Abstract
This study addressed the role of impairment of osteoblastic differentiation as a mechanism underlying pathophysiology of the osteogenesis imperfecta (OI). We hypothesized that combination of impaired osteogenic differentiation with increased bone resorption leads to diminished bone mass. By introducing visual markers of distinct stages of osteoblast differentiation, pOBCol3.6GFP (3.6GFP; preosteoblast) and pOBCol2.3GFP (2.3GFP; osteoblast/osteocytes), into the OIM model, we assessed osteoblast maturation and the mechanism of increased osteoclastogenesis. Cultures from oim/oim;2.3GFP mice showed a marked reduction of cells expressing GFP relative to +/+;2.3GFP littermates. No significant difference in expression of 3.6GFP between the +/+ and oim/oim mice was observed. Histological analysis of the oim/oim;3.6GFP mice showed an increased area of GFP-positive cells lining the endocortical surface compared with +/+;3.6GFP mice. In contrast GFP expression was similar between oim/oim;2.3GFP and +/+;2.3GFP mice. These data indicate that the osteoblastic lineage is under continuous stimulation; however, only a proportion of cells attain the mature osteoblast stage. Indeed, immature osteoblasts exhibit a stronger potential to support osteoclast formation and differentiation. We detected a higher Rankl/Opg ratio and higher expression of TNF-α in sorted immature osteoblasts. In addition, increased osteoclast formation was observed when osteoclast progenitors were cocultured with oim/oim-derived osteoblasts compared with osteoblasts derived from +/+ mice. Taken together, our data indicate that osteoblast lineage maturation is a critical aspect underlying the pathophysiology of OI.
Osteogenesis imperfecta (OI) is a genetic disorder resulting in most cases from a mutation within one of the genes that encode a collagen chain. This can cause underproduction of a normal collagen molecule or secretion of a defective collagen chain that can be the basis of a disorganized and weakened matrix. Histological studies were performed to understand the bone response to the underlying genetic defect. A state of high bone turnover in the human OI was observed especially in the more severe types of OI (type III and IV).1,2 Although the bone formation rate was higher in patients with OI, this was achieved by recruiting an increased number of osteoblasts. This histological finding would suggest that the bone formation rate per osteoblast is significantly lower in OI.1 Thus a cellular consequence of OI can be viewed as a limitation of bone forming activity at the per cell level that is compensated by a generalized activation of the lineage to produce a greater number of osteoblastic cells. Mesenchymal stem cells that reside within the bone marrow represent a major source of osteoblasts.3,4 Therefore, a mouse primary marrow stromal cell culture is a model most closely resembling the in vivo conditions and would allow the analysis of the osteoprogenitor cells ability to differentiate.5,6,7
To address these questions we crossed the oim/oim mouse with transgenic mice developed to identify different stages of osteoblastic differentiation. A visible marker was used to generate transgenic mice in which fragments of rat type I collagen promoter are used to drive green fluorescent protein (GFP) expression; (pOBCol3.6GFP contains 3.6-kb and pOBCol2.3GFP uses 2.3-kb region of rat Col1a1 promoter8). In marrow stromal cell cultures pOBCol3.6GFPtpz-positive cells first appeared at an early stage of differentiation, before nodule formation, while pOBCol2.3GFPemd fluorescence first appeared in nodules undergoing mineralization. These findings suggest that Col1a1GFP transgenes are marking different subpopulations of cells during differentiation of skeletal osteoprogenitors.8,9 To study the biology of OI as an in vitro model that would closely resemble in vivo state we have established cultures of primary bone marrow stromal cells derived from all +/+ and oim/oim mice.
In parallel to the activation of the osteoblast lineage in the oim/oim mice, the number of the osteoclasts has also been increased. In addition to increased number of osteoclasts/bone area, an increase in excretion of free DPD in urine was observed in oim/oim mice.10 A recent study by Zhang et al reported that osteoclasts in cultures derived from oim/oim mice have larger diameter, greater number of nuclei per cell, and more F-actin rings compared with +/+ osteoclasts. They also exhibited greater resorptive activity than osteoclast derived from +/+ mice.11 Regulation of osteoclast precursor commitment and osteoclastic activity is indirectly regulated by the osteoblast lineage cells through the expression of the receptor activator of NF-κB ligand (Rankl) and osteoprotegerin (Opg).12,13 In the case of OI, if much of the osteoblast lineage does not reach the stage of full differentiation this could be the underlying mechanism behind the higher induction and activity of the osteoclast lineage and the mechanism causing high bone turnover. Using previously described models, oim/oim and Col1a1GFP transgenic mice, we performed an evaluation of the in vitro and in vivo differentiation ability of osteoprogenitor cells in the oim/oim mice. We evaluated the osteoclast inductive potential of the osteoblasts derived from oim/oim mice and the ability of osteoblast lineage cells at different stages of maturation to support osteoclastogenesis. These analyses provide new insights into the mechanisms by which the genetic mutation causes a severe bone phenotype in the osteogenesis imperfecta murine (oim).
Materials and Methods
Mice, Breeding, and Genotyping
The original mouse strain that carries the oim mutation (frameshift mutation in the C-terminal propeptide) is maintained in the B6C3Fe-a/a (C57BL/6JLe X C3HeB/FeJLe) hybrid background as described by Chipman et al.14 Mice heterozygous for the mutation (oim/+) were bred with pOBCol3.6GFP or pOBCol2.3GFP mice, both of which are CD-1 strains. GFP transgene was introduced as homozygous and experimental mice were littermates obtained by breeding oim/+; GFP/GFP mice. The genotyping of mice was performed using the PCR method previously described by Saban et al.15
Preparation of Sections for Histological Analysis
The preparation and histological evaluation of frozen sections was performed as described previously.16 Briefly, femurs from 3-month oim/oim and wild-type mice were fixed in 4% paraformaldehyde for 3 days followed by decalcification in 14% EDTA for 3 to 4 days. The tissues were soaked overnight in 30% sucrose/PBS. These procedures were carried at 4°C under constant agitation. Samples were embedded in Frozen Embedding Medium (Thermo Shandon, Pittsburgh, PA) at −70°C and 5-μm full-length sections in the middle of the tissue marked by the central vein were collected with the assistance of the CryoJane tape transfer system (Instrumedics Inc., Hackensack, NJ).
