Abstract
Basal cell carcinoma (BCC) is the most common skin malignancy encountered worldwide. We hypothesized that CXC chemokines, small cytokines involved in inducing directed leukocyte chemotaxis, could play a key role in the modulation of BCC growth. In this study, quantitative RT-PCR revealed that the chemokines CXCL9, 10, 11, and their receptor CXCR3 were significantly upregulated by an average 22.6-fold, 9.2-fold, 26.6-fold, and 4.9-fold, respectively in BCC tissue samples as compared with nonlesional skin epithelium. Immunohistochemistry analysis revealed that CXCR3, CXCL10, and CXCL11, but not CXCL9, colocalized with cytokeratin 17 (K17) in BCC keratinocytes. In addition, CXCR3 and its ligands were expressed in cells of the surrounding BCC stroma. The chemokines and K17 were also expressed in cultured human immortalized HaCaT keratinocytes. Exposure of HaCaT cells or primary BCC-derived cells to CXCL11 peptides in vitro significantly increased cell proliferation. In primary BCC-derived cell cultures, addition of CXCL11 progressively selected for K17+/CXCR3+ co-expressing cells over time. The expression of CXCR3 and its ligands in human BCC keratinocytes, the enhancement of keratinocyte cell proliferation by CXCL11, and the homogeneity of K17+ BCC cells in human BCC-isolated cell population supported by CXCR3/CXCL11 signaling all suggest that CXCR3 and its ligands may be important autocrine and/or paracrine signaling mediators in the tumorigenesis of BCC.
Basal cell carcinoma (BCC), a type of nonmelanoma skin cancer, is the most prevalent neoplasm found in the population.1,2,3 Approximately one-third of the United States population developed nonmelanoma skin cancer from 1994 to 2002, and 83% of these patients presented with BCC.1,2,3 The BCC frequency in regional populations is increasing at a rate of 2% to 19% per year, with an annual rate increase of 4% in Canada.4,5,6 There are approximately 800,000 new cases of BCCs diagnosed in the United States and 70,000 in Canada each year.7 The age-adjusted incidence per 100,000 individuals is in the region of 100 to 2000 for men and 80 to 1500 for women, depending on the geographical location and specific genetic background of the individuals studied.8,9,10,11,12 The rising rate of BCC incidence is likely attributable to a combination of improved diagnosis and reporting, increased longevity, increased sun exposure, changes in clothing style, and increased UV radiation intensity due to ozone depletion.13,14
BCC tumors are composed of proliferating keratinocytes that derive from the basal layers of the epidermis and/or hair follicle bulge.15,16 There are several BCC subtypes, of which the most common one is nodular, followed by superficial, and morpheiform/infiltrating BCCs.17,18 Although BCCs rarely metastasize (rate of metastasis ranges from 0.003% to 0.55%) and cause death, they can result in significant patient morbidity.2,17 Because this cancer typically affects sun-exposed skin of the head and neck, cosmetic disfigurement is common. BCCs can behave aggressively with deep invasion, recurrence, and resistance to standard treatment. The presence of BCC is also associated with increased risk of developing other BCCs or skin cancers, such as malignant melanoma and squamous cell carcinoma.2,17
The fundamental initiation of BCC development is typically due to mutation of genes in the sonic hedgehog (Shh) signaling pathway.19,20,21 Mutation of the Shh pathway associated genes, SHH, PTCH, SMO, GLI1, or GLI2, can promote BCC tumorigenesis.21,22,23,24,25,26 Such genomic alteration has been found in >70% of BCC cases studied.19,20,21 SHH is a morphogene, and it is essential for the development of various organs including the skin and hair follicles.19,22 Shh plays a key role in regulating stem cell populations,22 and mutations in genes of this signaling pathway can result in inhibition of epithelial cell cycle arrest, neoplasia, and perturbation of other associated cell-cycle events.22,26,27
In addition to genetic defects, depletion of host immune responses and the development of a more permissive tissue environment may act as pivotal forces for BCC progression and transformation.2,28 Distinct epithelial-stromal-inflammatory patterns have been identified and correlated with specific BCC subtypes and tumor progression.28 However, the mechanisms that underlie these BCC tumorigenesis-enabling events are still not known, and the molecular profile of the immune response during BCC development has not been fully established.
The CXC chemokines, CXCL9, 10, and 11 are IFNγ–induced small secretory proteins that are synthesized and released by leukocytes, as well as epithelial, endothelial, and stromal cells.29 They interact with the heptahelical G protein complex receptor CXCR3 and exert signaling effects in a paracrine or autocrine fashion.30,31 They have been well established as chemoattractive for activated CXCR3+ T cells.29 Recently, increased expression of these chemokines, together with their receptor CXCR3, have been found to be associated with advanced-stage tumors, such as malignant melanoma, ovarian carcinoma, and B-cell lymphoma.32,33,34 In addition, previous literature has demonstrated that IFNγ-induced production of CXCL9, 10, and 11 in human neonatal foreskin keratinocytes is enhanced during inflammatory dermatoses such as psoriasis.35,36,37 However, little is known about the roles and mechanisms played by these CXCR3 ligands in the immunoregulation of other skin diseases and cancers.
Previously, our laboratory conducted a detailed microarray-based analysis of genes in superficial, nodular, and morpheiform BCC tissues to detect specific gene sets with significant differential expression as compared with normal skin.38 With further examination of gene ontology data from this study (Gene Expression Omnibus database series record GSE6520), we subsequently identified 27 immunoregulatory genes with significant differential expression in BCCs including several CXC chemokines and their receptors (unpublished data). We hypothesized that the upregulation of CXCR3 and its ligands could be a key BCC growth promotion event. In this study, we investigated the potential impact of CXCR3/ ligand signaling on BCC development. Our findings suggest that the chemokines CXCL9, 10, and 11 are potent signaling mediators via CXCR3 for BCC keratinocyte proliferation, and may be significantly involved in the regulation of BCC tumor development.
Materials and Methods
Tissues
BCC tumors and nonlesional skin samples were provided by the Moh’s Surgery Clinic, Department of Dermatology and Skin Science, the University of British Columbia. Tissue collection was approved by the University Clinical Research Ethics Board and according to the Declaration of Helsinki Principles. Specific patient consent was not required as tissues excised during surgery are considered as discarded materials according to Canadian law. In total, 31 human BCC tumor samples from 18 male (mean age 63.6 ± 16.2 years) and 13 female patients (mean age 62.2 ± 11.7 years) were used along with 13 nonlesional skin epithelium tissues excised from 6 male (mean age 53.7 ± 4.8 years) and 7 female patients (mean age 66.5 ± 15.6 years).
HaCaT Cell Culture
For initial studies, human immortalized HaCaT keratinocyte cells were maintained at 37°C in a humidified incubator with 5% CO2. They were propagated in Dulbecco’s Modified Eagle’s Medium (DMEM; Fisher Scientific, Ottawa, Ontario) with 5% fetal bovine serum (Fisher), 100 U/ml penicillin (Invitrogen, Burlington, Ontario), and 100 μg/ml streptomycin (Invitrogen). When the cells reached 80% confluence, they were transferred to 24-well plates for experimental assays.
Human BCC Cell Isolation and Culture
Human nodular BCC tissues from three patients were washed in Ca2+ and Mg2+-free Dulbecco’s phosphate-buffered saline (DPBS), cut into 2 mm3 pieces, and incubated with 25.0 caseinolytic units/ml dispase (Invitrogen) containing 1% antibiotic-antimycotic solution (Invitrogen) at 4°C overnight. Tissues were further incubated with 0.05% trypsin-EDTA at 37°C for 15 minutes, and a single cell suspension was obtained using a 40-μm cell strainer. The trypsin activity was stopped by adding an equal volume of 10 mg/ml soybean trypsin inhibitor (Invitrogen). After centrifugation and cell resuspension, the cells were seeded in flasks or multiwell-plates coated with collagen I (Invitrogen). Complete growth medium was prepared by mixing base medium M154 with the Human Keratinocyte Growth Supplement, 0.08 mmol/L CaCl2, 100 units Penicillin, 100 μg Streptomycin, and 0.25 μg/ml Amphotericin B (all Invitrogen).
CXCL11 Peptide Treatment
HaCaT cells were seeded at 4 × 104 cells in 24-well plates to which CXCL11 peptide was added (Cat. No. 300-46; Peprotech Inc., Rocky Hill, NJ) at 1 nmol/L, 5 nmol/L, 10 nmol/L or 20 nmol/L dissolved in phosphate-buffered saline (PBS) in triplicate wells. As a control, the same amount of PBS alone was added to the cell cultures. A minimum of 8 × 104 human BCC cells from each tissue sample were aliquoted for cytospin preparations on day 0 as controls for immunohistochemistry; the remainder were plated in 24-well plates (>2.9 × 104 cells per well). The seeded cells were treated with 0, 5, 10, or 20 nmol/L of CXCL11 peptide. All cell cultures were incubated at 37°C with 5% CO2. Cell populations from each tissue sample were investigated separately.
