Abstract
Polycomb group proteins (PcG) are required for proper developmental regulation and cell fate commitment in metazoans. Recently, four studies reported the identification of JARID2, a JmjC domain-containing protein, as a component of the Polycomb-repressive complex 2 (PRC2), which is involved in implementing histone H3 Lys 27 methylation and transcriptional repression during development. Here, we discuss the implications of these studies for an improved understanding of PcG function in development.
Keywords: Polycomb, chromatin, Histone methylation, Jarid2
Polycomb group genes (PcG) were initially identified based on genetic studies as pivotal regulators of Hox genes in Drosophila (Lewis 1978; Struhl 1981). Initial studies in Drosophila—and later in mammalian systems—have highlighted the importance of PcG genes as developmental regulators (Shumacher et al. 1996; Donohoe et al. 1999; O'Carroll et al. 2001; Voncken et al. 2003; Pasini et al. 2004). Increasing evidence also suggests that PcG proteins are necessary in the maintenance of proper tissue homeostasis. The absence or increase of certain PcG proteins has been linked to tumorigenesis in various contexts and model organisms (Varambally et al. 2002; Bracken et al. 2003; Kleer et al. 2003; Schlesinger et al. 2007; Kondo et al. 2008; Classen et al. 2009; Martinez et al. 2009). Therefore, the maintenance of proper levels of these proteins appears to be crucial in their respective developmental context. This is particularly true for highly regulated developmental events such as gastrulation, in which transcriptional fine-tuning of a larger number of genes is required for the onset of differentiation. In light of this, it is not surprising that PcG proteins play an essential role in regulating this process. In order to allow for more insight into the function and the mechanisms of the recruitment of PcG proteins to their respective target sites on chromatin, some research has focused lately on embryonic stem (ES) cells as a model for studying the transition that certain genes undergo during the process of cellular differentiation (Boyer et al. 2006; Lee et al. 2006; Pasini et al. 2007; Stock et al. 2007; Endoh et al. 2008; Ku et al. 2008).
Molecular function of Polycomb-repressive complexes (PRCs)
To date, molecular studies support the idea that PcG proteins in Drosophila exist in at least three major complexes, which are thought to be recruited sequentially to chromatin, and to result in the silencing of respective target genes (Wang et al. 2004; Schuettengruber et al. 2007; Schwartz and Pirrotta 2007; Simon and Kingston 2009). According to this model of sequential recruitment, the main members of the first complex, which include Pleiohomeotic (Pho) and dSfmbt, are targeted to chromatin. Pho, in turn, has been shown to act upstream of PRC2, a histone H3 Lys 27 methyltransferase (H3K27 HMTase). In Drosophila, PRC2 consists of the core components Enhancer of zeste [E(z)], Extra sex combs (Esc), Suppressor of zeste 12 [Su(z)12], and Nurf55 (EZH2, EED, SUZ12, and RBAP46/48 in humans). E(z)—the catalytic subunit of this complex—catalyzes the methylation of H3K27 (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). Trimethylation of H3K27 (H3K27me3) is, in turn, believed to provide a platform for Polycomb's (Pc's) interaction with chromatin, which, together with Polyhomeotic (Ph), dRING, and Posterior sex combs (Psc), constitute the core of PRC1 (CBX, RING1, BMI1, and PH in humans) (Schuettengruber et al. 2007; Schwartz and Pirrotta 2007; Simon and Kingston 2009). Even though much is already known about the basic core components of these three complexes in general, little is known as to how these individual complexes are targeted to chromatin (particularly in mammalian cells) and which proteins are involved in these processes. EED and AEBP2 in humans—and, more recently, Polycomb-like (Pcl) (PHF1, MTF2, and PHF19 in humans)—have been described as regulators of PRC2 that are required for optimal H3K27me3 activity of E(z) (O'Connell et al. 2001; Cao and Zhang 2004; Nekrasov et al. 2007; Cao et al. 2008; Sarma et al. 2008; Savla et al. 2008; Margueron et al. 2009). Others have suggested that Pcl not only may be involved in regulating PRC2's enzymatic activity (Nekrasov et al. 2007; Sarma et al. 2008), but rather could function in PRC2 recruitment as well (Savla et al. 2008).