Photography and Quantification of Fluorescent Microscopy Images
Acquisition and quantification of florescent images from tissue cultures or histology sections of mouse femurs were described in detail in previously published reports.16,17 Images were taken under computerized Zeiss Axiovert 200 microscope (Carl Zeiss, Thornwood, NY) operated by Openlab software (Improvision, Lexington, MA). A macro language was used to generate composite images of whole longitudinal bone or 63% of the 35-mm culture dish in a reproducible manner. Fluorescent expression of pOBCol3.6GFP and pOBCol2.3GFP was recorded using dual FITC/Texas red fluorescent filter cube. Quantification was done by analysis of independent GFP calculations generated for each sample in the metaphyseal region. The metaphyseal region was described as a 800-um boxed area located 2 mm distal to the growth plate. GFP images were analyzed using Openlab software by thresholding based on intensity values. The Openlab density slice function was used to generate binary maps of the selected GFP+ populations. Green fluorescence was scored positive over background if it had a level of 20 to 255 intensity units. The raw data were exported to Excel to quantify GFP expression.
Mouse Marrow Stromal Cultures, Northern Blot, and Histochemical Analysis of Cultures
Three-month- and twelve-month-old oim/oim mice and wild-type littermates were sacrificed by CO2 asphyxiation. Marrow stromal cell cultures (MSCs), RNA extraction, and histochemical staining of the cultures were carried using a previously described procedure.8 Briefly, for MSCs, the epiphyseal growth plates were removed and marrow was collected. The cells were plated at a density 10 × 106 cells per well in 6-well culture plates. On day 4, half of the medium was replaced with fresh αMEM; from day 7, total medium was changed into differentiation medium (αMEM freshly supplemented with 50 μg/ml ascorbic acid, 10−8 M dexamethasone and 8 mmol/L β-glycerophosphate). Medium was changed every two days for the duration of the experiment.
Total RNA was extracted from cultures using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. RNA pellets were redissolved in GTC buffer and further precipitated in isopropanol. 15 μg of RNA was separated on a 2.2 M formaldehyde/1% agarose gel and transferred onto a nylon membrane. Membranes were probed with (32P)dCTP labeled rat Col1a1, mouse OC, and BSP cDNA.8 The X-ray films were exposed and intensity of autoradiography signals was obtained using “Gene Tools” software (Syngene, Frederick, MD). ALP activity staining was performed by using a commercially available kit (86-R Alkaline phosphatase, Sigma Diagnostics Inc., St. Louis, MO) according to the manufacturer’s instructions.
Coculture of Osteoblast and Osteoclast Progenitor Cells
Calvarial osteoblast were isolated from 7-day-old oim/oim neonatal mice or wild-type littermates using a modification of the method described by Wong and Cohn.8,18 Calvaria were subjected to four sequential 30-minute digestions in an enzyme mixture containing 0.05% trypsin and 1.5 U/ml collagenase P (Roche Diagnostic Inc., Indianapolis, IN) at 37°C. Cell fractions two to four were pooled, and enzyme activity was terminated by addition of FCS. The cells were plated at a density of 1 × 106 cells per well in 100-mm culture dish in DMEM and switched to differentiation medium (αMEM containing 10% FCS, 50 μg/ml ascorbic acid, 4 mmol/L β-glycerophosphate) when confluent. After a 10-day culturing period the cells were replated at a density of 25 × 103 per well in 24-well plates in αMEM. The cells were allowed to attach for 5 hours before coculturing with bone marrow mononuclear cells. One day before the osteoblast secondary culture, two-month-old oim/oim and wild-type littermates were killed and bone marrow was flushed and plated. Twenty-four hours later nonadherent cells were harvested and mononuclear cells were isolated by Ficoll density centrifugation (GE Health care, Uppsala, Sweden). Purified mononuclear cells were plated at a density of 25 × 103 per well mixed with the secondary osteoblast culture. The cells were grown in αMEM supplemented with 10−8 M 1,25 dihydroxyvitamin D.
Osteoclast Resorption Assay
Bone marrow–derived nonadherent monocytes (5000 cells) were cocultured with osteoblast (5000 cells) with the addition of 1 × 10−8 M vitamin D in BD Biocoat osteologic microplates described as above. These plates contain precoated calcium, which functions as resorption substrate for mature osteoclasts. After 7 days in culture the plates were stained with 5% silver nitrate and imaged under Zeiss Axiovert 200 microscope. Resorption area was quantified using Adobe Photoshop (Adobe Systems Inc., San Diego, CA).
Preparation of Cells for Sorting, cDNA Amplification, and Real-Time PCR
Primary calvarial osteoblast cultures were prepared for sorting as previously described.9 Briefly, 7-day-old pOBCol3.6GFP calvarial osteoblast cultures were digested in 0.25% trypsin/1 mmol/L EDTA for 5 minutes. Seventeen-day-old cultures derived from pOBCol2.3GFP mice were digested in 0.2% collagenase/0.2% hyalouronidase/2.5% trypsin for 10 to 15 minutes. Cell sorting was done using a FACS Vantage (BD Biosciences) with 488 nm excitation and 530/30 emission filters. The cells were separated using a 100-μm nozzle and collected into DMEM/30% FCS media. Before, during, and after sorting, the cell suspensions were kept cold to minimize changes in gene expression. After a short centrifugation to pellet cells, RNA was isolated. DNase (DNase I, Invitrogen) digestion was performed on the extracted RNA to eliminate genomic DNA contamination. cDNA was synthesized using an Invitrogen Superscript First-strand Synthesis System for RT-PCR. TaqMan® Gene Expression Assays specific for Opg, Rankl, and TNF-α were obtained from ABI and analysis performed on the 7500 Real-Time PCR System. 18S ribosomal RNA was used as internal control. Before using the ΔΔCT method for quantification, validation experiments were performed to demonstrate that the amplification efficiencies of target genes and the reference gene were approximately equal.