Cell Proliferation Evaluation
Cell samples from each tissue sample exposed to each CXCL11 concentration were counted from three wells for each time-point, with evaluation repeated three times. Cell numbers were calculated as mean values ± SE of three independent experiments. Differences in cell numbers between the untreated and treated HaCaT cells were analyzed by Student t test, and a P value <0.05 was considered as statistically significant. Cell growth rate of BCC cells was calculated by dividing the number of treated cells at each time point by the cell number originally seeded on day 0. Values higher than 1 indicated a cell population increase, whereas a value between 0 and 1 indicated a reduction of cell numbers. The BCC cell growth rates of all of the treatment set-ups were compared with each other, and the statistical significance was analyzed by analysis of variance with a P value of 0.05.
Total RNA Isolation and cDNA Synthesis
For gene expression analysis, seven nodular, five superficial, and five morpheiform BCCs were excised and preserved in a RNA stabilization reagent (Qiagen, Mississauga, Ontario) and storage at −80°C before use. Samples were removed from the stabilization reagent, and homogenized using a QIAshredder kit (Qiagen) according to the manufacturer’s instructions for total RNA isolation. Total RNA of the lysates was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. DNase digestion was conducted using RNase-Free DNase Set (Qiagen) after the first buffer washing step of the RNA-bound RNeasy membrane. Total RNA was subsequently eluted in a volume of 25 μl RNase-free water.
The synthesis of cDNA was performed using the SuperScript First Strand Synthesis System for RT-PCR kit (Invitrogen). Briefly, 500 ng of each RNA sample was reverse transcribed in the presence of 50 ng/μL random hexamers, 10 mmol/L dNTP mix, 10× RT buffer, 25 mmol/L MgCl2, 0.1M DTT, and RNaseOUT recombinant ribonuclease inhibitor. The synthesis of cDNA reactions was catalyzed by SuperScript II reverse transcriptase at 42°C for 50 minutes, and was terminated at 70°C for 15 minutes. Any remaining RNAs were removed by incubating samples with RNase H at 37°C for 20 minutes. Reactions were programmed and performed in a Mastercycler Gradient (Eppendorf, Mississauga, Ontario).
Quantitative RT-PCR
Quantitative RT-PCR (qPCR) was performed using an MJ Research DNA Engine Opticon real-time cycler (Bio-Rad Laboratories, Mississauga, Ontario) using the QuantiTect SYBR Green RT-PCR kit (Qiagen). All reactions were performed in duplicate. The sequences of synthesized primers (Invitrogen) were obtained by using the online software Primer3 at http://frodo.wi.mit.edu/cgi-bin/primer3_www.cgi,38 and their specificities were determined by the online Blast program from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) and compared with the GenBank database. The forward (f) and reverse (r) primer sequences for each gene are as follows: CXCL11 f primer 5′-AGAGGACGCTGTCTTTGCAT-3′, r primer 5′- TGGGATTTAGGCATCGTTGT; XCL1 f primer 5′-GACTGCCGGTTAGCAGAATC-3′, r primer 5′-GTTGGCTTGGTCTGGATCAT-3′; CXCL9 f primer 5′-TTTTCCTCTTGGGCATCATC-3′, r primer 5′-GAACAGCGACCCTTTCTCAC-3′; CXCL10 f primer 5′-CCAATTTTGTCCACGTGTTG-3′, r primer 5′-TTCTTGATGGCCTTCGATTC-3′; CXCL14 f primer 5′-AAGCTGGAAATGAAGCCAAA-3′, r primer 5′-GGCGTTGTACCACTTGATGA-3′; CXCL4 f primer 5′-GCTGTTCCTGGGGTTGCT-3′, r primer 5′-CCAAAAGTTTCTTAATTATTTTCTTGT-3′; CXCL16 f primer 5′-TACACGAGGTTCCAGCTCCT-3′, r primer 5′-CACAATCCCCGAGTAAGCAT-3′; CXCL12 f primer 5′-AGAGCCAACGTCAAGCATCT-3′, r primer 5′-CTTTAGCTTCGGGTCAATGC-3′; CXCL5 f primer 5′-TCTGCAAGTGTTCGCCATAG-3′, r primer 5′-TTGTTTCCACCGTCCAAAAT-3′; XCR1 f primer 5′-AGCTGGGGTCCCTACAACTT-3′, r primer 5′-GACCCCCACGAAGACATAGA-3′; CXCR6 f primer 5′-CAGATGCCCTTCAACCTCAT-3′, r primer 5′-GGCTGACAAAGGCATAGAGC-3′; CXCR4 f primer 5′-GGTGGTCTATGTTGGCGTCT-3′, r primer 5′-CTCACTGACGTTGGCAAAGA-3′; CXCR3 f primer 5′-GTGGACATCCTCATGGACCT-3′, r primer 5′-CGGAACTTGACCCCTACAAA-3′; CXCR3-A f primer 5′-TGGTCCTTGAGGTGAGTGAC-3′, r primer 5′-AAGAGGAGGCTGTAGAGGGC-3′; CXCR3-B f primer 5′-AAGTACGGCCCTGGAAGACT-3′, r primer 5′-GGCGTCATTTAGCACTTGGT-3′; CXCR3-alt f primer 5′-TACAACTTCCCACAGGGGTC-3′, r primer 5′-CTCACAAGCCCGAGTAGGAG-3′; IL8Rβ f primer 5′-ACTTTTCCGAAGGACCGTCT-3′, r primer 5′-GTAACAGCATCCGCCAGTTT-3′; IL8 f primer 5′-GTGCAGTTTTGCCAAGGAGT-3′, r primer 5′-AAATTTGGGGTGGAAAGGTT-3′.
The relative quantization of each targeted gene was normalized to the 18S expression level in the samples and was determined by the comparative CT method. Gene expression levels are presented in terms of fold change using the formula 2−ΔΔCt. Each tissue sample was examined separately and analyses conducted in duplicate. Data are presented as the mean of individual samples within each tissue group. Statistical analysis was done by Student t test with a P value <0.05 considered as significant.
Immunohistochemistry
For immunohistochemistry (IHC), 11 BCC tumor and 6 nonlesional skin samples were embedded in Optimal Cutting Temperature (OCT) compound (Sakura, Torrance, CA), frozen in liquid nitrogen, and stored at −80°C before use. Frozen samples were processed by cryostat sectioning, air drying, and fixation in acetone at 4°C. Single color labeling for CXCL9, 10, 11, or CXCR3 was performed using the ABC System (Vector Laboratories, Burlington, Ontario). Dual labeling was similarly conducted for a specific BCC keratinocyte marker, cytokeratin 17 (K17) together with the CXCL9, 10, 11, or CXCR3 primary antibodies.
In brief, after washing the sections in PBS, endogenous peroxidase activities in the tissue samples were inhibited by incubating them in PBS with 0.3% H2O2 and 0.3% normal serum. The sections were treated with protein block serum-free solution (Dako, Mississauga, Ontario), followed by avidin/biotin blocking (Vector) and further treated with diluted normal blocking serum (Vector). Preliminary titration experiments indicated optimal dilutions for primary antibodies (data not shown). Monoclonal mouse anti-human K17 (Cat. no. sc-58726) (Santa Cruz Biotechnology, Santa Cruz, CA) was diluted 1:200 in DakoCytomation Antibody Diluent (Dako) with 5% normal serum, and was applied to the sections overnight at 4°C. After washes in Tris-buffered saline (TBS), the slides were exposed to diluted biotinylated secondary antibody (Vector) with 5% normal serum at room temperature. Subsequently, the labeling was revealed by peroxidase-based reaction using the Vectastain Elite ABC reagent and Vector NovaRed substrate solution (Vector).
For double labeling procedures, the sections were further rinsed in TBS and treated with protein block serum-free solution, avidin/biotin blocking reagent, as well as normal blocking serum again. The second primary antibodies were prepared in the antibody diluent (Dako) with 5% normal serum as follows; monoclonal mouse anti-human CXCL9 (Cat. no. MAB392; R&D Systems Inc., Minneapolis, MN) diluted 1:33, polyclonal goat anti-human CXCL10 (Cat. no. AF-266-NA; R&D Systems) diluted 1:200, CXCL11 (Cat. no. sc-34784; Santa Cruz) diluted 1:75, and CXCR3 (Cat. no. sc-9900; Santa Cruz) diluted 1:20. These primary antibodies were incubated with the sections overnight at 4°C, followed by treatment of diluted biotinylated secondary body with 5% normal serum at room temperature. The signals were detected by the alkaline phosphatase–based reaction of Vectastain ABC-AP reagent and Vector Blue substrate solution (Vector). Finally, the slides were dehydrated and mounted in Permount (Fisher).
For IHC analysis of cell cultures (both BCC cells and HaCaT cells), a sterilized glass coverslip was placed in each well, which was seeded with a minimum of 4 × 105 cells. In addition, BCC cells were used for cytospin preparations, in which 8 × 104 cells in 150 μl culture media were added in glass slide-loaded concentrator cuvettes and were centrifuged at 48.5 × g for 4 minutes. The cells were fixed with 4% v/v paraformaldehyde (Cedarlane Laboratories, Hornby, Ontario) in PBS and were permeabilized by 0.1% v/v triton X-100 (Cedarlane) in TBS. Subsequent IHC procedures were performed as per the double labeling protocol above.
