Abstract
RNA viruses are particularly vulnerable to RNAi-based defenses in the host, and thus have evolved specific proteins, known as viral suppressors of RNA silencing (VSRs), as a counterdefense. In this issue of Genes & Development, Azevedo and colleagues (pp. 904–915) discovered that P38, the VSR of Turnip crinkle virus, uses its glycine/tryptophane (GW) motifs as an ARGONAUTE (AGO) hook to attract and disarm the host's essential effector of RNA silencing. Several GW motif-containing cellular proteins are known to be important partners of AGOs in RNA silencing effector complexes in yeast, plants, and animals. The GW motif appears to be a versatile and effective tool for regulating the activities of RNA silencing pathways, and the use of GW mimicry to compete for and inhibit host AGOs may be a strategy used by many pathogens to counteract host RNAi-based defenses.
Keywords: Argonaute, GW motif, TCV, viral suppressor
Viruses use viral suppressors of RNA silencing (VSRs) to counteract host RNAi-based antiviral immunity
Small RNA-guided gene silencing is an important host defense strategy against viruses and other pathogens. During the replication of RNA viruses, viral dsRNA intermediates can be processed by Dicer or Dicer-like (DCL) proteins, giving rise to 21- to 24-nucelotide (nt) virus-derived small RNAs (viRNAs). Host RNA-dependent RNA polymerases (RDRs) amplify these primary viRNAs to generate secondary viRNAs that, together with the primary viRNAs, are then recruited by ARGONAUTE (AGO) proteins to target viral RNAs for degradation. To counteract this host defense strategy, viruses have evolved proteins to suppress RNA silencing. These proteins, known as VSRs, are essential for viruses to replicate in host cells and to achieve systemic infection (Ding and Voinnet 2007).
VSRs have been identified from almost all plant virus genera, and also from some animal viruses. They are extremely diverse in sequence and have ORFs within or overlapping with other conserved viral genes. More than 50 VSRs are known, and many of them are multifunctional and play important roles in viral replication, coating, movement, and pathogenesis, in addition to suppressing host RNA silencing-based antiviral immunity (Csorba et al. 2009). Evidence suggests that VSRs suppress RNA silencing pathways via two general mechanisms. The most common mechanism is through dsRNA binding to sequester small RNA duplexes, thus preventing small RNA loading into the AGO effector proteins to assemble the RNA-induced silencing complex (RISC). These dsRNA-binding VSRs differ in primary sequence and structure. The best-studied example is the P19 protein from the plant Tombusvirus (Tombusviridae), which forms a head-to-tail homodimer with two tryptophan residues from each monomer to precisely “measure” and bind siRNA duplexes (Vargason et al. 2003; Ye et al. 2003). In contrast, the tomato aspermy virus (TAV) 2b protein binds siRNA duplexes using a pair of hook-like structures generated between the long α helices of the 2b protein when it forms a dimer (Lucy et al. 2000; HY Chen et al. 2008). Whereas P19 and 2b represent VSRs that bind small RNA duplexes with defined lengths, some other VSRs, such as P21 from the plant closterovirus and B2 from the insect Flockhouse virus (FHV), can bind dsRNAs in a length-independent manner (Li et al. 2002; Ye et al. 2003; Chao et al. 2005).
Another common mechanism for VSR function is through interaction with and inhibition of the protein components of RNA silencing pathways. As the critical effector protein of RNA silencing, AGO is, not surprisingly, an easy target of VSRs. It has been reported that the 2b protein of cucumber mosaic virus (CMV) Fny strain interacts with the PAZ domain of AGO1 and inhibits AGO1 function (Zhang et al. 2006). The P0 protein of beet Polerovirus contains a minimal F-box motif, and appears to mediate the degradation of plant AGO1 (Pazhouhandeh et al. 2006; Baumberger et al. 2007). Furthermore, a recent study suggests that P0 preferentially targets unloaded AGO1 but does not affect preprogrammed, small RNA-loaded AGO1 (Csorba et al. 2009).