JARID2, a novel regulator of PRC2
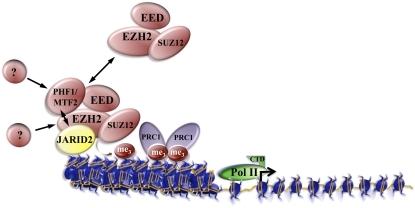
Given the high level of interest in understanding the molecular machinery required for proper PRC2 function and recruitment to chromatin, four recent studies—one in Genes & Development (Li et al. 2010), one in Nature (Pasini et al. 2010), and two in Cell (Peng et al. 2009; Shen et al. 2009)—have independently identified JARID2/JUMONJI, the founding member of the JmjC domain-containing protein family, as a novel component of PRC2 (Fig. 1). JARID2 has already been recognized as an important developmental regulator in mice and Drosophila (Takeuchi et al. 1995, 2006; Sasai et al. 2007) for its role in heart development (Lee et al. 2000; Kim et al. 2003; Toyoda et al. 2003; Jung et al. 2005), transcriptional regulation (Kim et al. 2003, 2004), cell cycle regulation (Jung et al. 2005), and Drosophila metamorphosis (Sasai et al. 2007). At the molecular level, JARID2 has been described as an important regulator of several cell cycle genes, including cyclin D1 (Jung et al. 2005). It has been demonstrated that JARID2 collaborates with the tumor suppressor Retinoblastoma by inhibiting E2F-mediated transcription (Jung et al. 2005), and is part of a G9a- and GLP-containing protein complex that silences cyclin D1 transcription by promoting H3K9 methylation on the cyclin D1 promoter (Shirato et al. 2009). In contrast to other JmjC domain-containing proteins, JARID2 does not share the conserved residues that are essential for histone demethylase activity. Therefore, JARID2 had been predicted to be catalytically inactive (Klose et al. 2006; Takeuchi et al. 2006). In light of this, it is even more exciting that another essential role for JARID2 has been unveiled. Now, four independent studies concur on JARID2's indispensability as an important developmental regulator by showing that it may constitute a substoichiometric subunit of PRC2 (Peng et al. 2009; Shen et al. 2009; Li et al. 2010; Pasini et al. 2010). Independently, the interaction of JARID2 with PRC2 was confirmed by purification of epitope-tagged JARID2, EZH2, EED, or SUZ12 complexes. Immunoprecipitation experiments against the endogenous proteins of the core PRC2 members or JARID2, and the comigration of JARID2 with PRC2 after gel filtration further confirm these findings. Considering the high interest in PcG-mediated repression, it is very surprising that such an essential factor of the PRC2 complex could have been missed for so long. Remarkably, the JARID2 chromatin-binding pattern displays a significant overlap with PRC2 target sites, and obviously correlates very well with the enrichment of these sites for H3K27me3 (Peng et al. 2009; Shen et al. 2009; Li et al. 2010; Pasini et al. 2010). The PCR2 subunits EZH2 and SUZ12 do not have the capacity to bind directly to chromatin (Margueron et al. 2009). Interestingly, JARID2 itself can bind directly to DNA with its C terminus, and appears to particularly prefer GC- and GA-rich motifs (Peng et al. 2009; Li et al. 2010). The identification of JARID2 as a novel component of PRC2 has shed new light on the issue of PRC2 targeting, as strong evidence by all groups reveals that localization of PRC2 to its respective target sites depends largely on JARID2, but that at the same time JARID2 recruitment also requires the presence of PRC2 (Peng et al. 2009; Shen et al. 2009; Li et al. 2010; Pasini et al. 2010). Importantly, several studies emphasize JARID2's requirement as a coordinator of early differentiation events in ES cells and gastrulation in Xenopus, thus confirming its function as an essential developmental regulator (Shen et al. 2009; Li et al. 2010; Pasini et al. 2010). It is very likely that this aspect of JARID2 function in development is also due to its interaction with PRC2, but this awaits further study. Despite this common theme, which depicts JARID2 as an integral modulator of PRC2's function, different laboratories emphasize certain aspects of the role of JARID2 more than others, thus providing us with a colorful image of the diversity of the processes that JARID2 is involved in. Below we attempt to discuss some of the highlights of the published work in a little bit more detail, suggest a model that could synthesize the diverse aspects of JARID2's function, and propose future experiments and strategies that might help us to understand how exactly JARID2 is involved in the regulation of PRC2 activity and its recruitment to target genes.