Immunohistochemistry Analysis
Ten randomly selected areas of BCCs in dual IHC labeled tissue sections, which coexpressed the chemokines (labeled blue) and K17 (labeled red), were evaluated for the chemokines’ label intensity using a protocol modified from previous publications.39,40,41 Each area had an average size of 131.57 μm2 containing approximately two cells. Optical color density in BCC keratinocytes was calculated and compared with equivalent regions of cells in the BCC-adjacent stroma within the same images. The immuno-label color of the chemokine antibodies was isolated from other colors in the images using Adobe Photoshop CS2 software (Adobe Systems, Etobicoke, Ontario). The immuno-labeling intensities in positive cells were then quantified using the software Scion Image (Scion, Frederick, MD). The analysis was done by converting the label color intensity of the images to gray scale (256 levels). The average label intensity for each antibody was calculated as mean pixel optical density (i.e., the sum of gray values of all pixels within a selected area divided by the number of pixels).
For cytospin cell culture preparations, the numbers of cells labeled as K17+/CXCR3−, K17+/CXCR3+, and K17−/CXCR3+ were counted in six random fields of view for each IHC dual label slide for each BCC cell culture. The percentage of cells with each expression presentation in the total cell population observed was calculated. Statistical significance was evaluated by analysis of variance.
Results
mRNAs for CXCR3/Ligands Are Upregulated in BCCs
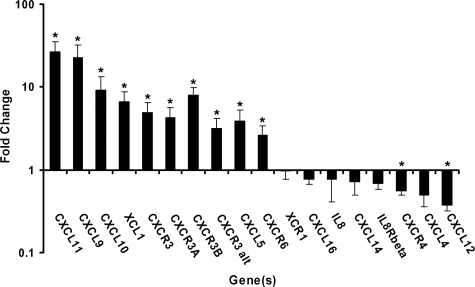
The expression levels of mRNA for 18 chemokine genes and their receptors in nodular, superficial, and morpheiform BCC tumors were analyzed by qPCR (Figure 1; Table 1). The expression levels for these chemokines in BCCs were compared with those in nonlesional skin epithelium samples. The analysis provides an overall indication of mRNA expression present in both keratinocytes and inflammatory cells in the tissue samples. Twelve of the 18 mRNAs exhibited significant differential expression (P value <0.05) in BCCs as compared with the controls. CXCL11, CXCL10, and CXCL9 were the 3 most upregulated genes in the tumor samples, and were increased by an average 26.6-fold, 9.2-fold, and 22.6-fold respectively. Their common receptor, CXCR3, was increased (4.9-fold) in the BCCs with statistical significance. Separate examination of specific CXCR3 isotypes CXCR3-A, CXCR3-B, and CXCR3-alternative (CXCR3-alt) revealed all types to be significantly upregulated in BCCs (4.3-fold, 8.0-fold, and 3.2-fold average, respectively).
Figure 1.
Real-time RT-PCR analysis of the chemokine expression levels in BCCs as compared with nonlesional skin epithelium. BCC tumors (seven nodular BCCs, five superficial BCCs, and five morpheiform BCCs) were studied. The gene expression levels in each BCC subtype were analyzed separately and were calculated in terms of fold change by using the 2−ΔΔCt equation. The average fold change values of the tissue samples in each of the three BCC subtypes are presented. Error bars represent the range factor difference (2−ΔΔCt−ΔCt SD and 2−ΔΔCt+ΔCt SD). Statistical significance was calculated by Student t test; *P < 0.05.
Table 1.
Expression Levels of Chemokines and Their Receptors in Nodular, Superficial, and Morpheiform BCCs
| All BCC
|
Nodular
|
Superficial
|
Morpheiform
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Fold change† | Range‡ | Fold change† | Range‡ | Fold change† | Range‡ | Fold change† | Range‡ | |
| Upregulation in mRNA expression | CXCL11 | 26.6* | ±6.5 | 35.5* | ±14.7 | 32.1* | ±11.8 | 15.4* | ±4.2 |
| CXCL9 | 22.6* | ±6.9 | 36.2* | ±17.1 | 16.5* | ±8.2 | 17.5* | ±5.9 | |
| CXCL10 | 9.2* | ±2.9 | 16.7* | ±7.7 | 9.9* | ±5.2 | 3.9* | ±1.4 | |
| XCL1 | 6.7* | ±1.6 | 12.2* | ±4.2 | 5.0* | ±1.4 | 3.6* | ±1.3 | |
| CXCR3 | 4.9* | ±1.2 | 7.1* | ±2.2 | 2.7 | ±1.1 | 5.1* | ±1.6 | |
| CXCR3-A | 4.3* | ±1.0 | 6.0* | ±2.3 | 3.3* | ±1.2 | 3.6* | ±1.3 | |
| CXCR3-B | 8.0* | ±1.5 | 9.3* | ±2.8 | 9.0* | ±2.7 | 6.0* | ±1.8 | |
| CXCR3-alt | 3.2* | ±0.7 | 4.4* | ±1.8 | 2.8* | ±0.9 | 2.4* | ±0.8 | |
| CXCL5 | 3.9* | ±1.0 | 4.8* | ±1.5 | 10.2* | ±5.2 | 1.0 | ±0.2 | |
| CXCR6 | 2.6* | ±0.6 | 2.7* | ±1.0 | 3.1* | ±1.2 | 1.8 | ±0.5 | |
| Downregulation in mRNA expression | XCR1 | 1.0 | ±0.2 | 0.7 | ±0.1 | 2.0 | ±0.6 | 0.9 | ±0.4 |
| CXCL16 | 0.8 | ±0.1 | 0.7 | ±0.1 | 0.7 | ±0.2 | 1.0 | ±0.2 | |
| IL8 | 0.8 | ±0.3 | 1.9 | ±1.2 | 0.9 | ±0.6 | 0.1 | ±0.1 | |
| CXCL14 | 0.7 | ±0.2 | 1.1 | ±0.4 | 1.3 | ±0.3 | 0.2 | ±0.1 | |
| IL8Rβ | 0.7 | ±0.1 | 0.6 | ±0.1 | 1.1 | ±0.3 | 0.6 | ±0.2 | |
| CXCR4 | 0.6* | ±0.1 | 0.6 | ±0.1 | 0.8 | ±0.1 | 0.3* | ±0.1 | |
| CXCL4 | 0.5 | ±0.1 | 0.4* | ±0.1 | 0.4 | ±0.2 | 0.8 | ±0.1 | |
| CXCL12 | 0.4* | ±0.1 | 0.3* | ±0.1 | 0.6 | ±0.2 | 0.3* | ±0.1 | |
Significant differential expression with a P value <0.05 by Student t test.
Fold change was evaluated using the 2−ΔΔCt equation.
The range of the fold change for each gene in BCCs relative to the nonlesional skin tissues was calculated by the formula ±2SD, where SD is the standard deviation of the ΔΔCt value.
Examination of differences in mRNA expression fold change associated with BCC subtypes revealed that all nodular, superficial, and morpheiform BCCs showed a uniform significant increase of the 12 upregulated genes (Table 1). In comparison with superficial and morpheiform BCCs, nodular BCCs exhibited the greatest levels of differential gene expression for CXCL11 at 35.46-fold, CXCL9 at 36.22-fold, and CXCL10 at 16.67-fold mean increase above control tissue levels. For the chemokines identified as down-regulated, there was a less consistent gene expression pattern associated with BCC subtypes (Table 1). However, statistically significant reductions in expression in some BCC subtypes were observed for CXCR4, CXCL4, and CXCL12.
CXCR3/Ligands Colocalize with K17 in BCC Tissues
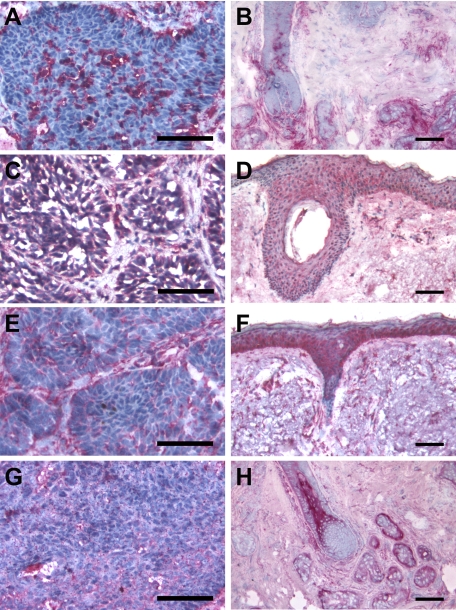
IHC serial staining of CXCL9, 10, 11, and CXCR3 indicated that these chemokines were more prevalent with wider distribution in nodular BCC tissue samples than in corresponding control skin tissues when processed in parallel (Figure 2, A–H). CXCL10, 11, and CXCR3 were mainly localized in the tumor masses, though some expression was detected in the surrounding stroma (Figures 2, C, E, and G). However, CXCL9 was largely not expressed specifically in the BCC tumor nests; its immuno-labeling was mainly distributed in the inflammatory infiltrate of the adjacent stromal region (Figure 2A).
Figure 2.
Serial immunohistochemistry of CXCL9, 10, 11, and CXCR3 in nodular BCCs and nonlesional skin. Five frozen sections of BCC tumors and normal skin tissues were analyzed for the protein expression patterns of CXCL9 (A and B, respectively), CXCL10 (C and D, respectively), CXCL11 (E and F, respectively), and CXCR3 (G and H, respectively). Positive labeling is indicated in red; the frozen sections were counterstained with hematoxylin (blue). Scale bar = 80 μm (A–H).