RNA Extraction from Calvaria and Real-Time PCR Gene Expression Analysis
Calvaria derived from 8-day-old wild-type and oim/oim mice were dissected free of surrounding tissue. Sutures were cut out and parietal bones were collected, minced, and homogenized directly in Trizol solution. RNA was extracted and cDNA generated as described above. Real-time PCR was performed using TaqMan® Gene Expression Assays specific for osteocalcin, bone sialoprotein, and osterix. Data obtained are normalized to the GAPDH expression levels and presented as relative gene expression between wild-type and oim/oim mice (supplemental Figure S1, see http://ajp.amjpathol.org).
Treatment with TNF-α–Neutralizing Antibody
Coculture of calvarial osteoblasts derived from oim/oim mice and BMMCs from wild-type mice was established as described above. After replating of osteoblasts (1 × 104 per well), BMMCs (1 × 104 per well) were added and cells were grown in αMEM supplemented with 10−8 M 1,25 dihydroxyvitamin D and with or without addition of TNF-α neutralizing antibody at concentration of 1 μg/ml or 5 μg/ml (e-Bioscience, San Diego, CA). Media was exchanged on day 3 and 5 with addition of VitD3 with TNF-α antibody or vehicle (PBS/150 mmol/L NaCl2). Cells were stained for the presence of osteoclast on day 7 of culture. Two independent biological experiments were completed.
Results
Decreased Ostegenic Differentiation of Osteoprogenitors Derived from Oim/Oim Mice
We have previously developed mice transgenic for pOBCol3.6GFP (3.6GFP) and pOBCol2.3GFP (2.3GFP) as visual markers for the identification of preosteoblast and mature osteoblast populations.8 The 3.6GFP is active early in the culturing period when cells express early markers of ostegenic differentiation, whereas the expression of 2.3GFP remains restricted to mature osteoblast/osteocyte located within the mineralized nodules. To examine whether the osteoblast differentiation is affected by the oim mutation the visual transgenes 3.6GFP and 2.3GFP were bred into oim mice (Figure 1A). MSCs were derived from oim/oim;GFP/GFP mice or +/+;GFP/GFP mice. Using repetitive imaging for GFP expression in the same culture plate throughout the culturing period we were able to record the tempo and magnitude of lineage differentiation as represented by the GFP expression. The oim/oim;3.6GFP cultures showed similar GFP expression patterns compared with +/+;3.6GFP cultures. However, cells derived from oim/oim;2.3GFP mice exhibited a marked impairment of the GFP expression relative to +/+;2.3GFP littermates (Figure 1B). These results indicated a defect in osteoblast lineage differentiation in the oim/oim mice. To confirm the results obtained with visual marker of osteblast differentiation, we evaluated the expression of conventional markers of osteoblast differentiation in these cultures. Alkaline phosphatase staining of marrow stromal cultures derived from 3-month- and 12-month-old mice showed lower ALP expression in oim/oim mice compared with wild-type littermates. The difference in ALP expression is more evident on day 17, when a higher proportion of cells should reach a stage of mature osteoblasts (Figure 2A). Similarly, decreased expression of mature bone marker osteocalcin became apparent in the oim/oim mice after 12 days of culturing as detected by Northern blot analysis (Figure 2B). Expression of earlier osteoblast lineage markers bone sialoprotein and Col1a1 shows smaller differences (supplemental Figure S2, see http://ajp.amjpathol.org). These in vitro data confirmed the presence of a defect in osteogenic differentiation into mature stage of in the oim/oim model. In addition we would like to indicate that at the age of 1 month we did not detect difference in the expression of bone markers using the primary culture system. At this stage (1 month), a high bone turnover is present in both wild-type mice as well as in oim/oim mice, and difference in the osteogenic differentiation was not detected under these high turnover conditions (Supplemental Figure S3, see http://ajp.amjpathol.org).
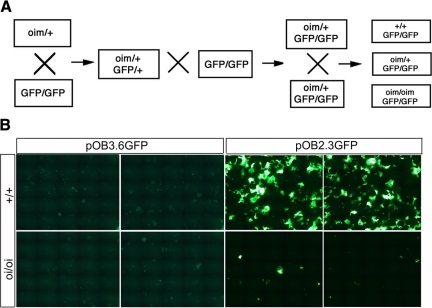
Figure 1.
Transgenic mice and GFP expression in vitro. A: Breeding approach to generate oim/oim;3.6GFP and oim/oim;2.3GFP mice. Heterozygous oim/+ mice were bred with mice homozygous for a GFP marker. The F1 progeny (heterozygous for both transgene [GFP] and for an OIM mutation) were selected to breed with homozygous GFP mice to obtain homozygous GFP mice with one allele harboring an OIM mutation. These mice were bred to obtain experimental mice either wild-type for the mutation (+/+) or homozygous for an OIM mutation (oim/oim) in a homozygous GFP background. B: GFP expression in marrow stromal cultures (MSCs) derived from 3-month-old wild-type or oim/oim mice. A series of images was obtained with a concatenated image covering 63% of the area of a 35-mm tissue culture well. The left panel shows 3.6GFP expression in 7-day-old cultures. No major differences were observed between wild-type cultures and oim/oim cultures. The right panel shows 2.3GFP expression in day 17 cultures. Large areas of GFP-expressing cells were detected in the +/+;2.3GFP cultures. In contrast, only a few GFP expressing colonies were observed in the oim/oim;2.3GFP cultures.
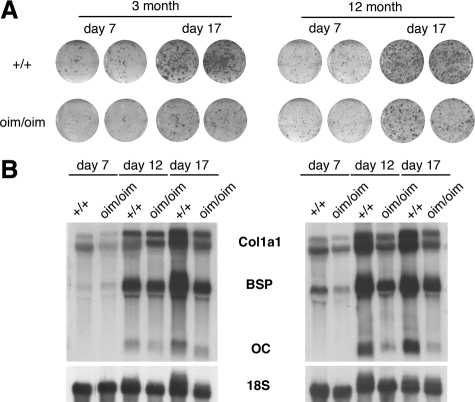
Figure 2.
