Abstract
Introduction
Cervical cancer is among the most common cancers in women worldwide. Discovery of biomarkers for the early detection of cervical cancer would improve current screening practices and reduce the burden of disease.
Objective
In this study, we report characterization of the human cervical mucous proteome as the first step towards protein biomarker discovery.
Methods
The protein composition was characterized using one- and two-dimensional gel electrophoresis, and liquid chromatography coupled with mass spectrometry. We chose to use this combination of traditional biochemical techniques and proteomics to allow a more comprehensive analysis.
Results and Conclusion
A total of 107 unique proteins were identified, with plasma proteins being most abundant. These proteins represented the major functional categories of metabolism, immune response, and cellular transport. Removal of high molecular weight abundant proteins by immunoaffinity purification did not significantly increase the number of protein spots resolved. We also analyzed phosphorylated and glycosylated proteins by fluorescent post-staining procedures. The profiling of cervical mucous proteins and their post-translational modifications can be used to further our understanding of the cervical mucous proteome.
Keywords: Cervical mucous, Cervical cancer biomarkers, Mass spectrometry, 2-D electrophoresis, LC/MS/MS, MS
Introduction
Advances in proteomic technologies have greatly accelerated the field of protein biomarker discovery. High-throughput technologies like surface-enhanced laser desorption and ionization-time of flight mass spectrometry (SELDI-TOF MS) or the combination of one- or two-dimensional gel electrophoresis (1-DE, 2-DE) methods with multidimensional chromatographic separation and MS provide a snapshot of the proteome. Validated protein biomarkers could be useful in early detection of disease, monitoring disease progression or monitoring response to treatment. Several studies have thus focused on biomarker discovery in body fluids which would be advantageous for eventual clinical implementation. Cervical mucous is potentially an ideal sample to screen for biomarkers for early detection of cervical cancer. As cervical mucous is produced in the microenvironment where cervical neoplasia arises, it is likely to include proteins produced by the lesion as well as by the host in response to the lesion.
Cervical mucous is a complex mixture of proteins consisting of an aqueous and glycoprotein phase [1], but is thought to have a restricted protein profile compared to other body fluids like serum or plasma. It is recognized that the cervix is hormonally responsive and differences in protein composition and concentration due to menopause, menstrual cycle, and hormonal contraceptives will impact sample to sample comparison. CVF includes, in addition to cervical mucous, contributions from vulvar secretions, vaginal wall transudate, exfoliated cells, endometrial and oviductal fluids, and metabolic products of vaginal microflora [2]. Thus, proteomic characterization of CVF is suited for exploring markers of genital tract infections or pregnancy status or overall evaluation of reproductive tract. However, protein contributions from multiple sites could significantly dilute the cervical contribution that would be most representative of cervical neoplasia localized to the cervix.
To date, there have been six studies of CVF [3–8], four of these were restricted to pregnant women [3–6]. These studies identified from 15 to 685 proteins using gel separation/MALDI-TOF MS ([4, 8]; 15 and 59 proteins respectively), LC-MS/MS ([5]; 39 proteins), gel separation and LC-MS/MS ([7]; 685 proteins), and gel separation and LC-LC-MS/MS ([3, 6]; 150 and 205 proteins respectively). Consistently, many CVF proteins were noted to be of plasma origin, suggesting CVF could largely be plasma transudate. Other major functional categories were represented by immune response, metabolism, and cellular transport. Only one study examined cervical mucous [9], and it focused on samples from normal women obtained during various points of the menstrual cycle. Using gel separation/LC-MS/MS they identified 194 proteins and again identified plasma proteins. Cervical mucous and CVF would be expected to share similarities in their proteome, but for reasons stated above, the cervical mucous is expected be more representative of cervical disease and the local host response to cervical disease.
In our study, the cervical mucous proteome was characterized by 2-DE and gel-based liquid chromatography–mass spectrometry (GeLC-MS/MS) techniques. Of the current proteomic tools, no single method can resolve an entire proteome. Combination of several pre-fractionation methods like gel separation and LC prior to mass spectrometry allowed for visualization of the proteins as well as the identification of proteins that could otherwise be missed by a single method [10]. Also, characterization of sub-proteomes like the glycoproteome or phosphoproteome and immunoaffinity depletion of high abundant proteins was attempted in order to reduce complexity and facilitate better resolution of proteins in cervical mucous.
Materials and Methods
Sample Collection and Processing
Women attending urban colposcopy clinics were enrolled as part of an ongoing study of cervical neoplasia [11]. Cervical mucous samples were collected at the time of colposcopy by absorption into two Weck-Cel® sponges (Xomed Surgical Products, Jacksonville, FL) placed, one at a time, into the cervical os. Samples were stored at −80°C until use. We processed 40 Weck-Cel® sponges (one or two sponges per subject depending on availability) with no visual blood contamination from 25 subjects who were randomly selected. The median age of the women in this subset was 27 years (range 18–57); 90% were black; 52% currently used hormonal contraceptive (oral, depo-provera, or both); 62% were HPV positive and 52% had pre-invasive cervical disease. No data was available on the menstrual cycle at the time of sample collection.
Total protein was extracted using M-PER® extraction reagent (M-PER, 0.15 M NaCl) (Pierce Biotechnology, Inc., Rockford, IL) as previously established [12]. All samples were combined and the pooled sample was aliquoted for storage at −80°C. A pooled sample was used in order to have sufficient volume that would allow for optimization of the 2-DE and GeLC-MS/MS methods described below. Total protein content was measured using the Coomasie PlusTM kit (Pierce Biotechnology Inc.) as per the manufacturer’s protocol.
Depletion of Albumin and IgG
Immunodepletion of the pooled mucous sample was carried out using IgY-C12 spin column (GenWay Biotech. Inc., San Diego, CA) following manufacturer’s protocol. In brief, an aliquot of protein extract (∼200 µg) was first mixed with 1× Tris-buffered saline (TBS, 10 mM Tris, 150 mM NaCl, pH 7.4) at 1:1 ratio. The mixture was added to IgY-C12 spin column and incubated at room temperature with shaking for 20 min. The unbound proteins were eluted and the column washed three times with 1× TBS buffer. The eluates from all the above steps were combined and dried down using a lyophilizer.
