Abstract
Wingless (Wg) signaling regulates expression of its target genes via Pangolin and Armadillo, and their interacting cofactors. In the absence of Wg, Pangolin mediates transcriptional repression. In the presence of Wg, Pangolin, Armadillo, and a cohort of coactivators mediate transcriptional activation. Here we uncover Coop (corepressor of Pan) as a Pangolin-interacting protein. Coop and Pangolin form a complex on DNA containing a Pangolin/TCF-binding motif. Overexpression of Coop specifically represses Wg target genes, while loss of Coop function causes derepression. Finally, we show that Coop antagonizes the binding of Armadillo to Pangolin, providing a mechanism for Coop-mediated repression of Wg target gene transcription.
Keywords: Pangolin, Coop, corepressor, signal transduction, Wingless
The Wnt/Wg signaling pathway controls diverse processes such as growth, patterning, tissue and energy homeostasis, and maintenance of somatic stem cells. Misregulation of the Wnt pathway contributes to human diseases including cancer and metabolic disorders (Logan and Nusse 2004; Clevers 2006; Prestwich and Macdougald 2007). Two transcriptional regulators, β-catenin/Armadillo (Arm) and TCF/Lef/Pangolin (Pan), play key roles in regulating the transcriptional outputs of the canonical Wnt/Wg pathway (MacDonald et al. 2009). In the absence of Wnt/Wg ligand, the function of β-catenin/Arm in transcription is inhibited. A “destruction complex” composed of APC, Axin, GSK3β, and CKI mediates phosphorylation of β-catenin/Arm and targets it for degradation via the proteasome pathway (Aberle et al. 1997; Pai et al. 1997). Also, during the off state, TCF/Lef/Pan is bound by corepressors and mediates repression of the Wnt/Wg target genes. Binding of Wnt/Wg ligand to its receptor at the cell surface primes a signaling cascade that inhibits the function of the destruction complex and results in β-catenin/Arm stabilization. Wnt/Wg target genes are activated when stabilized β-catenin/Arm enters the nucleus and recruits other coactivators to target loci through TCF/Lef/Pan (Mosimann et al. 2009).
TCF/Lef/Pan can mediate either transcriptional repression or activation, depending on the context of cofactors to which it is bound. Two universal corepressors, TLE/Groucho and CtBP, are known to function in TCF/Pan-mediated repression (Cavallo et al. 1998; Brannon et al. 1999). Both proteins are capable of recruiting histone deacetylases (HDACs), and bring about chromatin modifications that promote transcriptional silencing (Chen et al. 1999; Shi et al. 2003). In addition, oligomerization of TLE/Groucho may lead to chromatin remodeling over a wider region (Song et al. 2004; Sekiya and Zaret 2007). Following the binding of β-catenin/Arm to TCF/Lef/Pan, many transcriptional coactivators are recruited. One such coactivator is Bcl9/Legless (Lgs), whose primary function is believed to be recruiting Pygopus (Kramps et al. 2002; Thompson et al. 2002). Pygopus can interact with the Mediator complex subunits Med12 and Med13, and the TFIID complex component TAF4 (Carrera et al. 2008; Wright and Tjian 2009). Mammalian Pygopus also binds preferentially to dimethylated Lys 4 of histone H3; the functional importance of this interaction remains to be determined (Fiedler et al. 2008; Gu et al. 2009; Kessler et al. 2009). The C-terminal region of β-catenin/Arm functions as a transactivation domain and interacts with many coactivators involved in chromatin remodeling, including CBP/p300, Parafibromin/Hyrax, MLL1/SET1, Brg-1/Brahma, and the NURF complex (Hecht et al. 2000; Takemaru and Moon 2000; Barker et al. 2001; Mosimann et al. 2006; Sierra et al. 2006; Li et al. 2007; Song et al. 2009).
In this study, we report the identification, via a proteomic approach, of a novel corepressor acting in the Wg signaling pathway. This protein, encoded by CG1621, interacted with Pan in a series of independent assays. In addition, overexpression of CG1621 specifically repressed Wg target genes, while the knockdown of CG1621 derepressed them. Thus, we refer to it as Coop (corepressor of Pan). Similar to the function of TLE/Groucho in the Wnt/Wg pathway, we showed that the interaction between Coop and Pan obstructed the recruitment of Arm, suggesting that the competition between the corepressors and β-catenin/Arm could be a common mechanism to regulate the transcriptional outputs of the Wnt/Wg signaling pathway.