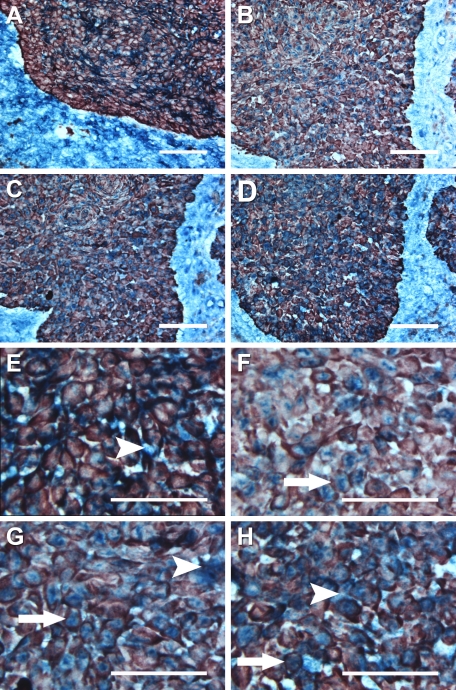
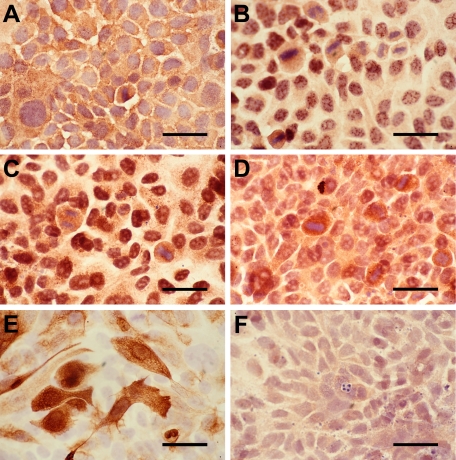
Double labeling of K17 and the chemokines using various BCC subtypes was subsequently performed (Figure 3). It has been shown that K17 is expressed in BCC subtypes, including nodular, superficial, and morpheiform BCCs,42,43,44,45,46 and it has been used as a histological diagnostic BCC marker.42 The co-expression patterns of the antibodies were consistent among the BCC subtypes. The IHC labeling revealed that K17 expression was positive in BCC keratinocytes and negative in non-follicular normal epithelium. K17 expression was localized close to the perimeter of the BCC cells consistent with a structural protein (Figure 3).
Figure 3.
Dual immunohistochemistry of K17 with CXCL9, 10, 11, and CXCR3 in nodular BCCs. Five frozen BCC biopsies were studied for the colocalization of K17 with CXCL9 (A, E), CXCL10 (B, F), CXCL11 (C, G), and CXCR3 (D, H). K17 is shown as red, whereas CXCR3 and its ligands are indicated as blue. Arrows indicate representative cells dual labeled with K17+ co-expression and CXCL10 (F), CXCL11 (G), or CXCR3 (H). Arrowheads indicate representative cells with CXCL9 (E), CXCL11 (G), or CXCR3 (H) labeling in the absence of K17 co-expression in the BCC tumor masses. Scale bar = 80 μm (A–D). Scale bar = 55 μm (E–H).
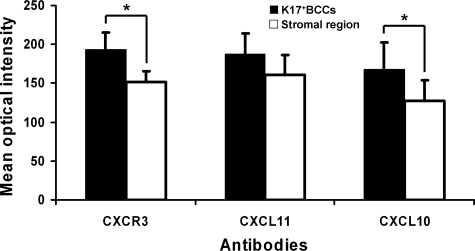
While CXCL9 was expressed by inflammatory cells in the interstitial area and stroma, CXCL9 was not expressed by K17+ BCC keratinocytes (Figure 3, A and E). In contrast, CXCL10 was positive in most K17+ BCC keratinocytes, and localized in the cell cytoplasm (Figure 3, B and F). Image analysis of label intensity demonstrated that K17+ BCC cells expressed a significantly higher level of CXCL10 than inflammatory cells (Figure 4). CXCL11 was present throughout the tissue sections with K17+ BCC cells exhibiting higher expression intensity than the inflammatory cells (Figures 3C and 4). CXCL11 was predominantly localized in cytoplasm of the keratinocytes, but some expression was detected in interstitial regions between the BCC cells and the stroma (Figure 3, C and G). CXCR3 was also labeled at a significantly higher intensity in K17+ BCC cells than in the inflammatory infiltrate (Figures 3D and 4). Unlike its chemokine ligands, CXCR3 was not only present throughout the whole cell, but was also detected at the BCC cell surface (Figure 3, D and H).
Figure 4.
Quantitative analysis of immunohistochemistry label intensities for CXCL10, 11, and CXCR3 in K17+BCC keratinocytes as compared with cells in the stromal area. The mean immuno-labeling intensities of the chemokines were evaluated using Scion Image software. The label intensities were derived from image pixel values (gray scale = 0 to 255) and are presented as mean optical intensity. In each of the 5 BCC biopsies, 10 randomly selected areas from each of the K17+ BCC cell nests and the stromal regions were measured and compared for each IHC label by Student t test. *P < 0.05.
CXCR3/Ligands and K17 Are Expressed by HaCaT Cells
Human immortalized HaCaT keratinocytes were examined by IHC analysis for K17, CXCR3, and its ligands. All of the factors evaluated were positively identified in the cells (Figure 5, A–F), though CXCL9 exhibited minimal expression. The localization of K17 was cytoplasmic as anticipated, though expression was variable from cell to cell. Cells overlying other cells were more likely to express K17 as compared with cells adherent to the glass substrate (Figure 5E). The presence of K17 was distributed with less coherence in the cultured HaCaT cells as compared with BCC keratinocytes (Figures 5E and 3).
Figure 5.
Immunohistochemistry analysis of CXCL9, 10, 11, CXCR3, and K17, in HaCaT cells. HaCaT cells were cultured on coverslips until 80% confluency. Cell fixation and permeabilization were conducted, followed by overnight incubation of primary antibodies against CXCL9 (A), CXCL10 (B), CXCL11 (C), CXCR3 (D), and K17 (E). HaCaT cells were counterstained with hematoxylin. The negative control was performed in parallel (F). Scale bar = 45 μm (A–F).
CXCL11 Enhances Proliferation of HaCaT Cells
Based on the observations of HaCaT cell expression of K17, CXCR3, and ligands above, an in vitro functional assay of CXCR3 ligand effect was performed with HaCaT cell cultures. Because previous qPCR analysis revealed that CXCL11 mRNA was expressed with greatest fold increase in BCC tissues, and given CXCL9, 10, and 11 all act through the same CXCR3 receptor,29,30,31 this study examined CXCL11-regulated HaCaT keratinocyte cell growth.
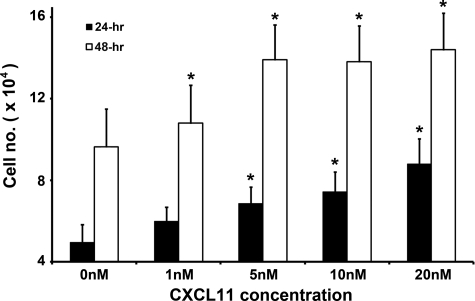
Growth rates of HaCaT cells incubated with 1 nmol/L, 5 nmol/L, 10 nmol/L, and 20 nmol/L of CXCL11 peptide for 24 and 48 hours were compared with that of nontreated cells. The keratinocytes proliferated in response to all concentrations of CXCL11 at both time points in a dose-dependent manner (Figure 6). HaCaT cells proliferated at a significantly faster rate as compared with controls (P < 0.05) at a minimum concentration of 5 nmol/L after 24-hour incubation. Significant difference in cell proliferation was observed with 1 nmol/L CXCL11 exposure after 48 hours (Figure 6). The 48-hour incubation yielded approximately double the number of cells as compared with the 24-hour time point (Figure 6).
Figure 6.
Growth rates of HaCaT cells incubated with CXCL11 for 24 or 48 hours. The y axis represents the mean cell numbers (± SE) of three individual experiments for each peptide concentration as shown on the x axis. Statistical significance of cell number increased relative to untreated controls (0 nmol/L) was calculated by Student t test. *P < 0.05.
Human BCC-Derived Cells Proliferated in Culture Media Supplemented with CXCL11 Peptide
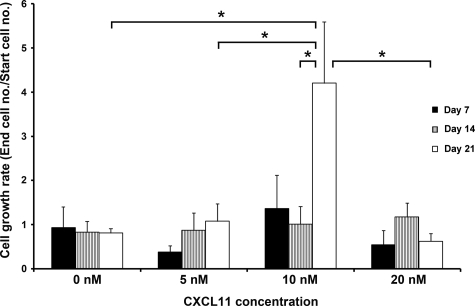
Single cell suspensions of human BCC-derived cells were treated with 0 nmol/L, 5 nmol/L, 10 nmol/L, and 20 nmol/L CXCL11 peptide for 7, 14, and 21 days. When no CXCL11 peptide was added to the culture media, cell numbers did not increase (Figure 7). When BCC cells were treated with 5 nmol/L CXCL11, the cell population exhibited a low, statistically insignificant, cell growth rate (Figure 7). With 10 nmol/L peptide treatment, increases in cell numbers at day 7 and day 14 were statistically insignificant, however on day 21 the cell growth rate was statistically significantly higher than with other peptide concentrations (Figure 7). Notably, the cell number was reduced when the concentration of CXCL11 was increased to 20 nmol/L, suggesting 10 nmol/L was the optimal concentration for BCC cell proliferation.