Two-Dimensional Gel Electrophoresis
For each gel, 80 µg of protein extract was prepared with the 2-D Clean Up Kit (GE Healthcare) to remove interfering components. All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. The samples (with or without depletion) were mixed in a rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 50 mM DTT, 1% Pharmalyte) and incubated for 12 h followed by focusing on an 11-cm immobilized pH gradient (IPG) strip (pH 3-11 NL, GE Healthcare) in a PROTEAN IEF cell (Bio-Rad, Hercules, CA). Focusing was performed under the condition: 500 V constant for 500 V h; linear gradient to 1,000 V for 800 V h; gradient to 6,000 V for 7,000 V h; gradient to 6,000 V for 3,700 V h. After the first-dimensional isoelectric focusing, IPG strips containing proteins were treated for 15 min with gentle shaking in the equilibration buffer (6 M urea, 30% glycerol, 2% SDS, 50 mM Tris, pH 8.8) containing 2.5% tributyl phosphine (TBP) for protein reduction followed by protein alkylation with 3% iodoacetamide (IAA) to replace TBP in the buffer for another 15 min. The second-dimensional SDS-PAGE was run on an 8–16% linear gradient criterion Tris–HCl gel (Bio-rad) at 110 V for 2 h. The gels were stained with colloidal Coomassie or SYPRO Ruby (Invitrogen, Carlsbad, CA) dyes as per the manufacturer’s protocol.
One-Dimensional Gel Electrophoresis
The sample (80 µg) was treated with the 2-D Clean Up kit as described above. The precipitate was solubilized in lysis buffer (1 M Tris–HCl, 8 M urea, and 4% CHAPS, pH 8.5) and run on an 8–16% linear gradient criterion Tris–HCl gel at 110 V for 1 h. After colloidal Coomassie staining, the entire lane was sliced into 40 pieces of equal size for further analysis.
Gel Staining
The 2-DE gel for the characterization of post-translational modifications on human cervical mucous proteins was stained using three different dyes: Pro-Q Diamond for phosphoproteins, Pro-Q Emerald 488 for glycoproteins, and SYPRO Ruby for total proteins (Invitrogen) following manufacturer’s protocol. In brief, the gels were fixed overnight at room temperature in 500 ml fixation solution (50% methanol, 10% acetic acid), followed by two 10-min wash steps with ultrapure water with gentle agitation. Staining was performed with 500 ml Pro-Q Diamond with gentle agitation in the dark for 90 min. The gel was destained by washing twice with 500 ml destain solution (20% acetonitrile, 50 mM sodium acetate, pH 4.0) with gentle agitation. The gel images were acquired on Typhoon 9400 (GE Healthcare) with 532 nm excitation and 560 nm long pass emission. After image scanning, gels were washed two times in 3% glacial acetic acid for 20 min each and then incubated in oxidizing solution (1% periodic acid in 3% acetic acid) for 1 h. The oxidizing solution was removed by washing three times with 3% glacial acetic acid for 20 min each. The gels were then incubated in fresh Pro-Q Emerald 488 staining solution for 3 h in the dark. Destaining was done by washing the gel twice in 3% acetic acid at room temperature for 30 min each. Gel images were acquired by Typhoon 9400 at 510 nm excitation and 520 nm emission wavelengths. Finally, the gels were visualized by colloidal Coomassie staining and the gel spots of interest were excised for further characterization.
In-Gel Trypsin Digestion
Sample in-gel digestion and micro-purification were carried out on a ZipPlate micro-SPE according to manufacturer’s In-Gel Digestion Protocol (Millipore, Billerica, MA) with minor modifications. Briefly, gel spots or bands were destained in buffer 1 (25 mM ammonium bicarbonate and 5% acetonitrile) for 30 min and then in buffer 2 (25 mM ammonium bicarbonate and 50% acetonitrile) for an additional 30 min. Following reduction with 10 mM dithiothreitol and alkylation with 55 mM IAA, the gel pieces were incubated in 15 or 30 µL of trypsin (165 ng) solution and incubated overnight at 37°C. The peptides were extracted, micro-purified, vacuum-dried and resuspended in 1% formic acid and 2% acetonitrile for mass spectrometric analysis.
Mass Spectrometry and Database Analysis
Nanocapillary LC-MS/MS analysis was performed in a Micromass Q-Tof Ultima mass spectrometer equipped with a nanospray ion source and coupled with an nanoAcquity ultraperformance liquid chromatography system (UPLC; Waters, Milford, MA). The protein digest (2 µl) was loaded onto an in-house packed reverse phase C18 capillary column (∼15 cm, 75 µm inner diameter) and separated using a linear gradient of 15% to 45% of buffer B (acetonitrile, 0.1% formic acid) in buffer A (water, 0.1% formic acid) in 60 min (2-DE gel samples) or 80 min (1-DE gel samples) at a flow rate of 0.5 µl/min.
All mass spectra were obtained in the positive-ion mode. An electric potential of 3.5 kV was applied to the emitter in the ion source. The acquisition of data was performed on a MassLynx data system (version 4.0; Waters) using a data-dependent mode where the four most intense precursors in a survey scan were isolated for collision-induced dissociation. Resulting MS/MS data were searched for protein candidates with automated database searching against NCBI nr database using MASCOT Daemon software (Matrix Sciences, Boston, MA). The peptide mass tolerance was set to ±50 ppm, and fragment mass tolerance was set to ±0.3 Da. Carboxyamidomethyl cysteine and oxidized methionine were set as variable modifications. All peptides identified with score less than 50 and one matched peptide during MASCOT searches were examined by manual inspection. Functional categories were annotated by literature surveys in PubMed or based on classification from the Database for Annotation, Visualization and Integrated Discovery (DAVID) [13].
