Abstract
Sushi domain-containing protein 4 (SUSD4) was a hypothetical cell surface protein whose tissue distribution and function were completely unknown. However, recent microarray-based studies have identified deletions of SUSD4 gene in patients with autism or Fryns syndrome, both of which are genetic diseases with severe abnormal neurological development and/or functions. In this article, we described the cloning, expression, refolding, tissue distribution, and functional analysis of this novel protein. Using polyclonal antibodies generated by immunizing chickens with the recombinant SUSD4, we found that SUSD4 is detectable in murine brains, eyes, spinal cords, and testis but not other tissues. In brains, SUSD4 is highly expressed in the white matter on oligodendrocytes/axons, and in eyes, it is exclusively expressed on the photoreceptor outer segments. In in vitro complement assays, SUSD4 augments the alternative but not the classical pathway of complement activation at the C3 convertase step. In in vivo studies, knocking down SUSD4 expression in zebrafish markedly increases ratios of mortality and developmental abnormality. These results provide the first insight into the important physiological roles of SUSD4 and could help to better understand the pathogenesis of autism and Fryns syndrome.
The Sushi domain is an extracellular motif commonly involved in protein-protein interactions.1 Each Sushi domain has ∼60 amino acid (Aa) residues with conserved tryptophan, glycine, proline, hydrophobic residues,2 and four invariant cysteines, the first of which forms a disulfide bond with the third, and the second forms another disulfide bond with the fourth. Because most complement control proteins, eg, decay-accelerating factor (DAF), membrane cofactor protein (CD46), factor H, and complement receptor 1 (CR1, CD35), contain continuous multiple Sushi domains, this motif is also called complement control protein domain.3 Although most proteins containing repetitive continuous Sushi domains possess complement regulatory activity, there are exceptions, eg, IL-15 receptor (IL-15R)4 and IL-2R (CD25).5
Sushi domain-containing protein 4 (SUSD4) was a hypothetic protein identified by previous genome-wide sequencing projects.6,7 It contains four continuous sushi domains according to its predicted Aa sequence. On the basis of the genomic sequencing results, the SUSD4 gene resides on chromosome 1q41 in humans, which spans ∼175 kb and contains eight exons and nine introns. Tissue-specific expressed sequence tag profiles indicated that SUSD4 is highly expressed in the central nervous system (CNS) in different species including human, mouse, and zebrafish. The homology of the Aa sequence is 95% between human and mouse SUSD4 and 63% between human and zebrafish counterparts. The highly conserved Aa sequences of SUSD4 proteins from zebrafish to humans and the abundant existence of its mRNAs in the CNS suggest that SUSD4 might have important roles related to neurological functions. In fact, previous microarray-based comparative genomic hybridization studies have identified deletions of SUSD4 gene in patients with Fryns syndrome.8,9 Fryns syndrome is an autosomal recessive multiple congenital anomaly syndrome that is usually lethal in the neonatal period.10 Patients that survive exhibit symptoms including severe development delay, mental retardation, diaphragmatic hernia, and distal limb hypoplasia.11 Ocular abnormalities including anophthalmia, microphthalmia, and retinal dysplasia are also noted in Fryns syndrome patients.12,13 Despite all these intriguing results, there is no evidence that the SUSD4 mRNA is translated in any tissues or any report on its possible function.
In this article, we expressed, purified, and refolded the four Sushi domains of human SUSD4 and raised antibodies to this protein in chickens. Using these new antisera, we characterized the tissue distribution of SUSD4 in mice. In functional studies, we found that SUSD4 augments the alternative but not the classical pathway of complement activation; and in zebrafish morpholino knockdown experiments, we found evidence indicating that Susd4 is critical for normal neurological development and/or functions.
Materials and Methods
Recombinant SUSD4 Sushi Domain Expression, Purification, and Refolding
cDNAs encoding the four Sushi domains of SUSD4 were amplified by RT-PCR from human brain total RNA (Ambion, Austin, TX) using proof reading Vent DNA polymerase (New England Biolabs, Beverly, MA) and primers (P1: 5′-AGTCCCATGGACCTTCAAGTGTGTGCTGAC-3′, P2: 5′-ACTGCTCGAGTCAATGATGATGATGATGATGCTC-3′). A 6× His tag coding region was introduced at the 3′ terminus by PCR primers. The cDNA was sequencing verified and cloned into the expression vector pET-14b (Novagen, Madison, WI), then transformed into host strain BL21 Rosetta (Novagen). The resultant expressing strain was cultured in the Overnight Express system (Novagen). Hereafter, cell pellets were lysed using the BugBuster Protein Extraction Reagent (Novagen), and inclusion bodies were collected by centrifugation at 10,000 rpm for 10 minutes, followed by three 2 M urea washes. The inclusion bodies were then dissolved in 8 M urea with 50 mmol/L β-mercaptoethanol, 0.5 mmol/L EDTA, and 20 mmol/L Tris and refolded by rapid dilution into refolding buffer [20 mmol/L ethanolamine and 1 mmol/L EDTA (pH 11.0)] in a drop-wise manner with stirring at 4°C as reported before.14,15,16 After 48 hours, refolded soluble recombinant SUSD4 Sushi domains were purified by nickel affinity chromatography (Qiagen, Valencia, CA) and dialyzed against PBS. The final concentration of the prepared SUSD4 was measured using a protein assay kit (Bio-Rad, Hercules, CA).