Results and Discussion
Coop interacts directly with Pan
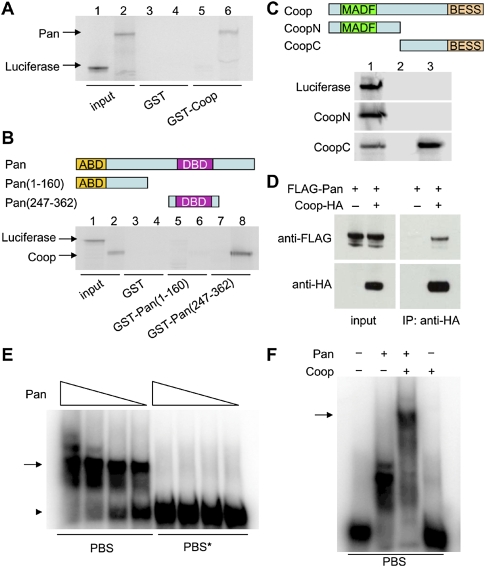
Coop (CG1621) was found in an attempt to identify novel Pan-interacting proteins by a mass spectrometry-based proteomic approach (Supplemental Fig. S1A). It contains an N-terminal MADF domain and a C-terminal BESS domain (Bhaskar and Courey 2002). This architecture is conserved in 16 Drosophila proteins, one C. elegans protein, and one zebrafish protein (Supplemental Fig. S1B). The putative dimeric BESS domain does not exist in mammals, and there is one uncharacterized MADF-containing protein in humans (ZSCAN29/ZNF690). Earlier studies on other members of this family indicated that they might be involved in transcriptional regulation (Cutler et al. 1998; Bhaskar and Courey 2002). We first confirmed the interaction between Coop and Pan in a series of pull-down experiments. In lysates of Drosophila Kc cells, Pan was coimmunoprecipitated by Coop (Fig. 1D). Consistent with a role in transcription, Coop was localized in the nucleus in these cells (Supplemental Fig. S1C). To determine if this interaction was direct, we performed GST pull-down experiments. A GST fusion of Coop could pull down in vitro translated Pan (Fig. 1A). In the reverse experiment, Coop was pulled down by a Pan central fragment (247–362) harboring its DNA-binding domain, but not by a Pan N-terminal fragment (1–160) (Fig. 1B). We further mapped this interaction to the C-terminal part of Coop (161–358), which contains its BESS domain (Fig. 1C).
Figure 1.
Coop and Pan interact with each other and form a complex on DNA. (A–C) In vitro translated samples were used for GST pull-down. (A) Control Luciferase and full-length Pan (lanes 1,2, 15% input) were pulled down by GST (lanes 3,4) or GST-Coop (lanes 5,6). (B) Control Luciferase and Coop (lanes 1,2, 5% input) were pulled down by GST (lanes 3, 4), GST-Pan (1–160) (lanes 5,6), or GST-Pan (247–362) (lanes 7,8). (C) Control Luciferase, CoopN (1–160), and CoopC (161–358) (lane 1, 5% input) were pulled down by GST (lane 2) or GST-Pan (247–362) (lane 3). (D) Kc cells were transfected with Flag-tagged Pan, or cotransfected with Flag-tagged Pan and HA-tagged Coop. Coop was immunoprecipitated by anti-HA antibody-conjugated beads and was analyzed by Western blotting. (E) Pan (247–362) caused a mobility shift (arrow) of a DNA probe harboring a Pan-binding site (PBS), but did not affect the mobility of a DNA probe harboring a mutated Pan-binding site (PBS*). Free probes are indicated by the arrowhead. (F) Coop alone did not bind to a DNA probe harboring a PBS, but caused a supershift in the presence of Pan (arrow).
Since the DNA-binding domain of Pan may be involved in the interaction with Coop, we asked whether Coop affected the DNA affinity of Pan. We addressed this question in a gel shift assay. Pan (247–362) induced the mobility shift of a DNA oligonucleotide containing a consensus Pan/TCF-binding site, but did not affect that of a control oligonucleotide in which the binding site was mutated (Fig. 1E). Coop alone did not bind to this DNA probe. However, in the presence of Pan, Coop induced a supershift of this oligonucleotide (Fig. 1F). This suggests that the interaction between Coop and Pan does not interfere with Pan binding to DNA. Instead, it suggests that Coop and Pan form a complex on Pan targets.