Results and Discussion
Identification of Cervical Mucous Proteins by Proteomic Techniques
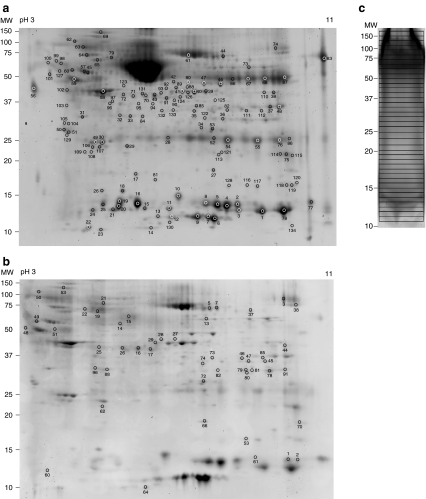
Complex protein mixtures can be resolved effectively according to their isoelectric points and molecular weights by 2-DE. While gel-based approaches provide good separation and direct visualization of individual proteins, highly accurate LC-MS/MS analysis on protein digests extracted from gel spots leads to high-confidence identification of these proteins. Therefore, the combination of these two techniques becomes a powerful tool in characterizing a proteome. The proteins extracted from pooled cervical mucous samples were first separated by 2-DE method (Fig. 1a). 183 spots were excised and subjected to in-gel trypsin digestion followed by liquid chromatography coupled to mass spectrometric analysis. High-confidence protein identification was accomplished by searching non-redundant protein databases with tandem mass spectra of the peptide mixtures. A total of 134 protein spots were successfully identified that represented 79 unique proteins (Table 1). Several spots resulted in identical protein identification, presumably due to the formation of protein isoforms bearing various modified residues introduced by post-translational modification or during sample processing.
Fig. 1.
Gel electrophoresis of the proteins extracted from cervical mucous. a 2-DE gel image (Coomassie staining); b 2-DE gel after depletion (SYPRO Ruby staining); and c 1-DE gel image (Coomassie staining). Only spots representing identified proteins are labeled on gel images
Table 1.
Cervical mucous proteins identified through proteomic analyses
| Protein name | Acc. no.a | MW (kDa) | pI | MASCOT score | Matched peptides | Sequence coverage (%) | Methodb | Spot no.c |
|---|---|---|---|---|---|---|---|---|
| 14-3-3 protein epsilon | P62258 | 29.1 | 4.63 | 92 | 3 | 10 | T | 51 |
| 14-3-3 protein sigma | P31947 | 27.7 | 4.68 | 71 | 3 | 10 | T | 51 |
| 14-3-3 protein zeta/delta | P63104 | 27.7 | 4.73 | 181 | 5 | 25 | T | 50 |
| 6-phosphogluconate dehydrogenase | P52209 | 53.1 | 6.80 | 51 | 2 | 5 | T,G | 46 |
| Alcohol dehydrogenase [NADP+]d | P14550 | 36.5 | 6.32 | 75 | 2 | 6 | T | 122 |
| Alpha-1-acid glycoprotein 1 precursore | P02763 | 23.4 | 4.93 | 194 | 6 | 34 | T,G | 56 |
| Alpha-1-antichymotrypsin precursore | P01011 | 47.6 | 5.33 | 285 | 7 | 21 | T | 98–100 |
| Alpha-1-antitrypsin precursore | P01009 | 46.7 | 5.37 | 1092 | 16 | 48 | T,G, D | 45, 57, 59, 97, 123, d19 |
| Alpha-enolase | P06733 | 47.1 | 7.01 | 338 | 9 | 19 | T,G | 46–47, 80 |
| Annexin A1 | P04083 | 38.7 | 6.57 | 1893 | 12 | 43 | T,G, D | 35–36, 85, 124, d78–82, d91 |
| Annexin A2 | P07355 | 38.5 | 7.57 | 573 | 13 | 43 | T,G, D | 82, 111–112, d37, d45–47, d73, d85 |
| Annexin A3 | P12429 | 36.4 | 5.63 | 758 | 12 | 42 | T,G, D | 32–33, 84, d88, d96 |
| Annexin A5 | P08758 | 35.9 | 4.94 | 167 | 7 | 25 | T,G | 31 |
| Antileukoproteinase 1 precursor | P03973 | 14.3 | 9.11 | 54 | 2 | 6 | G | |
| Anterior gradient protein 2 homolog precursor | O95994 | 19.9 | 9.03 | 126 | 4 | 26 | T,G | 118–119 |
| Apolipoprotein A-I precursore | P02647 | 30.8 | 5.56 | 1464 | 16 | 56 | T,G | 30, 49, 106–109 |
| Arginase-1 | P05089 | 34.7 | 6.72 | 36 | 3 | 10 | T | 125 |
| ATP synthase subunit beta | P06576 | 56.5 | 5.26 | 64 | 2 | 4 | D | d51 |
| Beta-actine | P60709 | 41.7 | 5.29 | 926 | 10 | 34 | T,G | 58, 96 |
| Calregulind | P27797 | 48.1 | 4.29 | 92 | 3 | 8 | T, D | 101, d48 |
| Carbonic anhydrase 1 | P00915 | 28.9 | 6.59 | 133 | 5 | 22 | T, D | 34, 53, 54, d72 |
| Catalase | P04040 | 59.7 | 6.9 | 38 | 1 | 1 | D | d13 |
| Cathelicidin antimicrobial peptide precursord | P49913 | 19.3 | 9.48 | 135 | 2 | 10 | T, G | 119–120 |
| Ceruloplasmin precursor | P00450 | 122.1 | 5.44 | 124 | 4 | 4 | T,G, D | 69, d53 |
| Complement C3 precursor | 1637 | 187.0 | 6.02 | 254 | 15 | 11 | T,G, D | 34, 43, d5, d7 |
| Complement C4-A precursor | 1618 | 192.7 | 6.65 | 60 | 2 | 1 | D | d38 |
| Complement factor B precursor | P00751 | 85.5 | 6.67 | 57 | 4 | 5 | G | |
| Cystatin-A | P01040 | 11.0 | 5.38 | 126 | 2 | 30 | T,G | 21, 24 |
| Cystatin-B | P04080 | 11.1 | 6.96 | 85 | 2 | 24 | G | |
| Elongation factor 1-alpha 1 | P68104 | 50.1 | 9.10 | 70 | 3 | 6 | T | 87 |
| Endoplasmin precursor | P14625 | 92.4 | 4.76 | 168 | 6 | 8 | T, D | 62, d50 |
| F-actin-capping protein subunit alpha-1 | P52907 | 32.9 | 5.45 | 43 | 1 | 3 | T | 32 |
