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea–a paradigm shift, Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895–1886. [DOI] [PubMed] [Google Scholar]
- Woodward WA, Sulman EP. Cancer stem cells: markers or biomarkers? Cancer Metastasis Rev. 2008;27:459–470. doi: 10.1007/s10555-008-9130-2. [DOI] [PubMed] [Google Scholar]
- Fillmore C, Kuperwasser C. Human breast cancer stem cell markers CD44 and CD24: enriching for cells with functional properties in mice or in man? Breast Cancer Res. 2007;9:303. doi: 10.1186/bcr1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M. ABC transporters, drug resistance, and cancer stem cells. J Mammary Gland Biol Neoplasia. 2009;14:3–9. doi: 10.1007/s10911-009-9109-9. [DOI] [PubMed] [Google Scholar]
- Donnenberg VS, Meyer EM, Donnenberg AD. Measurement of multiple drug resistance transporter activity in putative cancer stem/progenitor cells. Methods Mol Biol. 2009;568:261–279. doi: 10.1007/978-1-59745-280-9_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangemi R, Paleari L, Orengo AM, Cesario A, Chessa L, Ferrini S, Russo P. Cancer stem cells: a new paradigm for understanding tumor growth and progression and drug resistance. Curr Med Chem. 2009;16:1688–1703. doi: 10.2174/092986709788186147. [DOI] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells, Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal melanoma: evidence for a stem cell-like phenotype associated with metastasis. Melanoma Res. 2008;18:191–200. doi: 10.1097/CMR.0b013e3283005270. [DOI] [PubMed] [Google Scholar]
- Dick JE. Acute myeloid leukemia stem cells. Ann N Y Acad Sci. 2005;1044:1–5. doi: 10.1196/annals.1349.001. [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7:86–95. doi: 10.1186/bcr1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimeault M, Hauke R, Batra SK. Stem cells: a revolution in therapeutics-recent advances in stem cell biology and their therapeutic applications in regenerative medicine and cancer therapies. Clin Pharmacol Ther. 2007;82:252–264. doi: 10.1038/sj.clpt.6100301. [DOI] [PubMed] [Google Scholar]
- Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- Tang C, Ang BT, Pervaiz S. Cancer stem cell: target for anti-cancer therapy. FASEB J. 2007;21:3777–3785. doi: 10.1096/fj.07-8560rev. [DOI] [PubMed] [Google Scholar]
- Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R, Jr, Badve S, Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, Chieco P, Bonafe M. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilopoulos A, Wang RH, Petrovas C, Ambrozak D, Koup R, Deng CX. Identification and characterization of cancer initiating cells from BRCA1 related mammary tumors using markers for normal mammary stem cells. Int J Biol Sci. 2008;4:133–142. doi: 10.7150/ijbs.4.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- Kim H, Lapointe J, Kaygusuz G, Ong DE, Li C, van de Rijn M, Brooks JD, Pollack JR. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 2005;65:8118–8124. doi: 10.1158/0008-5472.CAN-04-4562. [DOI] [PubMed] [Google Scholar]
- Simeone AM, Tari AM. How retinoids regulate breast cancer cell proliferation and apoptosis. Cell Mol Life Sci. 2004;61:1475–1484. doi: 10.1007/s00018-004-4002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visus C, Ito D, Amoscato A, Maciejewska-Franczak M, Abdelsalem A, Dhir R, Shin DM, Donnenberg VS, Whiteside TL, DeLeo AB. Identification of human aldehyde dehydrogenase 1 family member A1 as a novel CD8+ T-cell-defined tumor antigen in squamous cell carcinoma of the head and neck. Cancer Res. 2007;67:10538–10545. doi: 10.1158/0008-5472.CAN-07-1346. [DOI] [PubMed] [Google Scholar]
- Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, Lako M. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- Kluger HM, Dolled-Filhart M, Rodov S, Kacinski BM, Camp RL, Rimm DL. Macrophage colony-stimulating factor-1 receptor expression is associated with poor outcome in breast cancer by large cohort tissue microarray analysis. Clin Cancer Res. 2004;10:173–177. doi: 10.1158/1078-0432.ccr-0699-3. [DOI] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- Kluger HM, Siddiqui SF, Angeletti C, Sznol M, Kelly WK, Molinaro AM, Camp RL. Classification of renal cell carcinoma based on expression of VEGF and VEGF receptors in both tumor cells and endothelial cells. Lab Invest. 2008;88:962–972. doi: 10.1038/labinvest.2008.65. [DOI] [PubMed] [Google Scholar]
- Looi LM, Cheah PL, Zhao W, Ng MH, Yip CH. CD44 expression and axillary lymph node metastasis in infiltrating ductal carcinoma of the breast. Malays J Pathol. 2006;28:83–86. [PubMed] [Google Scholar]
- Ouhtit A, Abd Elmageed ZY, Abdraboh ME, Lioe TF, Raj MH. In vivo evidence for the role of CD44s in promoting breast cancer metastasis to the liver. Am J Pathol. 2007;171:2033–2039. doi: 10.2353/ajpath.2007.070535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Sun X, Huo L, Wiley EL, Rao MS. Expression of cell adhesion molecules. CD44s and E-cadherin, and microvessel density in invasive micropapillary carcinoma of the breast. Histopathology. 2005;46:24–30. doi: 10.1111/j.1365-2559.2004.01981.x. [DOI] [PubMed] [Google Scholar]
- Auvinen P, Tammi R, Tammi M, Johansson R, Kosma VM. Expression of CD 44 s. CD 44 v 3 and CD 44 v 6 in benign and malignant breast lesions: correlation and colocalization with hyaluronan. Histopathology. 2005;47:420–428. doi: 10.1111/j.1365-2559.2005.02220.x. [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Fard SF, Paiwand FF, Tolg C, Veiseh M, Wang C, McCarthy JB, Bissell MJ, Koropatnick J, Turley EA. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J Biol Chem. 2007;282:16667–16680. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balic M, Lin H, Young L, Hawes D, Giuliano A, McNamara G, Datar RH, Cote RJ. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12:5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- Thanakit V, Sampatanukul P, Ruangvejvorachai P, Keelawat S. The association of co-expression of CD44v4/MMP-9 with different nodal status in high-grade breast carcinoma patients. J Med Assoc Thai. 2005;88 Suppl 4:S30–S35. [PubMed] [Google Scholar]
- Diaz LK, Zhou X, Wright ET, Cristofanilli M, Smith T, Yang Y, Sneige N, Sahin A, Gilcrease MZ. CD44 expression is associated with increased survival in node-negative invasive breast carcinoma. Clin Cancer Res. 2005;11:3309–3314. doi: 10.1158/1078-0432.CCR-04-2184. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Mylona E, Giannopoulou I, Fasomytakis E, Nomikos A, Magkou C, Bakarakos P, Nakopoulou L. The clinicopathologic and prognostic significance of CD44+/CD24(-/low) and CD44-/CD24+ tumor cells in invasive breast carcinomas. Hum Pathol. 2008;39:1096–1102. doi: 10.1016/j.humpath.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Grimshaw MJ, Cooper L, Papazisis K, Coleman JA, Bohnenkamp HR, Chiapero-Stanke L, Taylor-Papadimitriou J, Burchell JM. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, Tamaki Y, Noguchi S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15:4234–4241. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008;26:5630–5637. doi: 10.1200/JCO.2008.17.3567. [DOI] [PubMed] [Google Scholar]
- Milanes-Yearsley M, Hammond ME, Pajak TF, Cooper JS, Chang C, Griffin T, Nelson D, Laramore G, Pilepich M. Tissue micro-array: a cost and time-effective method for correlative studies by regional and national cancer study groups. Mod Pathol. 2002;15:1366–1373. doi: 10.1097/01.MP.0000036345.18944.22. [DOI] [PubMed] [Google Scholar]