Overexpression of Coop represses Wg targets
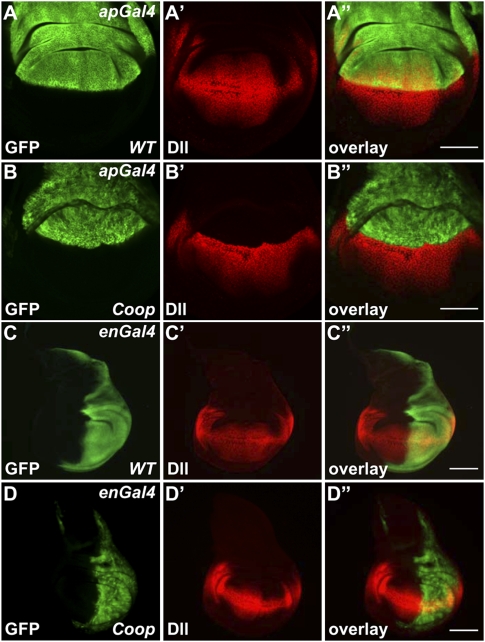
To test whether the interaction between Coop and Pan affects Wg signaling in vivo, we overexpressed Coop in wing imaginal discs and monitored expression of Wg targets, including Distal-less (Dll), senseless (sens), and Notum/wingful (wf) (Nolo et al. 2000; Gerlitz and Basler 2002; Giraldez et al. 2002). The expression of Coop in the dorsal compartment of wing discs by apterous-Gal4 (apGal4) resulted in the loss of Dll protein in this domain (Fig. 2A,B). This repression occurred at the transcriptional level, as the expression of a Dll-lacZ (DllZ) reporter transgene was similarly repressed in the same assay (Supplemental Fig. S2A–C). The expression of Coop in the posterior compartment of wing discs by engrailed-Gal4 (enGal4) also repressed Dll expression, but in a different pattern: Dll staining was lost in the domain away from the dorsal–ventral (D/V) boundary, and was only weakened in the domain close to the D/V boundary (Fig. 2C,D). The Wg morphogen is expressed at the D/V boundary and forms a concentration gradient in the wing disc. It is possible that our observations reflected a dosage-dependent effect of Coop on different levels of Wg signaling. To test this idea, we also expressed Coop at lower levels, either in clones or in the center of wing discs by spalt enhancer-Gal4 (salEGal4). Indeed, we observed the repression of Dll expression preferentially in the domain with lower levels of Wg signaling (Supplemental Fig. S2D–G). These results implied that Coop functions to antagonize incoming Wg signaling.
Figure 2.
Overexpression of Coop represses Wg target Dll in a dosage-dependent manner. (A–A″) The expression of apGal4 alone did not affect Dll expression. (B–B″) The expression of Coop by apGal4 abolished Dll expression. (C–C″) The expression of enGal4 did not affect Dll expression. (D–D″) The expression of Coop by enGal4 strongly reduced Dll expression. Bar, 100 μm.
sens is a high-threshold target of Wg in wing discs and is expressed in two to three rows of cells flanking the D/V boundary. The expression of sens starts at the center of the wing disc and extends to the periphery at late larval stage (Fang et al. 2006). The expression of Coop by enGal4 resulted in the repression of sens in the posterior compartment of wing discs, while the expression of Coop by apGal4 prevented Sens from extending to the periphery in the dorsal compartment (Supplemental Fig. S3A–D). Consistent with our observations with Dll, lower levels of Coop expression by act>CD2>Gal4 or salEGal4 did not affect sens expression (data not shown).
wf is another known Wg target gene. We previously isolated a 4-kb upstream element from the wf locus, which responded to Wg signaling in cell culture. A lacZ transgene under its control (wfZ) mimicked wf expression in wing discs (Supplemental Fig. S3E,F). The expression of Coop repressed this wfZ in wing discs and wf reporter in cultured Drosophila cells (Supplemental Fig. S3G,H). In addition to Wg targets in the wing disc, the expression of Coop also repressed H15, a Wg target gene in the leg disc (Supplemental Fig. S4; Brook and Cohen 1996). Thus, in the case of the four Wg target genes we analyzed, Coop functioned consistently as a negative transcriptional regulator.