| Fatty-acid-binding protein, epidermal | Q01469 | 15.2 | 6.60 | 315 | 6 | 40 | T,G | 10 |
| Flavin reductased | P30043 | 22.1 | 7.13 | 35 | 1 | 7 | T | 121 |
| Fructose-bisphosphate aldolase A | P04075 | 39.4 | 8.30 | 44 | 1 | 3 | D | d44 |
| Glutamine synthetased | P15104 | 42.0 | 6.43 | 99 | 5 | 19 | G | |
| Glutathione-S-transferase Pe | P09211 | 23.3 | 5.43 | 73 | 2 | 13 | T,G | 29 |
| Glyceraldehyde-3-phosphate dehydrogenase | P04406 | 36.0 | 8.57 | 168 | 6 | 25 | T,G | 37, 48 |
| Haptoglobin precursor | P00738 | 45.2 | 6.13 | 226 | 5 | 14 | T,G | 72 |
| Heat-shock 70 kDa protein 1 | P08107 | 69.9 | 5.48 | 311 | 11 | 17 | T, D | 126, d19 |
| Heat-shock 70 kDa protein 1 Ld | P34931 | 70.3 | 5.76 | 153 | 7 | 10 | T | 126 |
| Heat-shock 70 kDa protein 5 | P11021 | 72.3 | 5.07 | 261 | 10 | 20 | T | 64 |
| Heat-shock 70 kDa protein 6d | P17066 | 70.9 | 5.81 | 125 | 6 | 7 | T | 126 |
| Heat-shock 70 kDa protein 8 | P11142 | 70.9 | 5.37 | 240 | 7 | 16 | T, D | 79, 126, d22 |
| Heat-shock protein beta-1 | P04792 | 22.8 | 5.98 | 51 | 1 | 4 | G | |
| Heat-shock protein HSP 90-alpha | P07900 | 84.6 | 4.94 | 354 | 10 | 16 | T | 63 |
| Heat-shock protein HSP 90-beta | P08238 | 83.2 | 4.97 | 177 | 6 | 9 | T | 63 |
| Hemoglobin subunit alpha | P69905 | 15.2 | 8.72 | 1787 | 7 | 68 | G, D | 1–3, 13, 22, 78, d81 |
| Hemoglobin subunit beta | P68871 | 16.0 | 6.75 | 4704 | 12 | 95 | T,G, D | 1, 3–6, 8, 13, 34, 70, d61 |
| Hemoglobin subunit delta | P02042 | 16.0 | 7.85 | 2495 | 7 | 46 | G | |
| Hemopexin precursor | P02790 | 51.6 | 6.55 | 43 | 2 | 4 | T | |
| Histone H4 | P62805 | 11.4 | 11.40 | 40 | 2 | 17 | G | |
| Ig alpha-1 chain C region | P01876 | 37.6 | 6.08 | 251 | 6 | 25 | T,G | 70 |
| Ig gamma-1 chain C region | P01857 | 36.0 | 8.46 | 1881 | 10 | 37 | T,G | 38, 66, 67, 68 |
| Ig gamma-2 chain C region | P01859 | 35.9 | 7.66 | 745 | 6 | 21 | T,G | |
| Ig heavy chain V-III region BRO | P01766 | 13.2 | 6.45 | 49 | 1 | 15 | G | |
| Ig kappa chain V-I region Lay | P01605 | 11.8 | 7.96 | 46 | 1 | 8 | G | |
| Ig kappa chain C regione | P01834 | 11.6 | 5.58 | 3002 | 5 | 80 | T,G | 28, 30, 52–55, 76, 113 |
| Ig kappa chain V-I region OU | P01606 | 11.8 | 9.94 | 67 | 3 | 24 | G | |
| Ig kappa chain V-II region TEW | P01617 | 12.3 | 5.69 | 89 | 3 | 34 | G | |
| Ig kappa chain V-III region B6 | P01619 | 11.6 | 9.34 | 64 | 1 | 16 | G | |
| Ig kappa chain V-III region SIE | P01620 | 11.8 | 8.70 | 407 | 3 | 39 | T,G | 55, 76 |
| Ig kappa chain V-III region VG precursor (Fragment) | P04433 | 12.6 | 4.85 | 42 | 2 | 15 | G | |
| Ig kappa chain V-IV region precursor (Fragment) | P06312 | 13.4 | 5.09 | 45 | 1 | 7 | G | |
| Ig lambda chain C regions | P01842 | 11.2 | 6.92 | 597 | 5 | 75 | G | |
| Ig lambda chain V-I region HA | P01700 | 11.9 | 9.07 | 39 | 2 | 18 | G | |
| IgGFc-binding protein precursor | 4878 | 571.6 | 5.12 | 455 | 18 | 4 | T,G | 102, 127 |
| ITI heavy chain H4 | Q14624 | 103.3 | 6.51 | 29 | 2 | 2 | D | d53 |
| Isocitrate dehydrogenase [NAD] subunit alphad | P50213 | 39.6 | 6.47 | 51 | 1 | 2 | T | |
| Lactoferrin | P02788 | 78.1 | 8.50 | 110 | 5 | 8 | T,G, D | 74, d3 |
| Lysozyme C precursor | P61626 | 16.5 | 9.38 | 335 | 4 | 24 | T,G | 28, 70, 77 |
| Myeloblastin precursor | P24158 | 27.8 | 8.72 | 136 | 6 | 11 | T,G, D | 86, d21 |
| Neutrophil defensin 1 precursor | P59665 | 10.2 | 5.71 | 471 | 3 | 26 | G | |
| Neutrophil gelatinase-associated lipocalin precursor | P80188 | 22.5 | 9.02 | 494 | 5 | 32 | T,G | 75, 91, 92 |
| Peptidyl-prolyl cis–trans isomerase A | P62937 | 18.0 | 7.68 | 132 | 4 | 28 | T,G, D | 116, 117, d70 |
| Peroxiredoxin-1 | Q06830 | 22.1 | 8.27 | 52 | 1 | 5 | D | d62 |
| Peroxiredoxin-5 | P30044 | 22.0 | 8.85 | 57 | 3 | 18 | T | 128 |
| PH domain leucine-rich repeat protein phosphatased | O60346 | 185 | 5.88 | 43 | 2 | 43 | G | |
| Phosphoglycerate kinase 1 | P00558 | 44.6 | 8.30 | 190 | 8 | 26 | T,G | 38, 110 |
| Plastin-1d | Q14651 | 70.3 | 5.33 | 41 | 2 | 2 | G | |
| Plastin-2e | P13796 | 70.2 | 5.20 | 345 | 14 | 13 | T,G | 65 |
| Poly-Ig receptore | P01833 | 83.2 | 5.58 | 451 | 7 | 11 | T,G | 62, 63 |
| Profilin-1 | P07737 | 15 | 8.44 | 163 | 4 | 37 | G, D | d1–2 |
| Proteasome subunit alpha type 5d | P28066 | 26.3 | 4.74 | 66 | 2 | 10 | T | 129 |
| Protein disulfide isomerase precursor | P07237 | 57.1 | 4.76 | 197 | 6 | 12 | T, D | 60, d14, d49 |
| Protein S100-A11 | P31949 | 11.7 | 6.56 | 48 | 2 | 8 | T | 13 |
| Protein S100-A6d | P06703 | 10.1 | 5.33 | 50 | 3 | 46 | T,G | 23 |