Figure 1.
PRC2/JARID2 cocomplex and transcriptional regulation. JARID2 and PHF1/MTF2 are regulators of PRC2's activity. Copurification of PHF1/MTF2 together with JARID2 suggests that both candidates at least partially overlap in a subset of PRC2 complexes. Depending on the ratio of JARID2 to PHF1/MTF2 in the PRC2 complexes on each target gene, H3K27 methylation would be slightly up-regulated or down-regulated. As-yet-unknown factors might also be involved in recruiting PHF1/MTF2 and JARID2 to the PRC2 core complex in different developmental and gene-specific contexts, thus contributing to the fine-tuning of H3K27 HMTase activity of PRC2 on its target sites.
The diverse roles of JARID2 in regulating PRC2
Given the fact that PRC2 is well known for its role in epigenetic silencing, it is noteworthy that JARID2 has been reported to be an activator (Li et al. 2010), as well as an inhibitor (Peng et al. 2009; Shen et al. 2009), of PRC2-mediated H3K27 methylation. The positive effect on H3K27 methylation observed by Li et al. (2010) seems to mainly affect the H3K27 monomethylated and dimethylated states, whereas the inhibitory effect described by Shen et al. (2009) results mainly in changes of H3K27 trimethylation levels. The exact conditions appear to be paramount when trying to interpret this duality of JARID2's function. In all likelihood, the modification status of histone and nucleosomal substrates could play an intrinsic part in whether JARID2's incorporation into the PRC2 complex results in an increase or decrease of the H3K27 HMTase activity of EZH2. Furthermore, the individual PRC2 subunits used in these assays might have differed slightly between groups (aside from the required core components). Therefore, the presence or absence of certain PRC2 components could also be a vital determinant of whether JARID2 functions as an inhibitor or an activator.
Further in vitro characterization of JARID2's role in PRC2's HMTase function may not solve these differences, as the substrate usage (free histone vs. nucleosomes, mono- vs. oligonucleosomes, and purified vs. recombinant), enzyme preparations, and type of assays used could contribute to such differences. Ultimately, the in vivo analysis of the role of JARID2 in this process could be more informative. In this regard, the observations of JARID2's ability to function as both a repressor and activator of PRC2's activity are further confirmed, as knockdown of JARID2 results in increased H3K27me3 levels on some target genes (Peng et al. 2009; Shen et al. 2009) and decreased H3K27me3 on others (Li et al. 2010; Pasini et al. 2010). In this context, the question arises as to what could determine the differences in H3K27 methylation states at the JARID2 target sites. It would be worthwhile to identify additional factors that could account for the different effects of JARID2 on PRC2's function. Possible candidates for a PRC2-activating role of JARID2 could be G9a and GLP, as both have been described to interact with JARID2, and could help coordinate transcriptional repression by carrying out H3K9 methylation on target gene promoters (Shirato et al. 2009). Possibly, both H3K9 and H3K27 methylation marks might be involved coordinately in regulating JARID2 target gene expression. Alternatively, genes that are regulated by H3K9 and H3K27 methylation could represent two distinct sets of JARID2 targets. Genome-wide chromatin immunoprecipitation (ChIP) experiments for H3K9 methylation could resolve some of these questions in the future.