Figure 7.
Human BCC-derived cell proliferation with or without CXCL11. The y axis represents the cell growth rate, which was calculated by dividing the cell number at the end of the treatment by the cell number seeded initially. A value less than 1 indicates a reduction of the cell population, whereas a value greater than 1 represents the number of times more than the cell number started. The cell growth rates of all of the treatments were compared among each other over 7, 14, and 21 days. Statistical significance was evaluated by analysis of variance. *P < 0.05.
K17 and CXCR3 Were Detected in BCC Cell Cultures
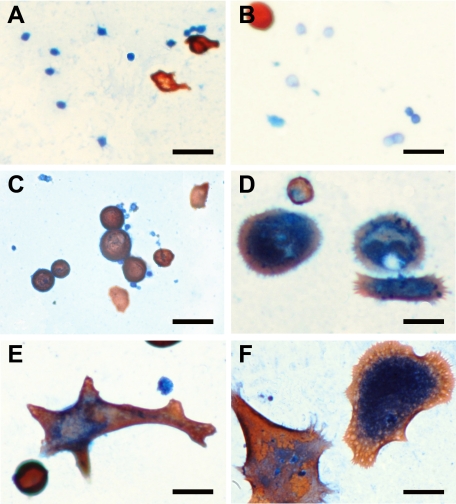
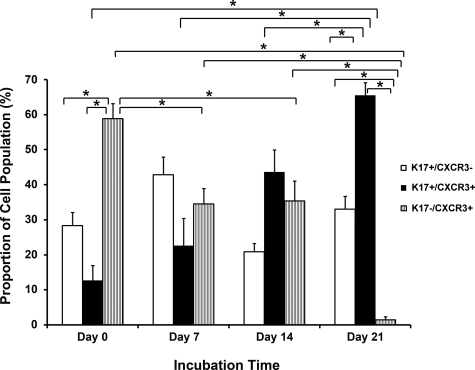
To characterize the BCC-derived cell cultures, cells incubated with 10 nmol/L CXCL11 were examined by dual IHC labeling for K17 and CXCR3 at day 0, 7, 14, and 21. The results revealed three different cell presentations; K17+/CXCR3−, K17+/CXCR3+, or K17−/CXCR3+ cells (Figures 8, A–F, and 9). On day 0, the proportion of K17−/CXCR3+ cells (58.9%) was significantly higher than the other two cell groups (Figure 9). On day 7, there was no significant difference among the cell numbers of the three marker groups. By day 14, K17+/CXCR3+ cells were more predominant (43.66%) in the cultures (Figures 8 and 9). On day 21, both K17+/CXCR3+ and K17+/CXCR3− cell groups were at significantly higher numbers (65.47% and 33.09%, respectively) than the K17−/CXCR3+ group, which was reduced to 1.44% of cells in the culture (Figures 8 and 9).
Figure 8.
Dual immunohistochemistry of K17 and CXCR3 in human BCC-derived cells incubated with CXCL11. The BCC cells were cultured with 10 nmol/L CXCL11 peptide and subsequently analyzed for the co-expression of K17 and CXCR3 at day 0 (A), 7 (B), 14 (C), and 21 (D–F). K17 labeling is indicated in red, whereas CXCR3 labeling is shown in blue. Scale bar = 50 μm (A–F).
Figure 9.
The proportion of cells presenting as K17+/CXCR3−, K17+/CXCR3+, K17−/CXCR3+ in the BCC cell population after incubation with CXCL11. Cells derived from human BCCs were cultured in the presence of 10 nmol/L CXCL11 and examined at day 0, 7, 14, and 21. The y axis represents the percentage of the three cell groups in the cell population at each time point. Statistical analysis was performed by analysis of variance. *P < 0.05.
Discussion
BCC Tumors Express CXCR3 and its Ligands
It has been shown that certain cytokines and chemokines, commonly associated with immune system activation, are involved in tumor cell growth and local immuno-suppression rather than exerting antitumor functions.32,33,34 As CXCL9, 10, and 11 have been suggested to play important roles in the development of other malignancies,29,32,34,47 we investigated the potential impact of chemokines, particularly the CXCR3 ligands, on the regulation of BCC etiopathology. In the current study, qPCR of 18 chemokines and their receptors in BCC tissues indicated significant increases in mRNA levels for selected chemokines. CXCL11, 9, and 10 exhibited the greatest fold increase in expression in BCCs, which led us to consider the possibility that these CXC chemokines may play a significant role in BCC tumorigenesis. We also observed significant reduced expression of CXCL12 and CXCR4 in BCCs. CXCL12–CXCR4 signaling has been shown to enhance the metastasis of solid tumors such as melanoma.48 The relative absence of CXCL12-CXCR4 expression may be consistent with the low rate of metastasis observed in BCCs.
CXCR3 and its Ligands are Coexpressed with K17 by BCC Keratinocytes
CXC chemokines of the superfamily of cytokines are characterized by four conserved cysteine residues.34,49 They bind to seven transmembrane, G-protein coupled receptors, and exert signaling effects on a wide spectrum of biological and physiological activities, including cell proliferation and cell migration.50 Their involvement in directing leukocyte migration has been extensively characterized.49,51,52 CXCL9, 10, and 11 are presumed to have antitumor functions attributable to their chemoattractive properties on CXCR3+ T cell migration into inflammatory and/or neoplastic sites.29,30 However, recent research has proposed that CXC chemokines and their receptors, particularly CXCR3 and its ligands CXCL9, 10, and 11, may participate in tumor progression and metastasis.49,51 CXCR3 and its ligands have been shown to enhance the progression of ovarian carcinoma and multiple myeloma,29,32,51,53 and to facilitate the metastasis of malignant melanoma.34,47,53,54 Also, on exposure to inflammatory cytokines, leukocytes typically lose their chemokine receptor expression.55,56 In the cases of malignant melanoma and ovarian carcinoma, CXCR3 is expressed at a much higher level in the tumor cells than in the infiltrating immunocompetent cells.34,47,53 Consequently, tumor cells may be relatively more receptive and responsive to chemokines, whether expressed by tumors or inflammatory cells.55
K17 is a marker of cell proliferation and follicular differentiation, and localizes in the lower hair follicle outer root sheath below the entry of the sebaceous duct.42,57,58 It has also previously been shown that K17 is expressed in nodular, superficial, morpheiform, micronodular, fibroepithelial, and nodulocystic BCCs.42,43,44,45,46,59 In this study, our IHC analysis reconfirmed that BCCs strongly expressed K17, consistent with K17 as a marker of keratinocyte cell proliferation, and a specific marker of BCC keratinocytes.
The dual IHC labeling indicated that CXCR3, CXCL10 and 11 proteins were represented at a high level in K17+ BCC cells but at a relatively low level in the stroma infiltrating inflammatory cells. The distribution and expression intensity patterns of these chemokines suggest that the BCC keratinocytes may respond to CXCR3/ligand signaling more readily and efficiently than the stromal infiltrating cells, and that the tumor keratinocytes may synthesize and use these chemokines to direct autocrine signaling effects for cell proliferation and migration. Thus, in BCCs, CXCR3 and its ligands may be involved in tumor promotion rather than anti-tumor functions as seen in some other malignancies.
Human BCCs Significantly Expressed all 3 CXCR3 Subtypes
The functional roles of CXCL9, 10, and 11 may depend on the subtype of CXCR3. There are at least 3 CXCR3 splice variants; CXCR3-A, CXCR3-B, and CXCR3-alt, which may be expressed at different levels depending on the observed cell type.60,61 CXCR3-A couples with Gαi to activate Erk1/2, p38/MAPK and PI3-kinase/Akt signaling pathways,50,62 and subsequently induces intracellular calcium influx, DNA synthesis, and cell proliferation (e.g., normal human bronchial epithelial cells, astrocytes, and glioma cells).50,62,63,64 CXCR3-B couples with Gαs to induce adenylyl cyclase, and results in inhibition of endothelial cell proliferation and migration61,65; this receptor subtype does not induce chemotaxis.61,62 CXCR3-alt co-expresses with CXCR3-A at a very low level (≈5% of CXCR3-A mRNA).50 It has been shown that all three variants are expressed in colorectal carcinoma, breast cancer, and multiple myeloma.61,66,67 Our qPCR results indicated that all CXCR3 splice variants were significantly upregulated in all of the BCC subtypes tested and that no strong bias in expression to a particular receptor variant was present. BCCs may use the signaling effects of these CXCR3 subtypes in a similar fashion as in other cancers.
CXCR3 and its Ligands may Modulate Tumor Growth
In this study, IHC revealed that CXCL9 proteins were present in cells of the interstitial area and stroma, but not in K17+ keratinocytes. Further, while CXCL10 and CXCL11 were present in K17+ keratinocytes, expression at a lower level was also distributed in cells surrounding the tumor nests. The distribution patterns of these 3 chemokines suggest inflammatory cells in the tumor tissue biopsies also express the CXCR3 ligands. It is possible that, in addition to autocrine activity within BCC tumors, chemokines produced by adjacent inflammatory cells may have a further paracrine mode of action on BCC keratinocytes.