| Protein S100-A7 | P31151 | 11.5 | 6.27 | 194 | 7 | 61 | T,G | 11 |
| Protein S100-A8e | P05109 | 10.8 | 6.51 | 1083 | 8 | 50 | T,G, D | 6, 7, 9, 12, 27, 130, d66 |
| Protein S100-A9e | P06702 | 13.2 | 5.71 | 1526 | 7 | 63 | T,G | 8, 15–16, 19–20, 25, 45 |
| Protein S100-A12 | P80511 | 10.6 | 41 | 2 | D | d64 | ||
| Pyruvate kinase isozymes M1/M2 | P14618 | 57.9 | 7.96 | 158 | 5 | 10 | T | 73 |
| Serpin B1 | P30740 | 42.7 | 5.90 | 707 | 14 | 43 | T,G, D | 42, 93, 131, d16, d26 |
| Serpin B13 | Q9UIV8 | 44.2 | 5.48 | 107 | 6 | 22 | T, D | 72, d25 |
| Serpin B3e | P29508 | 44.5 | 6.35 | 2296 | 19 | 46 | T,G, D | 39–42, 71, 88–89, 91–92, 132–134, d17, d26–29 |
| Serum albumin precursor | P02768 | 69.3 | 5.92 | 15383 | 119 | 71 | T,G, D | 58, 71, 83, 88, d15 |
| Small proline-rich protein 3 | Q9UBC9 | 18.1 | 8.86 | 94 | 5 | 23 | G | |
| Superoxide dismutase [Mn] | P04179 | 24.7 | 8.35 | 78 | 3 | 13 | T | 118 |
| Thioredoxin | P10599 | 11.7 | 4.82 | 49 | 1 | 12 | G, D | d60 |
| Transaldolase | P37837 | 37.5 | 6.36 | 154 | 2 | 6 | T | 90, 94–95 |
| Transferrine | P02787 | 77.0 | 6.81 | 855 | 20 | 32 | T,G | 61 |
| Transthyretin precursor | P02766 | 15.9 | 5.52 | 228 | 6 | 63 | T | 18, 26 |
| Triosephosphate isomerase | P60174 | 26.7 | 6.45 | 83 | 4 | 19 | T | 52 |
| Tropomyosin-1 alpha chaine | P09493 | 32.6 | 4.69 | 129 | 3 | 9 | T | 105 |
| Tropomyosin beta chaine | P07951 | 32.8 | 4.66 | 153 | 3 | 9 | T | 104 |
| Ubiquitin | P62988 | 8.6 | 6.56 | 84 | 2 | 32 | T | 14 |
| Vitronectin precursor | P04004 | 54.2 | 5.22 | 59 | 1 | 2 | T | 100 |
aSwiss-Prot protein accession number
b T 2D gel data, G GeLC-MS/MS data, D 2D-gel after depletion
c XX and dXX represent the numbers shown in Figs. 1a and 2b, respectively
dProteins not found in previously reported CVF and cervical mucous studies (Refs. [3–9])
ePTM proteins identified in this study. The region denoting heat-shock proteins and Immunoglobulins also showed glycosylation as seen in Fig. 4a
Applying different sample preparation and fractionation methods to a single sample could increase the total number of unique proteins identified. To get a broader proteome coverage, we analyzed the protein composition using two other techniques; 2-DE after immunoaffinity-based depletion of highly abundant proteins, and the combination of 1-DE and liquid chromatography coupled mass spectrometry (GeLC-MS/MS). Removal of highly abundant proteins in serum or other body fluid samples can effectively improve the number of proteins identified by decreasing the dynamic range of protein levels [14]. As shown in Fig. 1b, immunodepletion effectively removed albumin and immunoglobulins from the pooled mucous sample. The biggest spot corresponding to serum albumin on the previous gel (Fig. 1a) almost disappeared after depletion (Fig. 1b), allowing a few more protein spots to be resolved by 2-DE. Ninety-one gel spots were selected and LC-MS/MS analysis of these features allowed the identification of ten unique proteins in addition to those identified prior to immunodepletion.
In the GeLC-MS/MS method, the sample was first fractionated by 1-DE and the protein lane excised into 40 equally spaced gel slices (Fig. 1c). Nanocapillary LC-MS/MS analysis of the peptides generated from in-gel digestion of all gel bands resulted in the identification of 63 unique proteins from 7,963 tandem mass spectra.
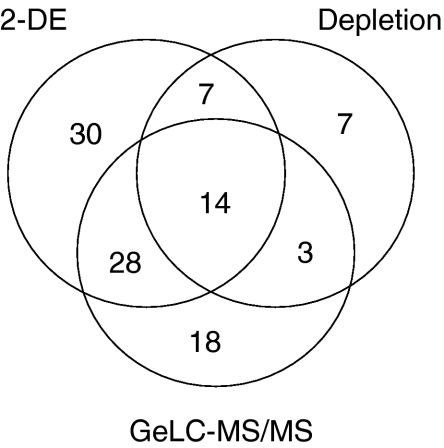
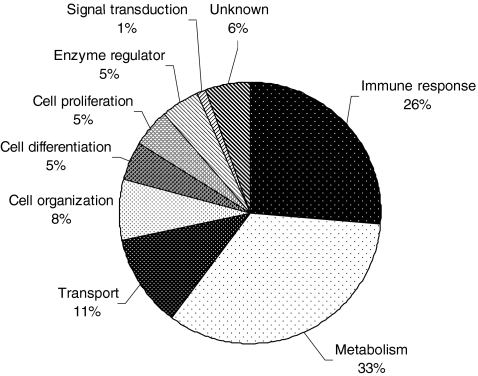
Overall, a total of 107 unique proteins were detected from the combination of the three approaches (Fig. 2). There was overlap in the proteins detected by all methods, but each identified some unique proteins, indicating the advantage of utilizing multiple techniques in sample preparation, fractionation, and analysis. Similar to the findings in CVF, the functional annotation of the majority of identified proteins are metabolism, immune response, and cellular transport process (Fig. 3). Based on the intensities of the protein spots on the 2-DE gels and the MASCOT scores of the proteins identified from GeLC-MS/MS method, the major high abundant proteins were plasma proteins including serum albumin, immunoglobulins, hemoglobins, alpha-1-antitrypsin, and transferrin. Also abundant were calcium-binding proteins S100-A8 and S100-A9. Interestingly, 14 proteins, identified with a superscripted letter “d” in Table 1, were not reported in previous descriptions of CVF and cervical mucous [3–9].