Polyclonal Anti-SUSD4 Generation and Purification
Polyclonal antibodies against SUSD4 were raised in chickens by QED Biosciences (San Diego, CA). In brief, each chicken was immunized intramuscularly with 400 μg of purified SUSD4 emulsified in complete Freund’s adjuvant and boosted three times every 2 weeks. Subsequently, enzyme-linked immunosorbent assay (ELISA) was performed to assess the titer of the generated antibody in blood after which both serum and eggs from the immunized chickens were collected.
Western Blot of Membrane Protein Extracts of Mouse Tissues
Membrane protein extracts from 8- to 10-week C57BL/6 mice brain, spinal cord, eye, liver, muscle, testis, thymus, kidney, stomach, lung, intestine and muscle were prepared by homogenizing the respective tissues in the extraction buffer (100 mmol/L Tris-HCl, 100 mmol/L NaCl, 10 mmol/L EDTA, 0.5% Nonidet P-40, and 0.5% sodium deoxycholate) together with protease inhibitor mixture (Roche, Indianapolis, IN). Five micrograms of total extracted protein from each tissue was separated by 10% SDS-PAGE and the cognate blots were probed with anti-SUSD4 serum (1/10,000) or the same dilution of serum collected before immunization. After incubation with 1/20,000 diluted horseradish peroxidase-labeled goat anti-chicken IgY (Jackson ImmunoResearch Laboratories, West Grove, PA), blots were developed using a ECL plus kit (GE Healthcare, PISCATAWAY, NJ).
Immunolabeling of SUSD4 in Mouse Tissues
Brain, spinal cord, eye, and testis were collected from 8- to 10-week-old C57BL/6 mice and snap-frozen in liquid nitrogen. Cryosections (7 micron) were prepared and used for immunohistochemical or immunofluorescent staining. In brief, for immunohistochemical staining, 10% hydrogen peroxide was first used to block endogenous peroxidase, then SUSD4 antiserum (1/1000) or the same concentration of control serum was applied on sections. After washing, sections were incubated with biotin conjugated anti-chicken IgY (Jackson ImmunoResearch Laboratories) and developed with an avidin-biotin complex kit (Vector Laboratories, Burlingame, CA) following manufacturer-provided protocols. For immunofluorescent staining of eye sections, Dylight 488-conjugated anti-biotin IgG was used for imaging, and micrographs were taken under a fluorescence microscope (Olympus, Melville, NY).
Complement Regulatory Activity Assays
For the classical pathway complement regulatory activity assessment, antibody-sensitized sheep erythrocytes (EshA) hemolytic assay was used. In brief, 2 × 106 EshA were washed three times with GVB++ buffer and resuspended in 100 μl of GVB++ buffer with 1/10 dilution of normal human serum and different concentrations of purified SUSD4, respectively. EDTA (1 mmol/L) was added as negative control, and the same numbers of EshA in water was included as positive control. For the alternative pathway complement regulatory activity assessment, rabbit erythrocytes hemolytic assay was performed. In brief, 2 × 106 rabbit erythrocytes were washed three times with GVB-Mg2+-EGTA buffer and resuspened in 100 μl of the same buffer with 1/20 or 1/10 dilution of normal human serum in GVB-Mg2+-EGTA buffer. EDTA was added as negative control, and rabbit erythrocytes in 100 μl of water were included as positive control. Both the classical and the alternative pathway assays were incubated at 37°C for 30 minutes, and the hemolysis were assessed by measuring OD541 after spinning at 2000 × g for 2 minutes. The average number of lytic sites/blood cell (z value) of condition x was calculated using following equation : z = −ln{1 − [(OD541 (x) − OD541 (0%)]/[OD541 (100%) − OD541 (0%)]}, in which the OD541 (0%) represents incubation of EshA with 1 mmol/L EDTA, and the OD541 (100%) represents incubation of EshA with H2O.17
SUSD4-Binding ELISAs
To study whether SUSD4 binds to C3b or factor B of the alternative pathway C3 convertase, 5 μg/ml purified human C3b or factor B (Complement Tech, Tyler TX) was coated on an ELISA plate overnight at 4°C. The same concentration of bovine serum albumin was coated as controls. After washing, different amounts of purified SUSD4 were added into the plate and incubated for another 2 hours. Following extensive washing with PBS plus 0.02% Tween 20, attached SUSD4 was detected using the SUSD4 anti-serum and a biotinylated anti-chicken IgY and an alkaline phosphotase-anti-biotin mAb following a standard ELISA protocol,18 and the OD405 was measured with a plate reader (Molecular Devices, Sunnyvale, CA).