CXCL11 Promotes Dose-Dependent HaCaT Cell Proliferation
Previous studies have used the HaCaT cell line to investigate BCC tumor formation and immunoregulation.68,69 The cells are spontaneously immortalized keratinocytes derived from human adult skin epithelium with UV-type specific mutations on both alleles of p53.19,70,71 HaCaT cells have been used extensively to study human skin carcinogenesis through overexpression or suppression of genes regulating cancer cell proliferation.19,72,73,74,75,76,77 In this study, HaCaT cells exhibited similar expression patterns of K17, CXCR3, and its ligands as observed in BCC keratinocytes. Our data demonstrated that CXCL11 significantly enhanced the proliferation rate of HaCaT cells in vitro. Such findings, combined with the positive IHC results for CXCR3 and its ligands in HaCaT cells, suggests that both exogenous and endogenously produced CXCL11 can interact with CXCR3+ HaCaT cells to activate cell growth pathways. Previous literature has suggested that the interaction of CXCL11 and CXCR3 can result in mitogenic effects and proliferation of human bronchial epithelial cells and pericytes through the Erk 1/2 and MAPK pathways.50,78
Human BCC-Derived Cells Require CXCL11 Supplementation for Survival and Proliferation in Vitro
To pursue the in vitro study of CXCR3-ligands in BCCs, we derived a modified BCC cell culture protocol from previous publications.79,80,81 We found that cells treated with 10 nmol/L CXCL11 peptide showed increased cell growth at all observed time points and achieved statistical significance on day 21. We observed that the primary BCC cells required up to 14 days to adhere to the substrate and begin forming colonies. Consequently, cell proliferation was delayed until substrate adherence was achieved. Notably, the BCC cells in the medium without CXCL11 peptide underwent apoptosis, and the remaining population number was progressively reduced as the incubation time increased. This suggests CXCL11 was needed for the survival of BCC cells, and that 10 nmol/L was the optimal concentration of the peptide for cell culture.
Because K17 is a marker of BCC,42,43,44,45,46,59 the cell groups in BCC cell cultures expressing K17+/CXCR3− and K17+/CXCR3+ were BCC keratinocytes. K17−/CXCR3+ cells are likely infiltrating dendritic cells, lymphocytes, or stromal cells.82 The number of K17−/CXCR3+ cells progressively reduced in number with time in culture such that by day 21, K17+ cells (with or without CXCR3 expression) accounted for ≥98% of the cells. Ligands for CXCR3 may preferentially select for BCC keratinocyte survival and proliferation over non-keratinocytes. Based on our IHC and qPCR results of CXCL10 and CXCL11 being highly upregulated in K17+ BCC cells, and the cell culture data here, it is possible that the chemokines produced by BCC keratinocytes and stroma infiltrating inflammatory cells may bind with CXCR3 expressed by BCC cells to induce tumor cell proliferating signals. CXCR3 and its ligands may act as BCC tumor promoters.
Conclusion
Taken together, CXCR3 and its ligands expressed by BCC keratinocytes, and to a lesser extent by inflammatory cells, may enable autocrine or paracrine signaling to induce BCC cell proliferation. Our results provide evidence for CXCR3 and its ligands being a novel mechanism for BCC growth and progression, and these chemokines may serve as potential targets in developing drug treatments against BCCs.
Acknowledgments
We thank Dr. Aziz Ghahary, Dr. Youwen Zhou, and Wency Ip for their advice and assistance in the completion of this study.
Footnotes
Address reprint requests to Kevin John McElwee, Ph.D., 835 West 10th Avenue, Vancouver, B.C., V5Z 4E8, Canada. E-mail: kevin@keratin.com.
See related Commentary on page 2088
Supported by the Canadian Institutes of Health Research (CIHR; No. MUS-94025) and the Canadian Dermatology Foundation.
None of the authors disclosed any relevant financial relationships.
References
- Aszterbaum M, Epstein J, Oro A, Douglas V, LeBoit PE, Scott MP, Epstein EH., Jr Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat Med. 1999;5:1285–1291. doi: 10.1038/15242. [DOI] [PubMed] [Google Scholar]
- Walling HW, Fosko SW, Geraminejad PA, Whitaker DC, Arpey CJ. Aggressive basal cell carcinoma: presentation, pathogenesis, and management. Cancer Metastasis Rev. 2004;23:389–402. doi: 10.1023/B:CANC.0000031775.04618.30. [DOI] [PubMed] [Google Scholar]
- Diepgen TL, Mahler V. The epidemiology of skin cancer. Br J Dermatol. 2002;146 Suppl 61:1–6. doi: 10.1046/j.1365-2133.146.s61.2.x. [DOI] [PubMed] [Google Scholar]
- Gallagher RP, Ma B, McLean DI, Yang CP, Ho V, Carruthers JA, Warshawski LM. Trends in basal cell carcinoma, squamous cell carcinoma, and melanoma of the skin from 1973 through 1987. J Am Acad Dermatol. 1990;23:413–421. doi: 10.1016/0190-9622(90)70234-9. [DOI] [PubMed] [Google Scholar]
- Marks R, Staples M, Giles GG. Trends in non-melanocytic skin cancer treated in Australia: the second national survey. Int J Cancer. 1993;53:585–590. doi: 10.1002/ijc.2910530410. [DOI] [PubMed] [Google Scholar]
- Urbach F. Incidence of nonmelanoma skin cancer. Dermatol Clin. 1991;9:751–755. [PubMed] [Google Scholar]
- Christenson LJ, Borrowman TA, Vachon CM, Tollefson MM, Otley CC, Weaver AL, Roenigk RK. Incidence of basal cell and squamous cell carcinomas in a population younger than 40 years. JAMA. 2005;294:681–690. doi: 10.1001/jama.294.6.681. [DOI] [PubMed] [Google Scholar]
- Roberts DL. Incidence of non-melanoma skin cancer in West Glamorgan. South Wales Br J Dermatol. 1990;122:399–403. doi: 10.1111/j.1365-2133.1990.tb08289.x. [DOI] [PubMed] [Google Scholar]
- Green A, Battistutta D. Incidence and determinants of skin cancer in a high-risk Australian population. Int J Cancer. 1990;46:356–361. doi: 10.1002/ijc.2910460303. [DOI] [PubMed] [Google Scholar]
- Buettner PG, Raasch BA. Incidence rates of skin cancer in Townsville. Australia Int J Cancer. 1998;78:587–593. doi: 10.1002/(sici)1097-0215(19981123)78:5<587::aid-ijc10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Holme SA, Malinovszky K, Roberts DL. Changing trends in non-melanoma skin cancer in South Wales, 1988–98. Br J Dermatol. 2000;143:1224–1229. doi: 10.1046/j.1365-2133.2000.03892.x. [DOI] [PubMed] [Google Scholar]
- Raasch BA, Buettner PG. Multiple nonmelanoma skin cancer in an exposed Australian population. Int J Dermatol. 2002;41:652–658. doi: 10.1046/j.1365-4362.2002.01573.x. [DOI] [PubMed] [Google Scholar]
- Jones RR. Ozone depletion and cancer risk. Lancet. 1987;2:443–446. doi: 10.1016/s0140-6736(87)90972-x. [DOI] [PubMed] [Google Scholar]
- Garssen J, Norval M, el-Ghorr A, Gibbs NK, Jones CD, Cerimele D, De Simone C, Caffieri S, Dall'Acqua F, De Gruijl FR, Sontag Y, Van Loveren H. Estimation of the effect of increasing UVB exposure on the human immune system and related resistance to infectious diseases and tumours. J Photochem Photobiol B. 1998;42:167–179. doi: 10.1016/s1011-1344(97)00122-x. [DOI] [PubMed] [Google Scholar]
- Hutchin ME, Kariapper MS, Grachtchouk M, Wang A, Wei L, Cummings D, Liu J, Michael LE, Glick A, Dlugosz AA. Sustained Hedgehog signaling is required for basal cell carcinoma proliferation and survival: conditional skin tumorigenesis recapitulates the hair growth cycle. Genes Dev. 2005;19:214–223. doi: 10.1101/gad.1258705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellheyer K, Krahl D. Basal cell (trichoblastic) carcinoma common expression pattern for epithelial cell adhesion molecule links basal cell carcinoma to early follicular embryogenesis, secondary hair germ, and outer root sheath of the vellus hair follicle: a clue to the adnexal nature of basal cell carcinoma? J Am Acad Dermatol. 2008;58:158–167. doi: 10.1016/j.jaad.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Wong CS, Strange RC, Lear JT. Basal cell carcinoma. BMJ. 2003;327:794–798. doi: 10.1136/bmj.327.7418.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein EH, Shepard JA, Flotte TJ. Case records of the Massachusetts General Hospital. Case 3–2008 An 80-year-old woman with cutaneous basal-cell carcinomas and cysts of the jaws. N Engl J Med. 2008;358:393–401. doi: 10.1056/NEJMcpc0707893. [DOI] [PubMed] [Google Scholar]