Fig. 2.
Total number of proteins identified using three different strategies for protein fractionation
Fig. 3.
Functional annotation of proteins identified in cervical mucous
While our preliminary study precludes comment on the role of proteins detected to serve as biomarkers of cervical disease, it is of interest that several identified proteins have been previously linked to cervical carcinoma and, in some cases, to pre-invasive disease. Specifically, Annexin, tropomyosin, 14-3-3 sigma, calreticulin and anterior gradient protein 2 will be discussed.
Protein profiling of tissue biopsies has shown Annexin A1, A2, and A5 to be upregulated in cancer when compared to normal tissue [15–17]. Annexins are a family of calcium and phospholipid binding proteins. Many members of the Annexin family are known to undergo alternate splicing yielding isoforms involved in cytoskeleton and cell motility. Other cytoskeletal proteins like tropomysoin 1 and 2 were reported to be downregulated in squamous cervical carcinoma tissue [15]. Isoforms of tropomyosin 1 are thought to prevent proper assembly of microfilaments thus promoting malignant transformation of the cells [18].
The 14-3-3 proteins are a family of ubiquitously expressed eukaryotic proteins that modulate an array of cellular functions including cell cycle control, metabolism, apoptosis and gene transcription. 14-3-3 sigma has been found to be a tumor suppressor [19]. Downregulation of 14-3-3 epsilon was reported in squamous cell carcinoma of the cervix [15, 19].
Alterations in the levels of calreticulin protein have been noted in several cancers, with the protein being downregulated in cervical cancer when compared to normal vaginal tissue [16]. While anterior gradient protein 2 (AGR2) has not been examined in relation to cervical cancer, it has been associated with pancreatic cancer [20]. AGR2 is a member of the protein disulfide isomerase family that is involved in intestinal mucus production, which could have implications for its presence in cervical mucous [21].
The fact that our study detected a lower number of proteins than in some of the studies of the CVF proteome could be attributed to the difference in the sample itself (sample collection and extraction), the need to study pooled rather than individual samples, and limits of our analysis methods. In addition, the absorbent wick used to focus collection at the cervix may not release all proteins [22]. At the same time the focused collection restricts sampling to a local region of the cervix where neoplastic changes occur. In the study by Shaw et al. [7], gauze was placed in the vagina for 1 h, a method quite different from the Weck-Cel and one that is unlikely to be clinically useful. In addition, the use of a pooled sample has the effect of diluting individual differences thereby limiting detection to proteins shared in common by several members in the pool. Both pooled and individual CVF samples were profiled by Shaw et al. [7]; 282 proteins were identified in the individual sample as opposed to 181 proteins in the pooled sample, with only 91 proteins represented in both sets. Finally, the number of proteins resolved could be expected to be increased with the use of gel-free fractionation methods such as polysulfoethyl strong cation exchange fractionation [3, 7, 23] as well as with a higher sensitivity mass spectrometer.
Characterization of Phosphorylation and Glycosylation of Cervical Mucous Proteins
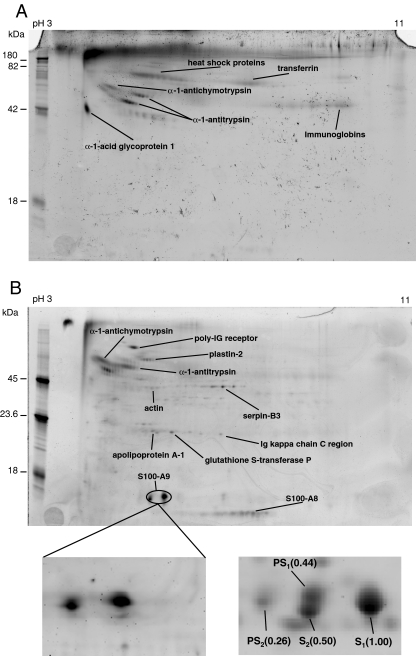
Post-translational modification (PTM) plays an important role in cellular events by altering the physical and chemical property, stability, activity and function of proteins. Among the hundreds of known PTMs, phosphorylation and glycosylation are two of the most common modifications that affect protein function. Aberrant modifications such as hyper-phosphorylation and changes in the glycosylation patterns are usually associated with human diseases and several cancers [24, 25]. A multiplexed gel staining method has been recently developed for the detection of these two popular PTMs together in which 1-DE, or 2-DE gels can be sequentially stained with glycol-specific Pro-Q Emerald, phosphor-specific Pro-Q Diamond, and SYPRO Ruby (for total protein) fluorescent dyes [26]. Unambiguous spot matching of phosphoproteins and glycoproteins to the total proteins can be made by direct comparison with the total protein profile in the same gel and low nanogram sensitivity can be achieved. In comparison with other techniques such as radioisotope labeling and immunodetection with antibodies, the fluorescent dye staining method provides a safe, simple, and streamlined way for highly sensitive detection of protein phosphorylation and glycosylation in a complex sample [27].
The existence of phospho- and glycoproteins in the cervical mucous proteome was implicated by prior detection of some glycoproteins, protein kinases and phosphatases in this study (Table 1). Identification of phosphoproteins and glycoproteins could provide information for understanding molecular mechanisms of protein function and regulation in the development of cervical cancer and for the discovery of post-translationally modified biomarkers. Utilizing the multiple-staining technique, we obtained 2-D gel images of glyco- and phosphoproteins within the cervical mucous proteome where the visualization of positive control bands on the lane of molecular weight markers indicated the efficient labeling of modified proteins (Fig. 4). The identities of modified proteins were determined by comparing gel spots on phospho-, glyco-, and total protein images with the knowledge of prior identification, while some spots were reanalyzed using LC-MS/MS for confirmation. As expected, several proteins were modified by either glycosylation, or phosphorylation, or both (Table 1; Fig. 4a and b). However, it should be noted that no individual phosphor- or glycol-peptides were detected in the MS/MS data, presumably due to the low quantity of PTM modified proteins. A larger amount of starting material and techniques to enrich for these peptides would be required for direct detection of modified peptides.