Oim/oim osteoblasts have decreased differentiation in vitro. A: Alkaline phosphatase (ALP) staining of MSC cultures derived from 3-month-old or 12-month-old mice. Lower expression of ALP was detected in oim/oim cultures derived from both young adult and aged mice. The phenotype was more obvious in day 17 cultures when mature osteoblasts were enriched. B: Northern blot analysis for the expression of osteoblast differentiation markers of the RNA extracted from the MSC cultures. Reduced expression of type I collagen (Col1a1), bone sialoprotein (BSP), and osteocalcin (OC) was detected in all time points of cultures from both age groups of the oim/oim mice.
To further explore our observations in vivo, frozen decalcified sections from 3-month-old mice were prepared using the CryoJane tape transfer system, and the entire bone area was imaged and reconstructed as an intact bone section. The bones from the oim/oim;3.6GFP mice showed a significant increase of endocortical surfaces area covered with GFP-positive cells when compared with +/+;3.6GFP mice. In contrast GFP expression was similar between oim/oim;2.3GFP and +/+;2.3GFP mice (Figure 3A). Because the oim/oim mice have lower trabecular bone parameters, the quantification of the area of GFP-expressing cells was carried on the endocortical surface (Figure 3B). The increased area covered by 3.6GFP-positive cells in oim/oim mice did not proportionally progress into mature osteoblasts represented by 2.3GFP expression. These data indicate that the lineage is under continuous stimulation; however, defects in osteoblast differentiation could result in lower numbers of mature osteoblasts.
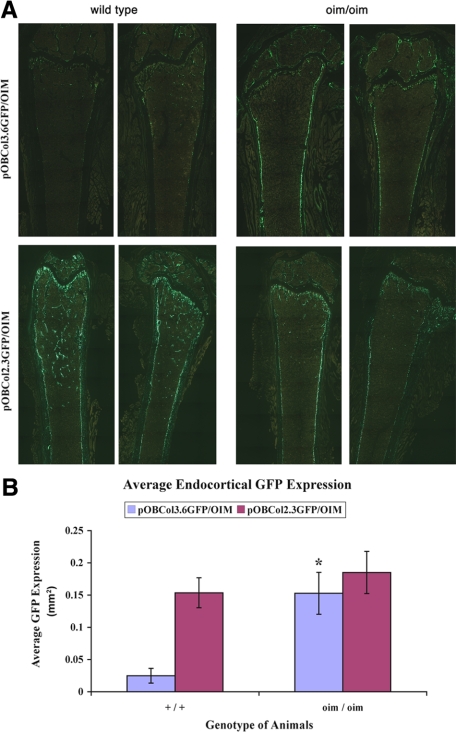
Figure 3.
Histological examination of GFP expression in vivo. A: Frozen sections of 3-month-old mouse femur. The upper panel shows 3.6GFP expression. Stronger and broader 3.6GFP expression can be detected on the endocortical surfaces of the femur of the oim/oim mice. The lower panel shows pOB2.3GFP expression. Note that the oim/oim mice have lower 2.3GFP expression in the trabecular area, whereas endocortical areas express GFP at similar levels in both +/+;2.3GFP and oim/oim;2.3GFP. B: Quantifications of the area of GFP-expressing cells in endocortical surfaces. Oim/oim;3.6GFP mice have a sixfold higher GFP expression area when compared with +/+;3.6GFP mice. No differences were detected in 2.3GFP expression on endocortical bone between +/+;2.3GFP and oim/oim;2.3GFP (*P < 0.05, n = 4 to 7).
Osteoblast Derived from Oim Mice Strongly Support Osteoclastogenesis
We previously reported that the oim/oim mice have increased bone turnover.10 To answer the question whether osteoblasts lineage cells derived from oim/oim mice can exhibit high osteoclastogenic support, we cocultured osteoblast lineage cells with bone marrow mononuclear osteoclast progenitors. In this experiment, osteoblasts derived from either +/+ or oim/oim mice were cocultured with bone marrow–derived nonadherent monocytes (Bmmc) obtained either from wild-type +/+ or oim/oim mice in the presence of Vitamin D. Significantly higher number of osteoclasts were formed when oim/oim osteoblasts were cocultured with either +/+ or oim/oim derived BMMCs (Figure 4, A and B). To examine the function of the osteoclasts formed in these cultures, an osteoclast resorption assay was used. We have evaluated the resorption pit area in precoated osteologic micro plates. Significantly larger resorption areas were observed in cultures where osteoclast progenitors from either genotype were cultured with oim/oim osteoblasts (Figure 4C). However, no differences were detected in osteoclast function between +/+ and oim/oim osteoclast progenitors when cocultured with osteoblasts derived from +/+ mice.
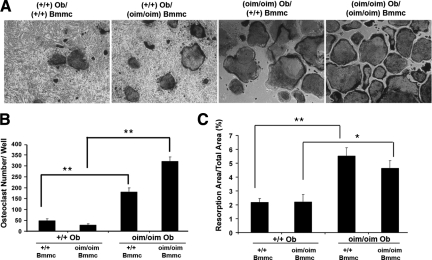
Figure 4.
Osteoclast formation and function in vitro from wild-type and oim mice. A: Representative images of cocultures of mouse calvaria osteoblasts (mCOB) with bone marrow–derived nonadherent cells (Bmmc) from +/+ or oim/oim mice. B: Quantifications of the Trap-positive multinucleated osteoclasts formed in these cultures. Significantly more osteoclasts were formed when the BMMC-derived osteoclast progenitors were cocultured with oim/oim mCOBs. C: Quantification of the area of osteoclast resorption pits in calcium precoated osteologic plates. A significantly greater change in resorption area was detected in cultures in which osteoclast formation was supported by oim/oim mCOB. Data are obtained from three independent biological experiments. *P ≤ 0.05, **P ≤ 0.01.