Despite the fundamental role of JARID2 in recruiting PRC2 to chromatin, global reductions in H3K27me3 levels are not observed when JARID2's function is removed in ES cells (Shen et al. 2009; Li et al. 2010). A modest increase in H3K27me3 upon JARID2 depletion can be detected in Xenopus (Peng et al. 2009). Generally, these relatively minor bulk changes in H3K27 methylation are difficult to reconcile given the strong accompanying depletion of EZH2, SUZ12, and EED at many PRC2 target genes upon JARID2 knockdown. All in all, these studies suggest that the levels of depletion of EZH2, SUZ12, and EED from chromatin under JARID2 knockdown conditions is comparable with the levels of depletion that can be observed in cells that are devoid of the core PRC2 members EZH2, SUZ12, or EED (Peng et al. 2009; Shen et al. 2009; Li et al. 2010; Pasini et al. 2010). If JARID2 does indeed represent a core component of PRC2, one would expect major changes in bulk H3K27me3 levels upon JARID2 knockdown, similar to what is found for EZH2-, SUZ12-, or EED-depleted cells. It also remains to be determined whether the PRC2 target genes presented in these studies are representative of a general mechanism of PRC2 recruitment by JARID2 on many genes, or whether the target genes analyzed reflect just a subset of PRC2 target genes that are JARID2-controlled. If JARID2 controls recruitment of PRC2 on just a subset of genes, then the modest changes in H3K27me3 upon PRC2 loss could be readily explained. Alternatively, the observed increase in H3K27me3 levels on some PRC2 target genes and the reduction of H3K27me3 on others upon JARID2 knockdown could cancel each other out in the measurement of bulk H3K27 methylation levels.
A model for JARID2 function
The noticeable gap between the minor bulk changes in H3K27me3 and the strong depletion of PRC2 components from their target genes in the absence of JARID2 might be reconciled by hypothesizing that JARID2 constitutes a substoichiometric subunit of PRC2 (Fig. 1). This also seems to be verified by biochemical data, and would allow for other JARID2-lacking PRC2 complexes to remain localized to chromatin and carry out H3K27 methylation (Fig. 1). However, as measured by PRC2 ChIP studies, most PRC2 components appear to be lost from chromatin after JARID2 depletion. One possible model that could provide an explanation for this paradox would allow for the existence of at least two independent PRC2 complexes: one that contains JARID2 (and PHF1/MTF2), which modulates H3K27 methylation; and one or several other JARID2-lacking PRC2 complexes that conduct bulk H3K27 methylation on chromatin (Fig. 1). The JARID2-containing PRC2 complex would be bound much more tightly to chromatin (perhaps through direct binding to DNA by JARID2) and represent a modulator of H3K27 methylation. The modulation toward increasing or reducing H3K27me3 on PRC2 target genes could be determined by the presence of factors such as JARID2 and either PHF1 or MTF2 or other uncharacterized factors, and could depend on the exact gene-specific chromatin context. Since this JARID2 complex can bind to DNA relatively strongly, it could represent the complex generally detected by ChIP experiments. A second PRC2 complex devoid of JARID2 performing bulk H3K27 methylation might interact with chromatin much more transiently, and thus would exist in a “phantom” state, scarcely being able to be tracked on chromatin. This model would be supported by data that has been reported previously, and shows that the H3K27me3 pattern on chromatin does not always correspond exactly to the pattern of PRC2 and PRC1 localization on Polycomb response elements (PREs) (Schwartz et al. 2006). Indeed, it appears that H3K27me3 exists in broad domains, often including the whole gene region, whereas PRC2 and PRC1 members are recruited much more locally (Schwartz et al. 2006). Alternatively, bulk H3K27 methylation by PRC2 might not occur on chromatin. In a first step, PRC2 could methylate H3K27 before it is assembled to form nucleosomes, and, subsequently, JARID2- and PHF1/MTF2-containing PRC2 complexes might access their target sites to fine-tune H3K27 methylation.