Morpholino Injection into Zebrafish
Morpholinos specific for zebrafish Susd4 mRNA (5′-CAGCATGATGAAACATCTTTCACTG-3′) and nonspecific controls (5′-CCTCTTACCTCAGTTACAATTTATA-3′) were designed and synthesized by Genetools (Philomath, OR). Morpholinos were dissolved in Danieau buffer [58 mmol/L NaCl, 0.7 mmol/L KCl, 0.4 mmol/L MgSO4, 0.6 mmol/L Ca(NO3)2, and 5 mmol/L HEPES (pH 7.6)]. Embryonic zebrafish of the Tübingen strain were injected at the one or two-cell stage with 2 ng of morpholinos directed to inhibit the translation of susd4 mRNA. The same amount of nonspecific control morpholinos were also injected as a control. Injected embryos were housed at 28.5°C with a daily change of fish water and were evaluated daily. All zebrafish experiments were performed under an approved protocol in accordance with the guidelines of the Institutional Animal Care and Use Committee of Case Western Reserve University.
Results
Production of Recombinant SUSD4 Sushi Domains
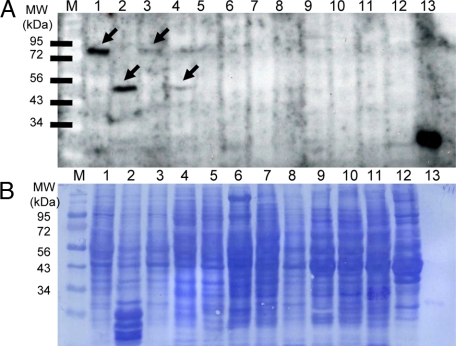
We amplified a ∼1.2-kb cDNA from human brain total RNA by RT-PCR and cloned it into the expression vector pET-14b for protein overexpression. A 6× histidine tag was added onto its C terminus to simplify downstream purification. Sequencing results verified that the amplified cDNA is the full-length SUSD4. We transformed the resultant expression construct into Escherichia coli host strain BL21 and induced expression. We found that after overnight induction, the recombinant SUSD4 was expressed as inclusion bodies. After 2 M urea washing, the purity of the inclusion bodies was >95% as assessed by SDS-PAGE (Figure 1). Following a previously developed refolding protocol16 for recombinant sushi domain proteins, we refolded the denatured SUSD4 by rapid dilution. We generally purified ∼10 mg of soluble SUSD4 Sushi domains from 200 ml of culture with purity >95% (Figure 1). The mol. wt. of the purified protein is ∼28 kDa, which is consistent with the mol. wt. predicted by the cDNA sequence. The refolded SUSD4 migrates faster than its reduced, denatured form (Figure 1), indicating the existence of intramolecular disulfide bonds.
Figure 1.
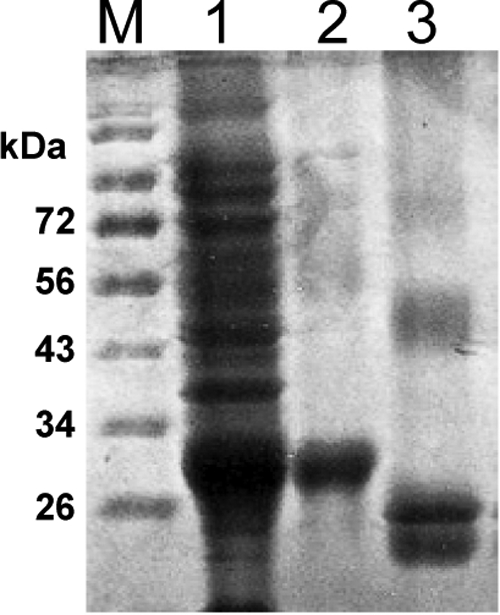
Expression, purification and refolding of SUSD4. After overnight induction, the E. coli total lysate was separated by SDS-PAGE, and it showed that recombinant SUSD4 was expressed at high levels (lane 1). Following purification, the resultant inclusion body reached >95% purity (lane 2). After refolding and dialysis against PBS, the soluble SUSD4 (lane 3) migrated faster than the reduced/denatured SUSD4 (lane 2), indicating the existence of intermolecular disulfide bounds. The two major bands of the soluble SUSD4 (lane 3) around the predicted mol. wt. (28 kDa) could be the refolded SUSD4 with different conformations, and the faint higher mol. wt. band could be the dimer form of the refolded SUSD4.