Glycine/tryptophane (GW) mimicry—a novel viral strategy to suppress RNA silencing in the host
In spite of the impressive progress in recent years in identifying the genetic and physical targets of VSRs, gaps remain in our understanding of the molecular mechanisms underlying the function of many VSRs. In this issue of Genes & Development, Azevedo et al. (2010) report exciting findings that help fill some of the gaps. They found that the capsid protein P38 of turnip crinkle virus (TCV), a well-known VSR, suppresses RNA silencing by a novel and fascinating mechanism that involves mimicry of cellular proteins. Previously, genetic evidence suggested that P38 suppresses DCL4 function, because DCL2-generated 22-nt viRNAs but not DCL4-generated 21-nt viRNAs accumulated during infection with P38-containing TCV (Deleris et al. 2006). P38-deficient TCV could successfully infect the dcl2–dcl4 double mutant but not the dcl4 single mutant, suggesting that P38 may also suppress 22-nt viRNA-mediated antiviral silencing. Given that DCL2-generated 22-nt viRNAs are associated preferentially with AGO1, and that P38 and hypomorphic ago1 mutation have similar effects on cellular miRNA accumulation, Azevedo et al. (2010) hypothesized that P38 suppresses AGO1 function. Indeed, they showed that P38 could interact with AGO1, as indicated by its cofractionation and coimmunoprecipitation with AGO1 in infected plants. Importantly, they noticed that P38 contains at its N- and C-terminal regions two GW motifs, which are well-known AGO-binding platforms used by some cellular proteins. The N-terminal GW motif is conserved and present in P38 proteins from almost all members of the Carmovirus genus, whereas the C-terminal GW motif is unique to the TCV P38. P38 carrying a GW → GA point mutation in each or both of the N-terminal and C-terminal GW motifs abolished its AGO-binding activity as well as its capacity for silencing suppression in transient assays in tobacco leaves, indicating an important role of the GW motifs in P38 function. TCV carrying the GW → GA double mutant (TCVGA2) version of P38 failed to infect wild-type plants, but the infectivity could be restored in ago1 mutant plants. These results demonstrate a critical role of the GW motifs of P38 in mediating the interaction with AGO1 and in suppression of AGO1-dependent RNA silencing in infected plants.
The inhibition of AGO1 function also has important consequences on cellular microRNA (miRNA) homeostasis. For example, Azevedo et al. (2010) also found that, in TCV-infected plants, the level of miR162 targeting DCL1 was reduced in a P38-dependent manner. Consequently, DCL1 protein level increased dramatically in TCV-infected plants, and DCL3 and DCL4 levels decreased, possibly as a result of the antagonism between these two DCLs and DCL1. Thus, the study by Azevedo et al. (2010) helps to explain the prominent contribution of DCL2 rather than DCL4 to anti-TCV silencing in wild-type plants, and suggests that the apparent targeting of DCL4 by TCV observed previously by the same group (Deleris et al. 2006) is only an indirect consequence of inhibiting AGO1.
The GW motif-mediated AGO1 binding of P38 mimics that of several cellular proteins that bind AGOs and are components of RNA silencing effector complexes. These cellular proteins have been identified from yeasts, plants, and animals, and contain various numbers (two to 40) of GW or tryptophane/glycine (WG) repeats. They interact directly with AGOs via the GW/WG repeats and play important roles in transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS). The GW/WG repeats can act as AGO hooks and are essential for AGO binding. The fission yeast Tas3 protein contains two GW/WG repeats and is a critical part of the RNA-induced transcriptional silencing complex (RITS), which also contains AGO1 and the Clr4 histone methyltransferase (Verdel et al. 2004; Partridge et al. 2007; Till et al. 2007; ES Chen et al. 2008; Zheng et al. 2009). The metazoan Processing body (P-body, also known as GW body) protein GW182 binds to AGO as well as to small RNA targeted transcripts, and thus serves as an adaptor protein in RISC and is important for small RNA-mediated mRNA degradation and translational inhibition (Ding and Voinnet 2007; Eulalio et al. 2009). In plants, the largest subunit of DNA-dependent RNA polymerase V (PolV), NRPE1, interacts with AGO4 of Arabidopsis via its 21 GW/WG repeats in the C-terminal region and functions in the RNA-directed DNA methylation (RdDM) pathway (El-Shami et al. 2007). PolV generates nascent uncapped and nonpolyadenylated RNA transcripts that act as scaffold molecules to assist AGO4-bound siRNAs in targeting homologous loci for de novo DNA methylation by the DNA methyltransferase DRM2 (Wierzbicki et al. 2009). Another GW/WG repeat protein, KTF1, a homolog of the transcription elongation factor SPT5, functions as an adaptor protein in a different step of the plant RdDM pathway by binding to both AGO4 and scaffold transcripts (He et al. 2009).