Immature Osteoblast Lineage Cells Exhibit Increased Osteoclastogenic Induction
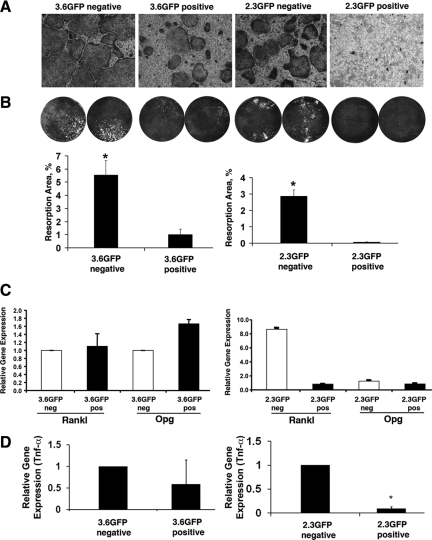
The osteoblast and osteoclast coculture experiments provided direct evidence that the oim/oim osteoblasts enchance osteoclastogenesis. We have also shown that the oim/oim osteoblasts have decreased differentiation in vitro and in vivo. Furthermore, analysis from the oim/oim;GFP/GFP mice indicated that this increased osteoclastogenesis may result from defects of oim/oim osteoblast lineage differentiation. To further pursue the potential mechanism of enhanced osteoclastogenesis, we hypothesized that different stages of osteoprogenitor cell differentiation have different abilities in supporting osteoclast formation and activity. To address this question, we isolated preosteoblasts and mature osteoblasts from cultures obtained from osteoblast lineage directed GFP marker mice. Cells obtained at different stages of maturation were cocultured with osteoclast progenitors derived from nontransgenic mice. Our results indicated that the population of 3.6GFP− cells that contains the most immature osteoblast progenitors exhibited the strongest support of osteoclast formation and function, whereas 2.3GFP+ mature osteoblasts showed the lowest level of osteoclastogenic support. These results are confirmed either by quantification of the TRAP staining–positive multinuclear cells (Figure 5A) or by the osteoclast resorption assay (Figure 5B).
Figure 5.
Osteoclastogenesis induced by osteoblast lineage cells at different stages of differentiation. A: Representative images of cocultures of bone marrow–derived osteoclast progenitor cells (BMMCs) from +/+ mice with FACS-sorted osteoblasts at different stages of osteoblast lineage differentiation. A significantly higher osteoclast number was observed when BMMCs were cocultured with a 3.6GFP− or 2.3GFP− cell population (earlier stages of osteoblast lineage) compared with GFP+ osteoblasts. The most mature osteoblast lineage cells (2.3GFP+) exhibited very limited potential to induce osteoclastogenesis. B: Images and quantification of the area of osteoclast resorption pits on precoated osteologic plates. Significantly more resorption areas were detected in the cultures where osteoclast progenitors were cocultured with GFP− cells compared with GFP+ osteoblasts. C: Quantative real-time PCR analysis of Rankl and Opg expression level in osteoblasts along the osteoblast lineage. A 2.3GFP− population has a 15-fold higher Rankl/Opg gene expression ratio when compared with the 2.3GFP+ cells. D: Lower levels of TNF-α expression were detected in 3.6GFP+ compared with a 3.6GFP− population. In addition TNF-α expression was much lower in the most mature cells of the osteoblast lineage (2.3GFP+) when compared with 2.3GFP− population. Data presented are obtained from two independent biological experiments derived from sorted population of osteoblasts (*P < 0.05).
Differentiation of osteoclast precursors is under the influence of factors produced by osteoblast lineage cells. The major regulators include Rankl, which positively affects differentiation of committed osteoclast precursors, and Opg, which can inhibit the process of osteoclastogenesis by binding to Rankl. TNF-α is an inflammatory cytokine that can exhibit osteoclastogenic potential. We have evaluated the expression of Rankl, Opg (Rankl/Opg gene expression ratio), and TNF-α using quantative real-time PCR analysis (Figure 5C). We observed a significantly higher Opg expression in 3.6GFP+ cells and no difference in Rankl expression between 3.6GFP− cells and 3.6GFP+ cells. Therefore, Rankl/Opg ratio was 50% higher in the 3.6GFP− population as compared with the 3.6GFP+ cells. The later stage of maturation, 2.3GFP+ cells exhibit a significantly lower expression levels of Rankl compared with 2.3GFP− cells, resulting in 15-fold higher ratio of Rankl/Opg in 2.3GFP− cells. Similar results were observed in the expression of TNF-α with a decrease in expression at the more mature stages of osteoblast differentiation (Figure 5D).
Interestingly the Rankl/Opg gene expression ratio in primary osteoblast cultures did not have a significant difference between the oim/oim and +/+ mice (Figure 6A). However, we have observed a threefold higher level of TNF-α expression in osteoblast cultures derived from oim/oim mice than in those derived from +/+ mice (Figure 6B). To test the potential role of increased TNF-α in osteoblast cell cultures derived from oim/oim mice, we have treated these cultures with TNF-α neutralizing antibody. A significant decrease in osteoclast number in cultures treated with TNF-α antibody was detected. These data are indicative of the active role of the TNF-α in the control of differentiation of the osteoclast lineage cells derived from oim/oim mice (Figure 6C).
Figure 6.