- Boukamp P. Non-melanoma skin cancer: what drives tumor development and progression? Carcinogenesis. 2005;26:1657–1667. doi: 10.1093/carcin/bgi123. [DOI] [PubMed] [Google Scholar]
- Gailani MR, Stahle-Backdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, Unden AB, Dean M, Brash DE, Bale AE, Toftgard R. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, Jr, de Sauvage FJ. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Athar M, Tang X, Lee JL, Kopelovich L, Kim AL. Hedgehog signalling in skin development and cancer. Exp Dermatol. 2006;15:667–677. doi: 10.1111/j.1600-0625.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- Aszterbaum M, Rothman A, Johnson RL, Fisher M, Xie J, Bonifas JM, Zhang X, Scott MP, Epstein EH., Jr Identification of mutations in the human PATCHED gene in sporadic basal cell carcinomas and in patients with the basal cell nevus syndrome. J Invest Dermatol. 1998;110:885–888. doi: 10.1046/j.1523-1747.1998.00222.x. [DOI] [PubMed] [Google Scholar]
- Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale AE. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- Ji J, Kump E, Wernli M, Erb P. Gene silencing of transcription factor Gli2 inhibits basal cell carcinomalike tumor growth in vivo. Int J Cancer. 2007;122:50–56. doi: 10.1002/ijc.23023. [DOI] [PubMed] [Google Scholar]
- Nilsson M, Unden AB, Krause D, Malmqwist U, Raza K, Zaphiropoulos PG, Toftgard R. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc Natl Acad Sci USA. 2000;97:3438–3443. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Khavari PA. Sonic hedgehog opposes epithelial cell cycle arrest. J Cell Biol. 1999;147:71–76. doi: 10.1083/jcb.147.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P, Mulvaney M, Carlson JA. Basal cell carcinoma progression correlates with host immune response and stromal alterations: a histologic analysis. Am J Dermatopathol. 2006;28:293–307. doi: 10.1097/00000372-200608000-00002. [DOI] [PubMed] [Google Scholar]
- Pellegrino A, Antonaci F, Russo F, Merchionne F, Ribatti D, Vacca A, Dammacco F. CXCR3-binding chemokines in multiple myeloma. Cancer Lett. 2004;207:221–227. doi: 10.1016/j.canlet.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Baggiolini M. Chemokines in pathology and medicine. J Intern Med. 2001;250:91–104. doi: 10.1046/j.1365-2796.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- Hall RA, Premont RT, Lefkowitz RJ. Heptahelical receptor signaling: beyond the G protein paradigm. J Cell Biol. 1999;145:927–932. doi: 10.1083/jcb.145.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya M, Suyama T, Usui H, Kasuya Y, Nishiyama M, Tanaka N, Ishiwata I, Nagai Y, Shozu M, Kimura S. Up-regulation of CXC chemokines and their receptors: implications for proinflammatory microenvironments of ovarian carcinomas and endometriosis. Hum Pathol. 2007;38:1676–1687. doi: 10.1016/j.humpath.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Jones D, Benjamin RJ, Shahsafaei A, Dorfman DM. The chemokine receptor CXCR3 is expressed in a subset of B-cell lymphomas and is a marker of B-cell chronic lymphocytic leukemia. Blood. 2000;95:627–632. [PubMed] [Google Scholar]
- Monteagudo C, Martin JM, Jorda E, Llombart-Bosch A. CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathological prognostic factors. J Clin Pathol. 2007;60:596–599. doi: 10.1136/jcp.2005.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda N, Shimizu T, Tada Y, Watanabe S. IL-18 enhances IFN-gamma-induced production of CXCL9. CXCL10, and CXCL11 in human keratinocytes. Eur J Immunol. 2007;37:338–350. doi: 10.1002/eji.200636420. [DOI] [PubMed] [Google Scholar]
- Kanda N, Watanabe S. Prolactin enhances interferon-gamma-induced production of CXC ligand 9 (CXCL9). CXCL10, and CXCL11 in human keratinocytes. Endocrinology. 2007;148:2317–2325. doi: 10.1210/en.2006-1639. [DOI] [PubMed] [Google Scholar]
- Mee JB, Johnson CM, Morar N, Burslem F, Groves RW. The psoriatic transcriptome closely resembles that induced by interleukin-1 in cultured keratinocytes: dominance of innate immune responses in psoriasis. Am J Pathol. 2007;171:32–42. doi: 10.2353/ajpath.2007.061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Zloty D, Cowan B, Shapiro J, Haegert A, Bell RH, Warshawski L, Carr N, McElwee KJ. Superficial, nodular, and morpheiform basal-cell carcinomas exhibit distinct gene expression profiles. J Invest Dermatol. 2008;128:1797–1805. doi: 10.1038/sj.jid.5701243. [DOI] [PubMed] [Google Scholar]
- Kokolakis G, Panagis L, Stathopoulos E, Giannikaki E, Tosca A, Kruger-Krasagakis S. From the protein to the graph: how to quantify immunohistochemistry staining of the skin using digital imaging. J Immunol Methods. 2008;331:140–146. doi: 10.1016/j.jim.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Sompuram SR, Kodela V, Zhang K, Ramanathan H, Radcliffe G, Falb P, Bogen SA. A novel quality control slide for quantitative immunohistochemistry testing. J Histochem Cytochem. 2002;50:1425–1434. doi: 10.1177/002215540205001101. [DOI] [PubMed] [Google Scholar]
- Robinson D, Hasharoni A, Evron Z, Segal M, Nevo Z. Synovial chondromatosis: the possible role of FGF 9 and FGF receptor 3 in its pathology. Int J Exp Pathol. 2000;81:183–189. doi: 10.1046/j.1365-2613.2000.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi E, Venegoni L, Fanoni D, Berti E. Cytokeratin profile in basal cell carcinoma. Am J Dermatopathol. 2008;30:249–255. doi: 10.1097/DAD.0b013e31816c828a. [DOI] [PubMed] [Google Scholar]
- Asada M, Schaart FM, de Almeida HL, Jr, Korge B, Kurokawa I, Asada Y, Orfanos CE. Solid basal cell epithelioma (BCE) possibly originates from the outer root sheath of the hair follicle. Acta Derm Venereol. 1993;73:286–292. [PubMed] [Google Scholar]
- Kurzen H, Esposito L, Langbein L, Hartschuh W. Cytokeratins as markers of follicular differentiation: an immunohistochemical study of trichoblastoma and basal cell carcinoma. Am J Dermatopathol. 2001;23:501–509. doi: 10.1097/00000372-200112000-00001. [DOI] [PubMed] [Google Scholar]
- Schirren CG, Rutten A, Kaudewitz P, Diaz C, McClain S, Burgdorf WH. Trichoblastoma and basal cell carcinoma are neoplasms with follicular differentiation sharing the same profile of cytokeratin intermediate filaments. Am J Dermatopathol. 1997;19:341–350. doi: 10.1097/00000372-199708000-00005. [DOI] [PubMed] [Google Scholar]
- Apaydin R, Gurbuz Y, Bayramgurler D, Bilen N. Cytokeratin contents of basal cell carcinoma, epidermis overlying tumour, and associated stromal amyloidosis: an immunohistochemical study. Amyloid. 2005;12:41–47. doi: 10.1080/13506120500032543. [DOI] [PubMed] [Google Scholar]
- Kawada K, Sonoshita M, Sakashita H, Takabayashi A, Yamaoka Y, Manabe T, Inaba K, Minato N, Oshima M, Taketo MM. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res. 2004;64:4010–4017. doi: 10.1158/0008-5472.CAN-03-1757. [DOI] [PubMed] [Google Scholar]
- Scala S, Giuliano P, Ascierto PA, Ierano C, Franco R, Napolitano M, Ottaiano A, Lombardi ML, Luongo M, Simeone E, Castiglia D, Mauro F, De Michele I, Calemma R, Botti G, Caraco C, Nicoletti G, Satriano RA, Castello G. Human melanoma metastases express functional CXCR4. Clin Cancer Res. 2006;12:2427–2433. doi: 10.1158/1078-0432.CCR-05-1940. [DOI] [PubMed] [Google Scholar]
- Chen GS, Yu HS, Lan CC, Chow KC, Lin TY, Kok LF, Lu MP, Liu CH, Wu MT. CXC chemokine receptor CXCR4 expression enhances tumorigenesis and angiogenesis of basal cell carcinoma. Br J Dermatol. 2006;154:910–918. doi: 10.1111/j.1365-2133.2006.07150.x. [DOI] [PubMed] [Google Scholar]
- Aksoy MO, Yang Y, Ji R, Reddy PJ, Shahabuddin S, Litvin J, Rogers TJ, Kelsen SG. CXCR3 surface expression in human airway epithelial cells: cell cycle dependence and effect on cell proliferation. Am J Physiol Lung Cell Mol Physiol. 2006;290:L909–L918. doi: 10.1152/ajplung.00430.2005. [DOI] [PubMed] [Google Scholar]
- Moller C, Stromberg T, Juremalm M, Nilsson K, Nilsson G. Expression and function of chemokine receptors in human multiple myeloma. Leukemia. 2003;17:203–210. doi: 10.1038/sj.leu.2402717. [DOI] [PubMed] [Google Scholar]
- Luster AD. Chemokines–chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- Oppenheim JJ, Murphy WJ, Chertox O, Schirrmacher V, Wang JM. Prospects for cytokine and chemokine biotherapy. Clin Cancer Res. 1997;3:2682–2686. [PubMed] [Google Scholar]