Tissue Distribution of SUSD4 in Mice
To confirm the existence of SUSD4 protein and to determine its tissue distribution, we probed membranes blotted with protein extracts from 8- to 10-weeks-old C57BL/6 mice brain, spinal cord, eye, liver, muscle, testis, thymus, kidney, stomach, lung, heart, intestine, and muscle with anti-SUSD4 or preimmunization sera. These assays (Figure 2, A and B) showed that among all tissues examined, an ∼80 kDa band is detectable only in brains and spinal cords, and a ∼50 kDa band is only detectable in eyes and testes. Identical membranes probed with the same dilution of preimmunization sera showed no band (data not shown). These results showed that SUSD4 protein does exist, and it is detectable in murine brains, eyes, spinal cords, and testes, in which brains and eyes are the primary expression sites.
Figure 2.
Tissue distribution of SUSD4 by Western blot. A: Membrane protein extracts of murine brain (lane 1), eye (lane 2), spinal cord (lane 3), testis (lane 4), thymus (lane 5), liver (lane 6), kidney (lane 7), stomach (lane 8), lung (lane 9), heart (lane 10), intestine (lane 11), and muscle (lane 12) and the purified SUSD4 protein (positive control, lane 13) were separated by 10% SDS-PAGE and probed with SUSD4 antiserum. An ∼80-kDa band was detected in brains and spinal cords, whereas an ∼50-kDa band was detected in eyes and testes (arrows). No band was detected in the other tissues. Control experiments using the same dilution of preimmunization serum showed no band (data not shown). B: After probing, the membrane was stained with Coomassie Blue to show relative amounts of loaded protein.
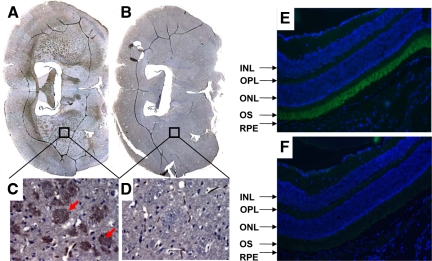
After confirming that SUSD4 protein is detectable in brains, eyes, spinal cords, and testes by Western blot analyses, we examined its distribution in these tissues by immunohistochemical or immunofluorescent stainings. These assays (Figure 3) showed that in brains, SUSD4 is mainly expressed on oligodendroglial–axonal units within the white matter, and in eyes, SUSD4 is exclusively expressed on outer segments of photoreceptors. Although we could detect weak bands on Western blots with spinal cords and testes extracts, immunohistochemical staining did not show any difference between the anti-SUSD4 and the preimmunization sera-stained sections from these tissues (data not shown).
Figure 3.
Distribution of SUSD4 in murine brains and eyes. SUSD4 is selectively expressed on oligodendroglial–axonal units in the brain (A–D, arrows) and outer segments of photoreceptors in the eye (E and F). Murine brain and ocular sections were staining with anti-SUSD4 Abs (A, C, and E) and preimmunization Abs (B, D, and F) using standard immunohistochemical or immunofluorescent staining protocols. In ocular sections, SUSD4 was detected with Alexa 488-anti-chicken IgY (green), and cell nucleus were labeled with 4′,6′-diamidino-2-phenylindole (blue). INL, inner nuclear layer; ONL, outer nuclear layer; OPL, outer plexiform layer; OS, outer segments, RPE, retinal pigment epithelium.
Complement Regulatory Activity of SUSD4
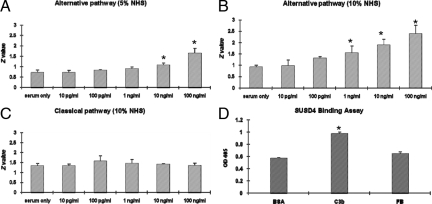
As indicated, proteins containing repetitive sushi domains are usually involved in complement activation regulation. To examine whether SUSD4 modulates complement activation, we used the conventional complement activation-mediated sheep or rabbit erythrocytes hemolytic assays. These assays (Figure 4, A–C) showed that SUSD4 augmented the alternative but not the classical pathway of complement activation at a concentration as low as 1 ng/ml. To determine whether SUSD4 binds to components of the alternative pathway C3 convertase (C3bBb) to augment complement activation, we incubated SUSD4 with plate-bound C3b and factor B. After extensive washing, we performed ELISA experiments to detect the bound SUSD4 on plates. These assays (Figure 4D) showed that SUSD4 binds to C3b but not factor B.
Figure 4.