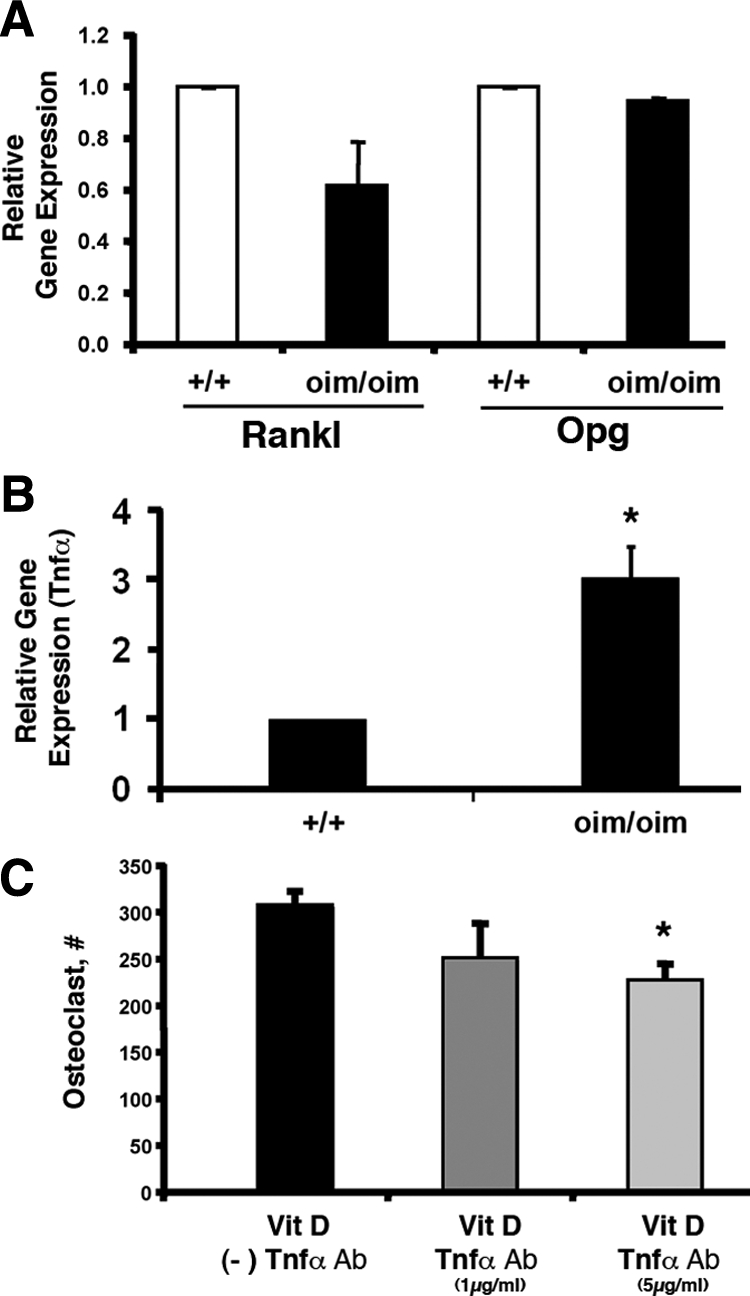
Expression of Rankl/Opg and TNF-α in cultured osteoblasts from OIM mice. A: Rankl/Opg ratio in samples derived from osteoblasts cultures. No significant difference was observed in a Rankl/Opg gene expression ratio between +/+ and oim/oim mice. B: TNF-α–relative gene expression was significantly higher in the oim/oim mice compared with +/+ controls. Data presented are obtained from three independent biological experiments. C: A representative experiment of coculture of oim/oim osteoblasts with or without addition of TNF-α–neutralizing antibody. Two concentrations were used, 1 μg/ml and 5 μg/ml along with 10−8 M vitamin D3 (*P < 0.01).
Discussion
The OI phenotype results from the accumulation of a defective bone matrix caused by underlying molecular mutation within the Col1A1 or Col1A2 genes. Recent studies suggest that high bone turnover is an important aspect of the pathophysiology of OI. This exaggerated process of bone remodeling may explain the clinical effectiveness of bisphosphonates in improving bone mass and decreasing fracture frequency. Although the success of the bisphosphonates in the treatment for the OI has been well documented and widely used, recent work by Uveges et al presented some of the potential negative effects of bisphosphonate treatment.19 This underlines the importance for better understanding the complexity of the bone cell responses (osteoblast and osteoclast) to the formation of a defective matrix.
The OIM model resembles the clinical and biochemical phenotype seen in severe type III of human OI and therefore has been widely used to study the pathophysiology of OI. Previous studies have characterized the phenotype of the oim/oim mouse by histomorphometric and molecular methods. Static histomorphometry showed lower values of trabecular and cortical bone parameters, whereas the bone formation rate was significantly increased with higher osteoblast surface per bone surface.10 Osteoblastic activity was significantly enhanced in oim/oim mice compared with wild-type mice when assessed by the Col3.6CAT (type I collagen promoter driven chloramphenicol acetyltransferase) expression in the oim/oim mouse. However, use of CAT transgenes does not allow for definition of the cell population expressing the transgene and, therefore, this model cannot determine which stage of osteprogenitor cells is responsible for higher Col1a1 transgene expression.10 Our study addressed whether the osteoprogenitor cells in the setting of a continued high bone turnover and stimulated recruitment toward osteoblast lineage can reach the fully differentiated stage or whether this setting generates a high proportion of undifferentiated osteoblasts, resulting in a lower per cell production of bone matrix.
To address these questions we crossed the OIM with transgenic mice developed to identify distinct stages of osteoblastic differentiation. A visible marker (GFP) was used to generate transgenic mice in which fragments of rat type I collagen promoter are used to drive transgene expression to preosteoblast (pOBCol3.6GFP) while its expression in vivo decreases with differentiation. The 2.3-kb promoter expressed the GFP mRNA in the mature osteoblasts and osteocytes with no expression in earlier stages of differentiation. Our analysis confirmed that osteoprogenitor cells derived from oim/oim mice can attain a stage of preosteoblasts as evaluated by expression of 3.6GFP, whereas they exhibit a block in differentiation into mature osteoblasts in vitro that is represented by presence of very few 2.3GFP-positive colonies (mature osteoblasts). In vivo data using GFP transgenic mice confirmed this observation and corroborated the possibility of expansion of the cells at the less mature stage of osteoblast lineage differentiation causing increased osteoblast surface in the oim/oim mice. To evaluate the maturation of osteoblast lineage cells in vivo we have also analyzed the gene expression of osteocalcin, bone sialoprotein, and osterix in bone samples derived from neonatal calvaria. Use of the calvarial bones avoids the potential differences in degree of contaminants (bone marrow and periosteum including ligaments) present in extracting the RNA from the long bone samples. In the samples of parietal bones we have observed significantly lower expression of osteocalcin in the oim/oim compared with wild-type mice (supplemental Figure S1, see http://ajp.amjpathol.org) No changes in the expression of BSP and Osterix were detected, indicating a block in the osteogenic differentiation process into the terminal stage of mature osteoblast.