- Robledo MM, Bartolome RA, Longo N, Rodriguez-Frade JM, Mellado M, Longo I, van Muijen GN, Sanchez-Mateos P, Teixido J. Expression of functional chemokine receptors CXCR3 and CXCR4 on human melanoma cells. J Biol Chem. 2001;276:45098–45105. doi: 10.1074/jbc.M106912200. [DOI] [PubMed] [Google Scholar]
- Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- Sica A, Saccani A, Bottazzi B, Bernasconi S, Allavena P, Gaetano B, Fei F, LaRosa G, Scotton C, Balkwill F, Mantovani A. Defective expression of the monocyte chemotactic protein-1 receptor CCR2 in macrophages associated with human ovarian carcinoma. J Immunol. 2000;164:733–738. doi: 10.4049/jimmunol.164.2.733. [DOI] [PubMed] [Google Scholar]
- Leigh IM, Navsaria H, Purkis PE, McKay IA, Bowden PE, Riddle PN. Keratins (K16 and K17) as markers of keratinocyte hyperproliferation in psoriasis in vivo and in vitro. Br J Dermatol. 1995;133:501–511. doi: 10.1111/j.1365-2133.1995.tb02696.x. [DOI] [PubMed] [Google Scholar]
- Moll R, Franke WW, Volc-Platzer B, Krepler R. Different keratin polypeptides in epidermis and other epithelia of human skin: a specific cytokeratin of molecular weight 46,000 in epithelia of the pilosebaceous tract and basal cell epitheliomas. J Cell Biol. 1982;95:285–295. doi: 10.1083/jcb.95.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Katagata Y, Kondo S. Biochemical and immunohistochemical analyses of keratin expression in basal cell carcinoma. J Dermatol Sci. 1998;17:15–23. doi: 10.1016/s0923-1811(97)00065-0. [DOI] [PubMed] [Google Scholar]
- Ehlert JE, Addison CA, Burdick MD, Kunkel SL, Strieter RM. Identification and partial characterization of a variant of human CXCR3 generated by posttranscriptional exon skipping. J Immunol. 2004;173:6234–6240. doi: 10.4049/jimmunol.173.10.6234. [DOI] [PubMed] [Google Scholar]
- Giuliani N, Bonomini S, Romagnani P, Lazzaretti M, Morandi F, Colla S, Tagliaferri S, Lasagni L, Annunziato F, Crugnola M, Rizzoli V. CXCR3 and its binding chemokines in myeloma cells: expression of isoforms and potential relationships with myeloma cell proliferation and survival. Haematologica. 2006;91:1489–1497. [PubMed] [Google Scholar]
- Ji R, Lee CM, Gonzales LW, Yang Y, Aksoy MO, Wang P, Brailoiu E, Dun N, Hurford MT, Kelsen SG. Human type II pneumocyte chemotactic responses to CXCR3 activation are mediated by splice variant A. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1187–L1196. doi: 10.1152/ajplung.00388.2007. [DOI] [PubMed] [Google Scholar]
- Loetscher M, Loetscher P, Brass N, Meese E, Moser B. Lymphocyte-specific chemokine receptor CXCR3: regulation, chemokine binding and gene localization. Eur J Immunol. 1998;28:3696–3705. doi: 10.1002/(SICI)1521-4141(199811)28:11<3696::AID-IMMU3696>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Maru SV, Holloway KA, Flynn G, Lancashire CL, Loughlin AJ, Male DK, Romero IA. Chemokine production and chemokine receptor expression by human glioma cells: role of CXCL10 in tumour cell proliferation. J Neuroimmunol. 2008;199:35–45. doi: 10.1016/j.jneuroim.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Kim S, Bakre M, Yin H, Varner JA. Inhibition of endothelial cell survival and angiogenesis by protein kinase A. J Clin Invest. 2002;110:933–941. doi: 10.1172/JCI14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta D, Flaxenburg JA, Laxmanan S, Geehan C, Grimm M, Waaga-Gasser AM, Briscoe DM, Pal S. Ras-induced modulation of CXCL10 and its receptor splice variant CXCR3-B in MDA-MB-435 and MCF-7 cells: relevance for the development of human breast cancer. Cancer Res. 2006;66:9509–9518. doi: 10.1158/0008-5472.CAN-05-4345. [DOI] [PubMed] [Google Scholar]
- Zipin-Roitman A, Meshel T, Sagi-Assif O, Shalmon B, Avivi C, Pfeffer RM, Witz IP, Ben-Baruch A. CXCL10 promotes invasion-related properties in human colorectal carcinoma cells. Cancer Res. 2007;67:3396–3405. doi: 10.1158/0008-5472.CAN-06-3087. [DOI] [PubMed] [Google Scholar]
- Boukamp P. UV-induced skin cancer: similarities–variations. J Dtsch Dermatol Ges. 2005;3:493–503. doi: 10.1111/j.1610-0387.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- Tanese K, Fukuma M, Yamada T, Mori T, Yoshikawa T, Watanabe W, Ishiko A, Amagai M, Nishikawa T, Sakamoto M. G-protein-coupled receptor GPR49 is up-regulated in basal cell carcinoma and promotes cell proliferation and tumor formation. Am J Pathol. 2008;173:835–843. doi: 10.2353/ajpath.2008.071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman TA, Modali R, Boukamp P, Stanek J, Bennett WP, Welsh JA, Metcalf RA, Stampfer MR, Fusenig N, Rogan EM, Harris CC. p53 mutations in human immortalized epithelial cell lines. Carcinogenesis. 1993;14:833–839. doi: 10.1093/carcin/14.5.833. [DOI] [PubMed] [Google Scholar]
- Billings SD, Southall MD, Li T, Cook PW, Baldridge L, Moores WB, Spandau DF, Foley JG, Travers JB. Amphiregulin overexpression results in rapidly growing keratinocytic tumors: an in vivo xenograft model of keratoacanthoma. Am J Pathol. 2003;163:2451–2458. doi: 10.1016/S0002-9440(10)63600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Peter W, Pascheberg U, Altmeier S, Fasching C, Stanbridge EJ, Fusenig NE. Step-wise progression in human skin carcinogenesis in vitro involves mutational inactivation of p53, rasH oncogene activation and additional chromosome loss. Oncogene. 1995;11:961–969. [PubMed] [Google Scholar]
- Mudgil AV, Segal N, Andriani F, Wang Y, Fusenig NE, Garlick JA. Ultraviolet B irradiation induces expansion of intraepithelial tumor cells in a tissue model of early cancer progression. J Invest Dermatol. 2003;121:191–197. doi: 10.1046/j.1523-1747.2003.12320.x. [DOI] [PubMed] [Google Scholar]
- Mueller MM, Fusenig NE. Constitutive expression of G-CSF and GM-CSF in human skin carcinoma cells with functional consequence for tumor progression. Int J Cancer. 1999;83:780–789. doi: 10.1002/(sici)1097-0215(19991210)83:6<780::aid-ijc14>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Mueller MM, Peter W, Mappes M, Huelsen A, Steinbauer H, Boukamp P, Vaccariello M, Garlick J, Fusenig NE. Tumor progression of skin carcinoma cells in vivo promoted by clonal selection, mutagenesis, and autocrine growth regulation by granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor. Am J Pathol. 2001;159:1567–1579. doi: 10.1016/S0002-9440(10)62541-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skobe M, Fusenig NE. Tumorigenic conversion of immortal human keratinocytes through stromal cell activation. Proc Natl Acad Sci USA. 1998;95:1050–1055. doi: 10.1073/pnas.95.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacchi A, Romagnani P, Romanelli RG, Efsen E, Annunziato F, Lasagni L, Francalanci M, Serio M, Laffi G, Pinzani M, Gentilini P, Marra F. Signal transduction by the chemokine receptor CXCR3: activation of Ras/ERK. Src, and phosphatidylinositol 3-kinase/Akt controls cell migration and proliferation in human vascular pericytes. J Biol Chem. 2001;276:9945–9954. doi: 10.1074/jbc.M010303200. [DOI] [PubMed] [Google Scholar]
- Boyce ST, Ham RG. Calcium-regulated differentiation of normal human epidermal keratinocytes in chemically defined clonal culture and serum-free serial culture. J Invest Dermatol. 1983;81:33s–40s. doi: 10.1111/1523-1747.ep12540422. [DOI] [PubMed] [Google Scholar]
- Brysk MM, Santschi CH, Bell T, Wagner RF, Jr, Tyring SK, Rajaraman S. Culture of basal cell carcinoma. J Invest Dermatol. 1992;98:45–49. doi: 10.1111/1523-1747.ep12494210. [DOI] [PubMed] [Google Scholar]
- Sneddon JB, Zhen HH, Montgomery K, van de Rijn M, Tward AD, West R, Gladstone H, Chang HY, Morganroth GS, Oro AE, Brown PO. Bone morphogenetic protein antagonist gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. Proc Natl Acad Sci USA. 2006;103:14842–14847. doi: 10.1073/pnas.0606857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, de Groot M, Kinsley D, Laverty M, McClanahan T, Arreaza M, Gustafson EL, Teunissen MB, de Rie MA, Fine JS, Kraan M. Expression of chemokine receptor CXCR3 by lymphocytes and plasmacytoid dendritic cells in human psoriatic lesions. Arch Dermatol Res. 2009;302:113–123. doi: 10.1007/s00403-009-0966-2. [DOI] [PubMed] [Google Scholar]