GW motif as a versatile tool for regulating the activities of RNA silencing pathways
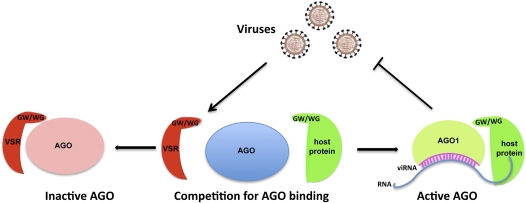
The cellular GW/WG repeat-containing proteins all have a positive role in TGS or PTGS. Their interaction with AGOs through the GW/WG repeats promotes the formation of silencing effector complexes. However, the viral P38 protein is a suppressor of gene silencing. How does the GW-mediated AGO1 binding by P38 lead to inhibition of silencing? Because P38 does not seem to affect AGO1 protein stability, it is possible that P38 binding to AGO1 interferes with the loading of viRNAs and other small RNAs onto AGO1, and/or may prevent loaded AGO1 from assembling into a functional RISC (Fig. 1). The GW/WG motifs of cellular proteins seem to have coevolved with RNA-binding activities because the cellular GW/WG proteins contain either an RNA recognition motif (e.g., GW182) or novel RNA-binding sequences (e.g., KTF1) (He et al. 2009). The RNA-binding activity probably helps the AGO-bound small RNAs to more efficiently search for and find complementary RNA targets for silencing. As a capsid protein, P38 also has RNA-binding activity. P38 might interact with the target RNAs and scaffold transcripts in silencing effector complexes, and thus compete with the RNA binding of the cellular GW/WG proteins. Identification of P38-bound RNAs in infected plants will help answer this question. Although a protein orthologous to the metazoan GW182 has not been reported in plants, AGO1 in plants probably requires association with a GW/WG repeat protein to form a functional RISC. The competition between the viral P38 and the expected cellular GW/WG repeat proteins for AGO1 will determine the balance between—and, ultimately, the coevolution of— RNA silencing-based host defense and viral counterdefense.
Figure 1.
Viral suppression of host RNA silencing by GW mimicry. The P38 VSR of TCV, and probably also other GW motif VSRs, mimic cellular GW/WG repeat-containing proteins in using the GW motif as an AGO hook to bind to AGOs. Whereas the VSR binding causes inactivation of AGOs, the binding by cellular GW/WG proteins leads to the assembly of a functional RISC that can silence viral RNAs. The extent of viral RNA silencing is determined by the competition for AGOs between VSR and cellular GW/WG proteins.
Suppression of silencing by deployment of a GW/WG protein that hooks up to AGOs might serve as a widespread strategy by pathogens and parasites to counteract RNAi-based host defenses. Indeed, another GW motif VSR, the P1 protein from sweet potato mild mottle virus (SPMMV, Potyviridae), has also been found to interact with AGO1 and to inhibit small RNA-programmed RISC (Josef Burgyan, pers. comm.). And, as discussed in the study by Azevedo et al. (2010), several suspected virulence factors of the bacterial pathogen Lysteria monocytogenes (Cabanes et al. 2002) contain GW repeats that may confer interactions with AGOs and interfere with RNA silencing in the mammalian host. It is also interesting to note that the infectious human prion protein contains five GW/WG repeats. Prion proteins can bind RNAs (for review, see Gomes et al. 2008), and prion protein conversion is stimulated by RNAs (Deleault et al. 2003). Thus, the potential interaction mediated by GW/WG repeats of prion protein with AGOs and prion protein's association with RNAs may play a role in the regulation of prion protein conversion and pathogenesis. The study by Azevedo et al. (2010) presents a striking example of viral mimicry of GW proteins to suppress gene silencing in the host. It is tempting to speculate that some cellular GW-based AGO hook proteins might also have evolved not to be part of the silencing effector complexes, but to positively or negatively regulate the activities of RNA silencing pathways.
Acknowledgments
The work in H.J.'s laboratory is supported by National Institutes of Health grant GM093008 and National Science Foundation Career grant MCB-0642843. The work in J.K.Z.'s laboratory is supported by grants from National Institutes of Health, National Science Foundation, and U.S. Department of Agriculture.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1927310.
References
- Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, et al. 2010. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes & Dev (this issue). doi: 10.1101/gad.1908710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC 2007. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17: 1609–1614 [DOI] [PubMed] [Google Scholar]
- Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol 10: 238–245 [DOI] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR 2005. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12: 952–957 [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI 2008. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Chen HY, Yang J, Lin C, Yuan YA 2008. Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep 9: 754–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Pantaleo V, Burgyán J 2009. RNA silencing: An antiviral mechanism. Adv Virus Res 75: 35–71 [DOI] [PubMed] [Google Scholar]
- Deleault NR, Lucassen RW, Supattapone S 2003. RNA molecules stimulate prion protein conversion. Nature 425: 717–720 [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao JS, Kasschau KD, Carrington JC, Voinnet O 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O 2007. Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T 2007. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Dev 21: 2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E 2009. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA 15: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MP, Cordeiro Y, Silva JL 2008. The peculiar interaction between mammalian prion protein and RNA. Prion 2: 64–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK 2009. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW 2002. Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321 [DOI] [PubMed] [Google Scholar]
- Lucy AP, Guo HS, Li WX, Ding SW 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J 19: 1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJP 2007. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell 26: 593–602 [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, et al. 2006. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci 103: 1994–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG 2007. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol 14: 897–903 [DOI] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyán J, Hall TM 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS 2009. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Malinina L, Patel DJ 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426: 874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes & Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu J, Chen X 2009. Intergenic transcription by RNA Polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes & Dev 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]