Future studies on JARID2 and PRC2-containing complexes
The many aspects of JARID2's function that have been revealed in these studies (Peng et al. 2009; Shen et al. 2009; Li et al. 2010; Pasini et al. 2010) could depict a microcosm of JARID2's and PRC2's function in the regulation of development. The in vitro methyltransferase assays, which seem to give varying results depending on substrate or complexes used in these studies, may not accurately clarify the regulatory steps that occur on chromatin in the process of PRC2 recruitment in the context of a living organism, where the dynamic interplay of many chromatin remodeling activities determines the readout. The present studies clearly emphasize the importance of JARID2 as a developmental regulator for genes that are involved in differentiation (Peng et al. 2009; Shen et al. 2009; Li et al. 2010; Pasini et al. 2010). Further analyses of the role of JARID2 during development and differentiation are likely to yield additional unexpected findings. Ultimately, questions of this nature can be fully addressed only in a living organism. In the past, many aspects of Polycomb group gene function have been addressed convincingly in Drosophila because it has been shown to operate with less redundancy than the PcG counterparts in mammalian systems. In order to answer at least some of the questions that have been raised above concerning the function of JARID2, it would be essential to not only determine the localization of JARID2, PRC2 core components, and their respective readouts (H3K27me3) on chromatin in a wild-type setting, but also to perform these experiments in JARID2 mutant tissue. Changes in the H3K27 methylation patterns in JARID2 mutant tissue would provide a direct readout of JARID2 activity on target genes. Further answers could be provided regarding the increase of H3K27me3 levels on some, and the decrease of H3K27me3 levels on other, PRC2 target genes, and the effects those alterations might have on the transcriptional response of these genes. Ultimately, ChIP-seq experiments in JARID2 mutant tissue would offer another mode of tackling the question of whether JARID2 is required globally or locally for PRC2 recruitment in vivo. Therefore, future studies focusing on the in vivo aspects of JARID2 biology will allow us to gain a better understanding of this fascinating molecular regulator of gene expression.
Acknowledgments
We thank Dr. Edwin Smith for helpful comments on the manuscript, and Laura Shilatifard for ediotrial assistance. H.M.H. is a fellow of the Jane Coffin Childs Memorial Fund. Studies in Shilatifard's laboratory are supported in part by NIH grants R01CA89455, R01CA150265, and R01GM069905 to A.S.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1921610.
References
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K 2003. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22: 5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol Cell 15: 57–67 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y 2008. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol 28: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D 2009. A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK–STAT signaling. Nat Genet 41: 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol 19: 7237–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. 2008. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135: 1513–1524 [DOI] [PubMed] [Google Scholar]
- Jung J, Kim TG, Lyons GE, Kim HR, Lee Y 2005. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J Biol Chem 280: 30916–30923 [DOI] [PubMed] [Google Scholar]
- Kim TG, Kraus JC, Chen J, Lee Y 2003. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem 278: 42247–42255 [DOI] [PubMed] [Google Scholar]
- Kim TG, Chen J, Sadoshima J, Lee Y 2004. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol Cell Biol 24: 10151–10160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci 100: 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y 2006. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 40: 741–750 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE 2000. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res 86: 932–938 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes & Dev 24: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AM, Schuettengruber B, Sakr S, Janic A, Gonzalez C, Cavalli G 2009. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat Genet 41: 1076–1082 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J 2007. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J 26: 4078–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell S, Wang L, Robert S, Jones CA, Saint R, Jones RS 2001. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J Biol Chem 276: 43065–43073 [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K 2010. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D 2008. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 28: 2718–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Kato Y, Kimura G, Takeuchi T, Yamaguchi M 2007. The Drosophila jumonji gene encodes a JmjC-containing nuclear protein that is required for metamorphosis. FEBS J 274: 6139–6151 [DOI] [PubMed] [Google Scholar]
- Savla U, Benes J, Zhang J, Jones RS 2008. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development 135: 813–817 [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G 2007. Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH 2009. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T 2009. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem 284: 733–739 [DOI] [PubMed] [Google Scholar]
- Shumacher A, Faust C, Magnuson T 1996. Positional cloning of a global regulator of anterior–posterior patterning in mice. Nature 383: 250–253 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: Knowns and unknowns. Natl Rev 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Struhl G 1981. A gene product required for correct initiation of segmental determination in Drosophila. Nature 293: 36–41 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T 1995. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes & Dev 9: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S 2006. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn 235: 2449–2459 [DOI] [PubMed] [Google Scholar]
- Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T 2003. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell 5: 85–97 [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629 [DOI] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M 2003. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci 100: 2468–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 14: 637–646 [DOI] [PubMed] [Google Scholar]