The explanation for the apparent discrepancy in the activity of the reporters from an in vitro to an in vivo setting can be explained by the requirement for a critical number of mature osteoblasts to make a skeletal structure sufficient for survival. For this to happen in vivo, the animal must activate its osteogenic lineage to produce a sufficient number of 2.3GFP-expressing osteoblasts to make enough matrix to overcome the high rate of bone loss. Thus in vivo there is excessive 3.6GFP relative to normal bone, whereas there is less total bone but normal mature (2.3GFP) osteoblast cell density. In contrast, there is no need for the lineage to make mature osteoblasts for the cells to grow in culture. Additionally, the in vitro model is deprived from strong osteogenic signals provided by mechanical stimulation of bone cells in vivo.20 Instead, in vitro progenitors develop at a normal rate, but the inherent defect in maturation to a 2.3GFP expressing osteoblast is revealed. This altered relationship between progenitor and mature cell is seen in other disease of high turnover especially in hemolytic anemia of multiple causes.21
Pediatric OI disease (type III and IV) exhibits high bone turnover, increased osteoblast and osteoclast surfaces, increased bone formation rate, and decreased mineral apposition rate resembling OIM phenotype except for mineral apposition rate that was not decreased in OIM mice.1,10 In addition, osteoclastic activity is increased in the children with OI as well as in the OIM mice. Therefore we aim to test whether the osteoblast lineage that does not achieve a stage of full differentiation could explain enhanced induction and activity of osteoclast lineage causing high bone turnover. Cocultures of osteoblast lineage cells derived from oim/oim with bone marrow–derived mononuclear cells confirmed the ability of oim/oim osteoblasts to induce formation of greater number of osteoclasts. Osteoclast precursor differentiation and osteoclastic activity are regulated through the expression of Rankl and Opg by osteoblast lineage cells. Use of Col1a1 transgenes (pOBCol3.6GFP and pOBCol2.3GFP) enabled us to determine the expression of these factors at different stages of the lineage maturation. This was a critical element to evaluate functional interactions between osteoblast and osteoclast lineages that occur mainly through osteoblast production of osteoclast-regulating factors.22 Our results revealed that less mature osteoblast lineage cells exhibit increased osteoclast inductive potential. Furthermore, Rankl/Opg ratios were much higher in less mature GFP− cells than in corresponding GFP+ cells (3.6GFP− versus 3.6GFP+ and 2.3GFP− versus 2.3GFP+). The most mature population of osteoblast lineage cells (2.3GFP+) exhibits the lowest level of osteoclastogenic support paralleled by 15-fold lower Rankl/Opg ratio compared with 2.3GFP− population. Based on those results and in line with the oim/oim osteoblasts defective maturation, we expected the oim/oim osteoblasts to have increased Rankl/Opg ratio. However, we did not observe a difference in Rankl/Opg ratio between the osteoblast cultures derived from oim/oim and +/+ mice. In addition to Rankl, other factors, specifically inflammatory cytokines such as TNF-α and IL-1, can stimulate osteoclastogenesis.23,24 This mechanism may be relevant in the OIM model because oim/oim mice can develop spontaneous fractures that, because of the tissue injury, are followed by inflammation and proinflammatory cytokine production.25 Defective collagen present in osteogenesis imperfecta may also trigger the secretion of inflammatory cytokines.19 This hypothesis is supported by the finding of approximate threefold increase of TNF-α expression in oim/oim osteoblasts compared with +/+ osteoblasts. In addition, we also found that less mature GFP− populations in wild-type mice have increased TNF-α expression compared with corresponding GFP+ populations. Because OIM is marked by the block in osteoblast differentiation, accumulation of immature osteoblast lineage cells may induce changes in bone cellular microenvironment toward enhanced osteoclast differentiation.19 Our experiments using TNF-α antibody showed a partial block of the osteoclastogenesis in the coculture system. Therefore, the higher number and enhanced resorptive activity of osteoclasts formed in coculture with oim/oim osteoblasts can be explained by the synergistic effect of TNF-α, highly expressed by immature osteoblasts, along with Rankl and M-CSF also produced by osteoblasts.19,26 In this context it is likely that imbalance in bone cell number and functioning will occur, marked by poorly differentiated osteoblasts and highly activated osteoclasts, leading to enhanced osteoresorption in OI.11 Further investigations should reveal intracellular mechanisms of the synergistic action of TNF-α and Rankl in enhanced osteoclastogenesis27 in OI. Future studies should also aim at evaluating the effects of blocking TNF-α on induction of osteoclasts using in vivo animal models.
Footnotes
Address reprint requests to Ivo Kalajzic, M.D., Ph.D., Department of Reconstructive Sciences, MC 3705, University of Connecticut Health Center, 263 Farmington Ave., Farmington, CT 06032. E-mail: ikalaj@neuron.uchc.edu.