Complement regulatory activity of SUSD4. A and B: Erab-based hemolytic assays with different concentrations of SUSD4 added, showing SUSD4 augments the alternative pathway complement activation mediated hemolysis in different serum dilutions (A: 1/20 and B: 1/10); C, EshA-based hemolytic assays with different concentrations of SUSD4 added, showing SUSD4 has no effect on the classical pathway complement activation mediated hemolysis; D: SUSD4 binding assays, showing that SUSD4 binds to C3b but not factor B. (Representative results of more than three experiments; *P < 0.05).
Role of SUSD4 in Development
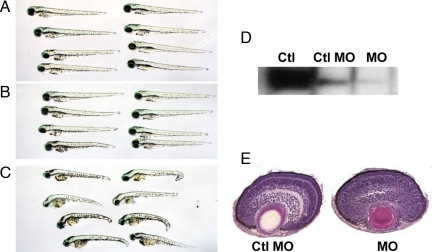
We microinjected with 2 ng of SUSD4 specific or nonrelevant morpholinos into 150 one- or two-cell stage zebrafish embryos and monitored them daily for 7 days. We counted fish with apparent abnormal head, smaller eyes, curved tails, or difficulty in swimming as “abnormal” (Figure 5). The numbers of survived fish and the percentages of fish with developmental/locomotive abnormalities are summarized in Table 1. Western blots verified that Susd4 protein levels are significantly reduced in SUSD4 morphants (Figure 5). Histological analysis of fish at 3 days postfertilization (dpf) showed that SUSD4 morphants have abnormally (delayed) developed eyes. These results suggest that SUSD4 could be integrally involved in zebrafish neurological development and/or locomotive functions.
Figure 5.
Representative pictures of zebrafish developed from noninjected (A), control morpholino (Ctl MO) injected (B), and SUSD4-specific morpholino-injected embryos (C) on 7 dpf. Fish developed from control morpholino-injected embryos showed little if any difference from the noninjected. Fish with SUSD4 knockdown showed smaller heads, eyes and curved tails (representative results from three independent experiments). Western blot using whole fish extracts at 3 dpf verified that SUSD4 expression is knocked down in SUSD4–morpholino-injected embryos with brain extracts from normal adult fish as a positive control (D); H&E staining of ocular sections at 3 dpf showed abnormal retina in SUSD4 morphants compared with control morphants (E).
Table 1.
SUSD4 Knockdown Induces Significantly Higher Percentages of Mortality and Abnormality in Zebrafish
| Time | Category | Noninjected | MO - Control | MO - SUSD4 |
|---|---|---|---|---|
| 3 dpf | Mortality | 2.6% (4 of 150) | 6.0% (9 of 150) | 8.0% (12 of 150) |
| Abnormality | 1.3% (2 of 146) | 3.5% (5 of 141) | 28.7% (43 of 138) | |
| 5 dpf | Mortality | 2.6% (4 of 150) | 6.0% (9 of 150) | 12.7% (19 of 150) |
| Abnormality | 1.4% (2 of 146) | 4.0% (6 of 141) | 27.5% (36 of 131) | |
| 7 dpf | Mortality | 2.6% (4 of 150) | 6.7% (10 of 150) | 14.0% (21 of 150) |
| Abnormality | 1.4% (2 of 146) | 5.7% (8 of 140) | 28.7% (37 of 129) |
Two nanograms of nonspecific morpholinos (MOs) or SUSD4-specific MOs were injected into 150 zebrafish embryos at one- or two-cell stage; surviving fish with morphological or locomotive abnormalities were counted at 3, 5, and 7 dpf (representative results of multiple experiments).
Discussion
In this article, we cloned, expressed, and refolded the human SUSD4 extracellular sushi domains. Using the anti-SUSD4 polyclonal antibodies raised in chickens, we detect SUSD4 only on oligodendrocytes/axons in brains and on outer segments of photoreceptors in eyes but not in other tissues examined in mice by immunohistochemical staining. SUSD4 augments the alternative but not the classical pathway of complement activation at the C3 convertase step. Blocking SUSD4 expression in zebrafish leads to retarded development and abnormal locomotive movement, indicating that SUSD4 is integrally involved in normal development or neurological functions.