Fig. 4.
2-DE gel images of cervical mucous proteins stained for phosphoproteins and glycoproteins. a Staining with Pro-Q Emerald for detection of glycosylated proteins; b Staining with Pro-Q diamond for detection of phosphorylated proteins. The enlarged area (bottom left) was compared with the corresponding region in the total protein image stained with SYPRO Ruby fluorescent dye (bottom right). The relative intensities of four labeled spots representing S-100 A9 protein are listed in parenthesis
Acute-phase plasma proteins like α-1-antichymotrypsin and α-1-antitrypsin, were both phosphorylated and glycosylated (Fig. 4a and b). These serine protease inhibitors have been implicated in several human diseases including cancer, and the effect of glycosylation in different cancers is being studied [28, 29]. Heat-shock proteins (HSP), which serve as molecular chaperones, were another group that were overexpressed and are generally linked to poor prognosis in several cancers [30]. HSP 40, HSP 60, and HSP 70 have been shown to be upregulated in pre-invasive lesions of the cervix [31]. Mucins, which are common glycoproteins in mucous [32], were not identified in the present study. Since all three of our approaches include one step of separation/fractionation in SDS/polyacrylamide gel electrophoresis, mucins could have failed to migrate at all into the gels, presumably due to the combination of large size and lack of charged residues [33].
Apart from serpins, several phosphorylated proteins like apolipoprotein A1, actin, plastin 2, glutathione-S-transferase P, and immunoglobulins were identified (Fig. 4b). Two high-intensity spots (PS1 and PS2) on the phosphorprotein image were identified as protein S100-A9 by unambiguous spot matching with unphosphorylated spots (S1 and S2) on the total protein gel (Fig. 4b). While two separated spots (S1 and S2) identified as one protein could be caused by some unknown modification(s), increased mass and negative charges due to partial phosphorylation of the protein/isoform could lead to the formation of new spots shifting to high mass and low pI areas as indicated on the 2-DE gel (PS1 and PS2). If this is the case, the phosphorylation level of S100-A9 could be relatively high (>20%) based on the ratios of the relative intensities of these spots on the total protein 2-D gel. S100-A9 is a member of the S100 family of calcium-binding proteins and has been demonstrated to be a tumor suppressor in some cancers but a tumor promoter in others [34]. Phosphorylation of S100-A9 is thought to play a role in mediating MAPK-dependent functional responses in human neutrophils [35]. A few proteomic studies have also detected differential abundance of this protein in relation to intra-amniotic inflammation, preterm labor/birth, as well as breast and prostate cancer [6, 36–38]. Biological meaning of the high level expression and phosphorylation of protein S100-A9 in the cervical mucous proteome with respect to cervical disease will be investigated in a future study.
Conclusion and Future Work
The expression profiling of human cervical mucous proteins was investigated by a combination of proteomic techniques in this study. One hundred eighty-three of the 2-DE gel spots were analyzed and 79 proteins were identified. An additional 28 proteins were identified from GeLC-MS/MS and depletion experiments. Fourteen of these 107 proteins were determined to be modified with phosphorylation and/or glycosylation. Our data indicated that plasma proteins were abundant in cervical mucous. The majority of proteins identified could be categorized under metabolism and immune-response functional groups. This study found 14 proteins not previously identified in studies of CVF or cervical mucous. Further studies will determine the proteins differentially expressed in disease samples and investigate their potential as protein biomarkers for early detection of cervical neoplasia.
Acknowledgments
This work was supported in part by the National Cancer Institute’s Early Detection Research Network (EDRN), Interagency Agreement Y1-CN-0101-01, Y1-CN-5005-01 and Oak Ridge Institute of Science and Education. The authors have declared no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the funding agency.
Gitika Panicker, Yiming Ye, and Dongxia Wang contributed equally to this paper.
References
- 1.Elstein M. Cervical mucus: its physiological role and clinical significance. Adv Exp Med Biol. 1982;144:301–18. doi: 10.1007/978-1-4615-9254-9_50. [DOI] [PubMed] [Google Scholar]
- 2.Huggins GR, Preti G. Vaginal odors and secretions. Clin Obstet Gynecol. 1981;24:355–77. doi: 10.1097/00003081-198106000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Dasari S, Pereira L, Reddy AP, et al. Comprehensive proteomic analysis of human cervical–vaginal fluid. J Proteome Res. 2007;6:1258–68. doi: 10.1021/pr0605419. [DOI] [PubMed] [Google Scholar]
- 4.Di Quinzio MK, Oliva K, Holdsworth SJ, et al. Proteomic analysis and characterisation of human cervico-vaginal fluid proteins. Aust NZ J Obstet Gynaecol. 2007;47:9–15. doi: 10.1111/j.1479-828X.2006.00671.x. [DOI] [PubMed] [Google Scholar]
- 5.Klein LL, Jonscher KR, Heerwagen MJ, Gibbs RS, McManaman JL. Shotgun proteomic analysis of vaginal fluid from women in late pregnancy. Reprod Sci. 2008;15:263–73. doi: 10.1177/1933719107311189. [DOI] [PubMed] [Google Scholar]
- 6.Pereira L, Reddy AP, Jacob T, et al. Identification of novel protein biomarkers of preterm birth in human cervical-vaginal fluid. J Proteome Res. 2007;6:1269–76. doi: 10.1021/pr0605421. [DOI] [PubMed] [Google Scholar]
- 7.Shaw JL, Smith CR, Diamandis EP. Proteomic analysis of human cervico-vaginal fluid. J Proteome Res. 2007;6:2859–65. doi: 10.1021/pr0701658. [DOI] [PubMed] [Google Scholar]
- 8.Tang LJ, De SF, Odreman F, et al. Proteomic analysis of human cervical–vaginal fluids. J Proteome Res. 2007;6:2874–83. doi: 10.1021/pr0700899. [DOI] [PubMed] [Google Scholar]