Supported by grants from the Children Brittle Bone Foundation, National Institutes of Health, NIAMS (R03-AR053275), and Institutional support to I.K. through NIH-5U24-DE016495-02.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Rauch F, Travers R, Parfitt AM, Glorieux FH. Static and dynamic bone histomorphometry in children with osteogenesis imperfecta. Bone. 2000;26:581–589. doi: 10.1016/s8756-3282(00)00269-6. [DOI] [PubMed] [Google Scholar]
- Baron R, Gertner JM, Lang R, Vignery A. Increased bone turnover with decreased bone formation by osteoblasts in children with osteogenesis imperfecta tarda. Pediatr Res. 1983;17:204–207. doi: 10.1203/00006450-198303000-00007. [DOI] [PubMed] [Google Scholar]
- Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Friedenstein AJ. Marrow stromal fibroblasts, Calcif Tissue Int. 1995;56:S17. [Google Scholar]
- Aubin JE. Bone stem cells, J Cell Biochem Suppl. 1998;30–31:73–82. [PubMed] [Google Scholar]
- Dacic S, Kalajzic I, Visnjic D, Lichtler AC, Rowe DW. Col1a1-driven transgenic markers of osteoblast lineage progression. J Bone Miner Res. 2001;16:1228–1236. doi: 10.1359/jbmr.2001.16.7.1228. [DOI] [PubMed] [Google Scholar]
- Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- Kalajzic I, Staal A, Yang WP, Wu Y, Johnson SE, Feyen JH, Krueger W, Maye P, Yu F, Zhao Y, Kuo L, Gupta RR, Achenie LE, Wang HW, Shin DG, Rowe DW. Expression profile of osteoblast lineage at defined stages of differentiation. J Biol Chem. 2005;280:24618–24626. doi: 10.1074/jbc.M413834200. [DOI] [PubMed] [Google Scholar]
- Kalajzic I, Terzic J, Rumboldt Z, Mack K, Naprta A, Ledgard F, Gronowicz G, Clark SH, Rowe DW. Osteoblastic response to the defective matrix in the osteogenesis imperfecta murine (oim) mouse. Endocrinology. 2002;143:1594–1601. doi: 10.1210/endo.143.5.8807. [DOI] [PubMed] [Google Scholar]
- Zhang H, Doty SB, Hughes C, Dempster D, Camacho NP. Increased resorptive activity and accompanying morphological alterations in osteoclasts derived from the oim/oim mouse model of osteogenesis imperfecta. J Cell Biochem. 2007;102:1011–1020. doi: 10.1002/jcb.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer LC, Khosla S, Dunstan CR, Lacey DL, Boyle WJ, Riggs BL. The roles of osteoprotegerin and osteoprotegerin ligand in the paracrine regulation of bone resorption. J Bone Miner Res. 2000;15:2–12. doi: 10.1359/jbmr.2000.15.1.2. [DOI] [PubMed] [Google Scholar]
- Roodman GD. Regulation of osteoclast differentiation. Ann NY Acad Sci. 2006;1068:100–109. doi: 10.1196/annals.1346.013. [DOI] [PubMed] [Google Scholar]
- Chipman SD, Sweet HO, McBride DJ, Jr, Davisson MT, Marks SC, Jr, Shuldiner AR, Wenstrup RJ, Rowe DW, Shapiro JR. Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: a model of human osteogenesis imperfecta, Proc Natl Acad Sci USA. 1993;90:1701–1705. doi: 10.1073/pnas.90.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saban J, King D. PCR genotyping of oim mutant mice. Biotechniques. 1996;21:190. doi: 10.2144/96212bm03. 192. [DOI] [PubMed] [Google Scholar]
- Jiang X, Kalajzic Z, Maye P, Braut A, Bellizzi J, Mina M, Rowe DW. Histological analysis of GFP expression in murine bone. J Histochem Cytochem. 2005;53:593–602. doi: 10.1369/jhc.4A6401.2005. [DOI] [PubMed] [Google Scholar]
- Wang YH, Liu Y, Buhl K, Rowe DW. Comparison of the action of transient and continuous PTH on primary osteoblast cultures expressing differentiation stage-specific GFP. J Bone Miner Res. 2005;20:5–14. doi: 10.1359/JBMR.041016. [DOI] [PubMed] [Google Scholar]
- Wong GL, Cohn DV. Target cells in bone for parathormone and calcitonin are different: enrichment for each cell type by sequential digestion of mouse calvaria and selective adhesion to polymeric surfaces. Proc Natl Acad Sci USA. 1975;72:3167–3171. doi: 10.1073/pnas.72.8.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uveges TE, Collin-Osdoby P, Cabral WA, Ledgard F, Goldberg L, Bergwitz C, Forlino A, Osdoby P, Gronowicz GA, Marini JC. Cellular mechanism of decreased bone in Brtl mouse model of OI: imbalance of decreased osteoblast function and increased osteoclasts and their precursors. J Bone Miner Res. 2008;23:1983–1994. doi: 10.1359/JBMR.080804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam I, Mantila SM, Gluhak-Heinrich J, Bellido TM, Harris SE, Turner CH. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J Biol Chem. 2008;283:5866–5875. doi: 10.1074/jbc.M705092200. [DOI] [PubMed] [Google Scholar]
- Kremer JP, Datta T, Pretsch W, Charles DJ, Dormer P. Mechanisms of compensation of hemolytic anemia in a lactate dehydrogenase mouse mutant. Exp Hematol. 1987;15:664–670. [PubMed] [Google Scholar]
- Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40:251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Lam J, Takeshita S, Barker JE, Kanagawa O, Ross FP, Teitelbaum SL. TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J Clin Invest. 2000;106:1481–1488. doi: 10.1172/JCI11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaura H, Sands MS, Aya K, Zhou P, Hirayama T, Uthgenannt B, Wei S, Takeshita S, Novack DV, Silva MJ, Abu-Am Y, Ross FP, Teitelbaum SL. Marrow stromal cells and osteoclast precursors differentially contribute to TNF-alpha-induced osteoclastogenesis in vivo. J Immunol. 2004;173:4838–4846. doi: 10.4049/jimmunol.173.8.4838. [DOI] [PubMed] [Google Scholar]
- Kon T, Cho TJ, Aizawa T, Yamazaki M, Nooh N, Graves D, Gerstenfeld LC, Einhorn TA. Expression of osteoprotegerin, receptor activator of NF-kappaB ligand (osteoprotegerin ligand) and related proinflammatory cytokines during fracture healing. J Bone Miner Res. 2001;16:1004–1014. doi: 10.1359/jbmr.2001.16.6.1004. [DOI] [PubMed] [Google Scholar]
- Kitaura H, Zhou P, Kim HJ, Novack DV, Ross FP, Teitelbaum SL. M-CSF mediates TNF-induced inflammatory osteolysis. J Clin Invest. 2005;115:3418–3427. doi: 10.1172/JCI26132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Yao Z, Li F, Zhang Q, Badell IR, Schwarz EM, Takeshita S, Wagner EF, Noda M, Matsuo K, Xing L, Boyce BF. NF-kappaB p50 and p52 regulate receptor activator of NF-kappaB ligand (RANKL) and tumor necrosis factor-induced osteoclast precursor differentiation by activating c-Fos and NFATc1. J Biol Chem. 2007;282:18245–18253. doi: 10.1074/jbc.M610701200. [DOI] [PubMed] [Google Scholar]