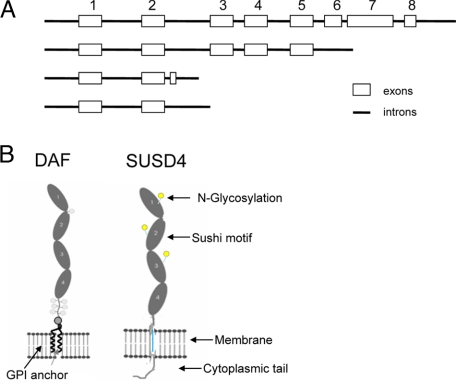
We became interested in SUSD4 during our ongoing studies on the cell surface complement regulator, DAF (CD55).19,20,21,22,23 Previous sequencing results indicate that the SUSD4 gene has at least four different transcriptional isoforms, probably due to mRNA alternative splicing (Figure 6A). The full-length SUSD4 protein, as predicted by its DNA sequence, has a very similar extracellular structure as DAF (Figure 6B) with four head-to-tail linked Sushi domains (21). The putative SUSD4 protein has a 41-Aa leader sequence (as predicted by SignalP 3.0)24 at the N terminus, followed by four Susi domains. After the extracellular domains are a 20-Aa transmembrane region (Aa 322-342) and a 149-Aa cytoplasmic tail (Aa 343-492), which contains multiple potential phosphyratation sites, including sites for PKA, PKC, P38MAPK, and CDC2 kinases.
Figure 6.
A: SUSD4 has different transcription isoforms, probably because of alternative splicing. B: Diagram of DAF and putative SUSD4 molecules. Both have an identical extracellular structure of four continuous Sushi domains.
Previous attempts to express recombinant Sushi domains of DAF,25 Crry,15 or CR114,16 in E. coli all resulted in inclusion bodies, probably due to the incorrect formation of disulfide bonds within and among different Sushi domains. Fortunately, refolding procedures using excessive dilution in ethanolamine buffer at low temperatures have been successful in refolding the denatured Sushi domain proteins to their correct conformations, restoring their full bioactivities.14,15,16,25 Our results showed that the protocol established for refolding recombinant DAF, Crry, or CR1 sushi domains is also effective in renaturing recombinant SUSD4.
Because sequencing results predicted that SUSD4 is highly conserved (>95%) in all sequenced mammals, we chose to immunize chickens instead of rodents or rabbits to raise antibodies (chicken and human SUSD4 share only 65% homology). Our results indicated that antibodies can be efficiently raised in chickens and the raised anti-human SUSD4 antibodies cross-react with zebrafish and mouse SUSD4.
Knowledge of the tissue distribution of SUSD4 protein should facilitate future studies on its physiological roles. Using the newly developed antibody, we found that SUSD4 gene products exist in the CNS as predicted by previous sequencing projects. The results that SUSD4 protein in spinal cords and testis can be detected by Western blot analyses but not immunohistochemistry could be explained by the markedly lower expression levels of SUSD4 in the spinal cord and testis (Figure 2) and the different sensitivities of these two technologies. The protein-protein interaction nature of Sushi domains and the selective high expression levels of SUSD4 on oligodendrocytes/axons in the brain white matter suggest that SUSD4 could be involved in normal neurological functions. Moreover, its exclusive expression on photoreceptor outer segments in the eye indicates that SUSD4 could also be important in retinal development and/or functions.
In view of previous studies on complement regulatory proteins containing repetitive sushi domains (3), it is interesting that SUSD4 augments rather than inhibits complement activation. The results from C3b uptake assays and binding assays indicate that SUSD4 functions at the C3 convertase step by directly interacting with C3b but not factor B of the alternative pathway C3 convertase C3bBb. In view of previous reports that C3−/−,26 factor B−/−,27 or factor D−/−28 mice appear to develop normally, it is unlikely that the modest complement regulatory activity of SUSD4 could play a significant role in the CNS. In support of this, a recent publication indicates that a similar sushi domain-containing protein in Caenorhabditis elegans is directly involved in the organization/function of the synapse, independently from its complement regulatory function.29 However, more studies are required to fully dissect the mechanism underlying the complement regulatory activity of SUSD4 and to understand its impacts in the CNS.
Because injecting high concentrations of morpholinos into zebra-fish embryos can cause nonspecific developmental impairments, we used small doses of Susd4 specific or nonspecific morpholinos. In our experiments, injection of 2 ng of nonrelevant morpholino only resulted in ∼5.7% nonspecific developmental abnormalities and ∼6.7% mortalities. In contrast, injection of the same amount of Susd4 specific morpholino led to significantly higher rates of developmental retardation (28.7%) and mortality (14.0%). These results, together with the above data that SUSD4 is selectively expressed in the CNS, strongly argue that SUSD4 could be integrally involved in neurological development and/or normal neurofunctions. These results are also consistent with the previously published results that deletions of Susd4 gene occur in autism and Fryn’s syndrome, genetic diseases in both of which patients have severe neurological developmental/functional irregularities,11,30 and at least in Fryns syndrome, patients suffer high mortality during gestation.10
Although more work are needed to fully understand the importance of SUSD4 under normal conditions and in disease states, the data obtained in this study provide the first insight into the potential critical roles of SUSD4 in neurological development and/or function and could help to better understand the pathogenesis of both autism and the Fryn’s syndrome.