- 9.Andersch-Bjorkman Y, Thomsson KA, Holmen Larsson JM, Ekerhovd E, Hansson GC. Large scale identification of proteins, mucins, and their O-glycosylation in the endocervical mucus during the menstrual cycle. Mol Cell Proteomics. 2007;6:708–16. doi: 10.1074/mcp.M600439-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Marshall J, Jankowski A, Furesz S, et al. Human serum proteins preseparated by electrophoresis or chromatography followed by tandem mass spectrometry. J Proteome Res. 2004;3:364–82. doi: 10.1021/pr034039p. [DOI] [PubMed] [Google Scholar]
- 11.Rajeevan MS, Swan DC, Nisenbaum R, et al. Epidemiologic and viral factors associated with cervical neoplasia in HPV-16-positive women. Int J Cancer. 2005;115:114–20. doi: 10.1002/ijc.20894. [DOI] [PubMed] [Google Scholar]
- 12.Panicker G, Lee DR, Unger ER. Optimization of SELDI-TOF protein profiling for analysis of cervical mucous. J Proteomics. 2009;71:637–46. doi: 10.1016/j.jprot.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 14.Liu T, Qian WJ, Mottaz HM, et al. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5:2167–74. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae SM, Lee CH, Cho YL, et al. Two-dimensional gel analysis of protein expression profile in squamous cervical cancer patients. Gynecol Oncol. 2005;99:26–35. doi: 10.1016/j.ygyno.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 16.Hellman K, Alaiya AA, Becker S, et al. Differential tissue-specific protein markers of vaginal carcinoma. Br J Cancer. 2009;100:1303–14. doi: 10.1038/sj.bjc.6604975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson-Bennett BL, Deford J, az-Arrastia C, et al. Implications of tyrosine phosphoproteomics in cervical carcinogenesis. J Carcinog. 2008;7:2. doi: 10.1186/1477-3163-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 19.Lodygin D, Hermeking H. The role of epigenetic inactivation of 14-3-3sigma in human cancer. Cell Res. 2005;15:237–46. doi: 10.1038/sj.cr.7290292. [DOI] [PubMed] [Google Scholar]
- 20.Ramachandran V, Arumugam T, Wang H, Logsdon CD. Anterior gradient 2 is expressed and secreted during the development of pancreatic cancer and promotes cancer cell survival. Cancer Res. 2008;68:7811–8. doi: 10.1158/0008-5472.CAN-08-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SW, Zhen G, Verhaeghe C, et al. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc Natl Acad Sci USA. 2009;106:6950–5. doi: 10.1073/pnas.0808722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohan LC, Edwards RP, Kelly LA, Colenello KA, Bowman FP, Crowley-Nowick PA. Optimization of the weck-Cel collection method for quantitation of cytokines in mucosal secretions. Clin Diagn Lab Immunol. 2000;7:45–8. doi: 10.1128/cdli.7.1.45-48.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gravett MG, Thomas A, Schneider KA, et al. Proteomic analysis of cervical–vaginal fluid: identification of novel biomarkers for detection of intra-amniotic infection. J Proteome Res. 2007;6:89–96. doi: 10.1021/pr060149v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem. 2001;268:5001–10. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 25.Orntoft TF, Vestergaard EM. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis. 1999;20:362–71. doi: 10.1002/(SICI)1522-2683(19990201)20:2<362::AID-ELPS362>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 26.Steinberg TH, Agnew BJ, Gee KR, et al. Global quantitative phosphoprotein analysis using Multiplexed Proteomics technology. Proteomics. 2003;3:1128–44. doi: 10.1002/pmic.200300434. [DOI] [PubMed] [Google Scholar]
- 27.Steinberg TH, Pretty On Top, Berggren KN, et al. Rapid and simple single nanogram detection of glycoproteins in polyacrylamide gels and on electroblots. Proteomics. 2001;1:841–855. doi: 10.1002/1615-9861(200107)1:7<841::AID-PROT841>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Saldova R, Royle L, Radcliffe CM, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17:1344–56. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 29.Tajiri M, Ohyama C, Wada Y. Oligosaccharide profiles of the prostate specific antigen in free and complexed forms from the prostate cancer patient serum and in seminal plasma: a glycopeptide approach. Glycobiology. 2008;18:2–8. doi: 10.1093/glycob/cwm117. [DOI] [PubMed] [Google Scholar]
- 30.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castle PE, Ashfaq R, Ansari F, Muller CY. Immunohistochemical evaluation of heat shock proteins in normal and preinvasive lesions of the cervix. Cancer Lett. 2005;229:245–52. doi: 10.1016/j.canlet.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 32.Lagow E, DeSouza MM, Carson DD. Mammalian reproductive tract mucins. Hum Reprod Updat. 1999;5:280–92. doi: 10.1093/humupd/5.4.280. [DOI] [PubMed] [Google Scholar]
- 33.Tytgat KM, Swallow DM, Van Klinken BJ, Buller HA, Einerhand AW, Dekker J. Unpredictable behaviour of mucins in SDS/polyacrylamide-gel electrophoresis. Biochem J. 1995;310(Pt 3):1053–4. doi: 10.1042/bj3101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34:357–64. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Lominadze G, Rane MJ, Merchant M, Cai J, Ward RA, McLeish KR. Myeloid-related protein-14 is a p38 MAPK substrate in human neutrophils. J Immunol. 2005;174:7257–67. doi: 10.4049/jimmunol.174.11.7257. [DOI] [PubMed] [Google Scholar]
- 36.Goncalves A, Charafe-Jauffret E, Bertucci F, et al. Protein profiling of human breast tumor cells identifies novel biomarkers associated with molecular subtypes. Mol Cell Proteomics. 2008;7:1420–33. doi: 10.1074/mcp.M700487-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermani A, Hess J, De SB, et al. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin Cancer Res. 2005;11:5146–52. doi: 10.1158/1078-0432.CCR-05-0352. [DOI] [PubMed] [Google Scholar]
- 38.Ruetschi U, Rosen A, Karlsson G, et al. Proteomic analysis using protein chips to detect biomarkers in cervical and amniotic fluid in women with intra-amniotic inflammation. J Proteome Res. 2005;4:2236–42. doi: 10.1021/pr050139e. [DOI] [PubMed] [Google Scholar]