Acknowledgments
We thank Dr. Brian McDermott (Department of Otolaryngology) and members of his laboratory for use of their fish facility and guidance with the zebrafish-based experiments. We also thank Catherine Doller at the Histology core for excellent histology service, and Dr. Scott Howell at the Vision Science Research Center imaging core for digital imaging analysis.
Footnotes
Address reprint requests to Feng Lin, Ph.D., Assistant Professor, Institute of Pathology, Case Western Reserve University School of Medicine, 2085 Adelbert Rd., Room 306, Cleveland, OH 44106; or Hong Bu, M.D., Professor, Department of Pathology, West China Hospital Sichuan University, Chengdu 610041, China. E-mail: feng.lin@case.edu or hongbu@scu.edu.cn.
Supported by National Institutes of Health (grants NS052471 to F.L. and EY11373 to Vision Science Research Center), National Multiple Sclerosis Society (grant RG3664 to F.L.), Natural Science Foundation of China (grant 30671988 to H.B.), and the Chinese Oversea Postgraduate Program Fellowship (grant 3019-2008624057 to Z.T.).
References
- Ichinose A, Bottenus RE, Davie EW. Structure of transglutaminases. J Biol Chem. 1990;265:13411–13414. [PubMed] [Google Scholar]
- Reid KB, Day AJ. Structure-function relationships of the complement components. Immunol Today. 1989;10:177–180. doi: 10.1016/0167-5699(89)90317-4. [DOI] [PubMed] [Google Scholar]
- Kirkitadze MD, Barlow PN. Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol Rev. 2001;180:146–161. doi: 10.1034/j.1600-065x.2001.1800113.x. [DOI] [PubMed] [Google Scholar]
- Wei X, Orchardson M, Gracie JA, Leung BP, Gao B, Guan H, Niedbala W, Paterson GK, McInnes IB, Liew FY. The Sushi domain of soluble IL-15 receptor α is essential for binding IL-15 and inhibiting inflammatory and allogenic responses in vitro and in vivo. J Immunol. 2001;167:277–282. doi: 10.4049/jimmunol.167.1.277. [DOI] [PubMed] [Google Scholar]
- Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. The structure of interleukin-2 complexed with its α receptor. Science. 2005;308:1477–1480. doi: 10.1126/science.1109745. [DOI] [PubMed] [Google Scholar]
- Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, Chow B, Chui C, Crowley C, Currell B, Deuel B, Dowd P, Eaton D, Foster J, Grimaldi C, Gu Q, Hass PE, Heldens S, Huang A, Kim HS, Klimowski L, Jin Y, Johnson S, Lee J, Lewis L, Liao D, Mark M, Robbie E, Sanchez C, Schoenfeld J, Seshagiri S, Simmons L, Singh J, Smith V, Stinson J, Vagts A, Vandlen R, Watanabe C, Wieand D, Woods K, Xie MH, Yansura D, Yi S, Yu G, Yuan J, Zhang M, Zhang Z, Goddard A, Wood WI, Godowski P, Gray A. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13:2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Kantarci S, Casavant D, Prada C, Russell M, Byrne J, Haug LW, Jennings R, Manning S, Blaise F, Boyd TK, Fryns JP, Holmes LB, Donahoe PK, Lee C, Kimonis V, Pober BR. Findings from aCGH in patients with congenital diaphragmatic hernia (CDH): a possible locus for Fryns syndrome. Am J Med Genet A. 2006;140:17–23. doi: 10.1002/ajmg.a.31025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Theisen A, Bejjani BA, Ballif BC, Aylsworth AS, Lim C, McDonald M, Ellison JW, Kostiner D, Saitta S, Shaikh T. The discovery of microdeletion syndromes in the post-genomic era: review of the methodology and characterization of a new 1q41q42 microdeletion syndrome. Genet Med. 2007;9:607–616. doi: 10.1097/gim.0b013e3181484b49. [DOI] [PubMed] [Google Scholar]
- Fryns JP, Moerman F, Goddeeris P, Bossuyt C, Van den Berghe H. A new lethal syndrome with cloudy corneae, diaphragmatic defects and distal limb deformities. Hum Genet. 1979;50:65–70. doi: 10.1007/BF00295591. [DOI] [PubMed] [Google Scholar]
- Slavotinek AM. Fryns syndrome: a review of the phenotype and diagnostic guidelines. Am J Med Genet A. 2004;124A:427–433. doi: 10.1002/ajmg.a.20381. [DOI] [PubMed] [Google Scholar]
- Pierson DM, Subtil A, Taboada E, Butler MG. Newborn with anophthalmia and features of Fryns syndrome. Pediatr Dev Pathol. 2002;5:592–596. doi: 10.1007/s10024-002-2106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson DM, Taboada E, Butler MG. Eye abnormalities in Fryns syndrome. Am J Med Genet A. 2004;125A:273–277. doi: 10.1002/ajmg.a.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossakowska D, Dodd I, Pindar W, Smith RA. Structure-activity relationships within the N-terminal short consensus repeats (SCR) of human CR1 (C3b/C4b receptor, CD35): SCR 3 plays a critical role in inhibition of the classical and alternative pathways of complement activation. Eur J Immunol. 1999;29:1955–1965. doi: 10.1002/(SICI)1521-4141(199906)29:06<1955::AID-IMMU1955>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Fraser DA, Harris CL, Smith RA, Morgan BP. Bacterial expression and membrane targeting of the rat complement regulator Crry: a new model anticomplement therapeutic. Protein Sci. 2002;11:2512–2521. doi: 10.1110/ps.0212402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I, Mossakowska DE, Camilleri P, Haran M, Hensley P, Lawlor EJ, McBay DL, Pindar W, Smith RA. Overexpression in Escherichia coli, folding, purification, and characterization of the first three short consensus repeat modules of human complement receptor type 1. Protein Expr Purif. 1995;6:727–736. doi: 10.1006/prep.1995.0003. [DOI] [PubMed] [Google Scholar]
- Roos A, Nauta AJ, Broers D, Faber-Krol MC, Trouw LA, Drijfhout JW, Daha MR. Specific inhibition of the classical complement pathway by C1q-binding peptides. J Immunol. 2001;167:7052–7059. doi: 10.4049/jimmunol.167.12.7052. [DOI] [PubMed] [Google Scholar]
- Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA): quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- Lin F, Fukuoka Y, Spicer A, Ohta R, Okada N, Harris CL, Emancipator SN, Medof ME. Tissue distribution of products of the mouse decay-accelerating factor (DAF) genes: exploitation of a Daf1 knock-out mouse and site-specific monoclonal antibodies. Immunology. 2001;104:215–225. doi: 10.1046/j.0019-2805.2001.01287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Kaminski HJ, Conti-Fine BM, Wang W, Richmonds C, Medof ME. Markedly enhanced susceptibility to experimental autoimmune myasthenia gravis in the absence of decay-accelerating factor protection. J Clin Invest. 2002;110:1269–1274. doi: 10.1172/JCI16086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Immormino RM, Shoham M, Medof ME. Bulk production and functional analyses of mouse CD55’s native and deglycosylated active domains. Arch Biochem Biophys. 2001;393:67–72. doi: 10.1006/abbi.2001.2488. [DOI] [PubMed] [Google Scholar]
- Lin F, Emancipator SN, Salant DJ, Medof ME. Decay-accelerating factor confers protection against complement-mediated podocyte injury in acute nephrotoxic nephritis. Lab Invest. 2002;82:563–569. doi: 10.1038/labinvest.3780451. [DOI] [PubMed] [Google Scholar]
- Uhrinova S, Lin F, Ball G, Bromek K, Uhrin D, Medof ME, Barlow PN. solution structure of a functionally active fragment of decay-accelerating factor. Proc Natl Acad Sci USA. 2003;100:4718–4723. doi: 10.1073/pnas.0730844100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- White J, Lukacik P, Esser D, Steward M, Giddings N, Bright JR, Fritchley SJ, Morgan BP, Lea SM, Smith GP, Smith RA. Biological activity, membrane-targeting modification, and crystallization of soluble human decay accelerating factor expressed in E. coli. Protein Sci. 2004;13:2406–2415. doi: 10.1110/ps.03455604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels MR, Butko P, Ma M, Warren HB, Lage AL, Carroll MC. Studies of group B streptococcal infection in mice deficient in complement component C3 or C4 demonstrate an essential role for complement in both innate and acquired immunity. Proc Natl Acad Sci USA. 1995;92:11490–11494. doi: 10.1073/pnas.92.25.11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman JM, Ljubanovic D, Edelstein CL, Gilkeson GS, Holers VM. Lack of a functional alternative complement pathway ameliorates ischemic acute renal failure in mice. J Immunol. 2003;170:1517–1523. doi: 10.4049/jimmunol.170.3.1517. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ma M, Ippolito GC, Schroeder HW, Jr, Carroll MC, Volanakis JE. Complement activation in factor D-deficient mice. Proc Natl Acad Sci USA. 2001;98:14577–14582. doi: 10.1073/pnas.261428398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M, Rapti G, Richmond JE, Bessereau JL. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 2009;461:992–996. doi: 10.1038/nature08430. [DOI] [PubMed] [Google Scholar]
- Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest. 2009;119:747–754. doi: 10.1172/JCI37934. [DOI] [PMC free article] [PubMed] [Google Scholar]