The specificity of Coop in the Wg signaling pathway
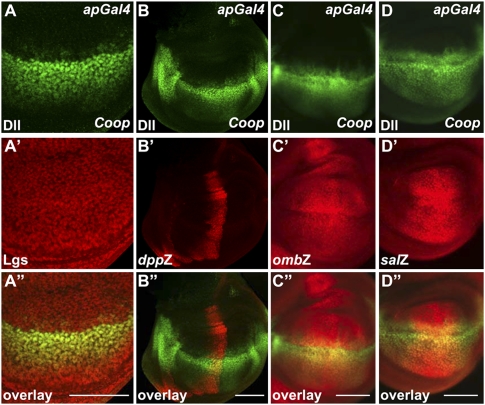
Next we tested whether Coop generally affected transcription, or specifically affected Wg signaling. We first examined the effect of Coop on the expression of Notch targets wg and cut. The expression of Coop by enGal4 or other drivers did not affect expression of either gene (Supplemental Fig. S5A,B; data not shown), confirming that the effect we observed on Wg targets was not due to the interference in the upstream level of Wg signaling. We then expressed Coop by apGal4, which strongly repressed Wg targets, and monitored the expression of other non-Wg targets. Lgs plays a key role in transducing Wg signal. We observed no effect on lgs expression by Coop (Fig. 3A). Similarly, the expression of Coop had no effect on the Hedgehog (Hh) target gene decapentaplegic (dpp), or the Dpp targets optomotor blind (omb) and spalt (sal) (Fig. 3B–D; Supplemental Fig. S6). Similarly, hh, patched (ptc), and en expression were unaffected (Supplemental Fig. S5C–E). In our study we noticed that overexpression of Coop had a weak negative effect on the Gal4/UAS system (Supplemental Fig. S7). It is therefore possible that Coop expression was reduced, leading to an underestimation of the strength of Coop's activity. Whatever the case, the effect does not compromise any of our conclusions, as none of the targets we monitored in this study was driven by the Gal4/UAS system. Taken together, our results suggest that the repressive effect of Coop is probably mediated by its interaction with Pan, and not an indirect effect on wg expression or components of the Wg signaling cascade. Importantly, Coop does not appear to affect Hh or Dpp signaling, suggesting its effect on the Wg pathway is fairly specific.
Figure 3.
Coop is not a universal repressor. (A–D) The expression of Coop by apGal4 strongly repressed Dll, but had no effect on lgs (A′), dpp (B′), omb (C′), or sal (D′) expression. Dll staining is in green, and staining of Lgs or the lacZ reporters is in red. Bar, 100 μm.
Loss of Coop derepresses Wg targets
In the next step, we asked whether Coop was required for the proper regulation of Wg target gene expression. We first examined the effect of Coop RNAi on the basal transcriptional levels of Wg target genes in Drosophila Kc cells. For comparison, we also performed RNAi against Groucho or CtBP, two known corepressors of the Wg pathway. To our surprise, the knockdown of Coop mRNA in Kc cells by dsRNA treatment was very inefficient. In our hands, the levels of most mRNAs could be knocked down to 10%–15% after 4 d (data not shown); however, the levels of Coop mRNA remained at 60%–70% after 4 d and 40%–50% after 7 d (Supplemental Fig. S8A–C). Even so, under this condition, the knockdown of Coop mRNA caused a twofold to threefold increase in Arm-independent basal expression of Wg target gene nkd and CG6234 (Zeng et al. 2000; Fang et al. 2006; Chang et al. 2008), similar to the effect observed with Groucho RNAi, but slightly weaker than the effect seen with CtBP RNAi. In addition, the combination of Coop and CtBP RNAi showed an additive effect (Supplemental Fig. S8D–F).
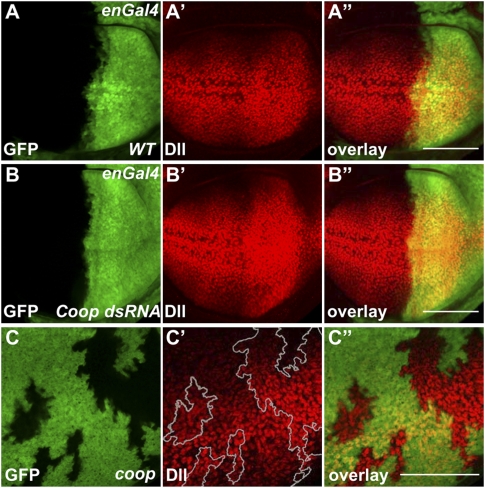
We next examined whether reducing Coop function affected the activation of Wg target genes in vivo. We first tested this by expressing dsRNA against Coop in the wing disc and monitoring expression of Wg targets. Coop is expressed ubiquitously in imaginal discs (Supplemental Fig. S9A–C). The knockdown of Coop enhanced expression of Dll and wf (Fig. 4A,B; Supplemental Fig. S10A,B), suggesting that endogenous Coop also affects Wg-mediated target activation. Starting from an enhancer P element (EP) line, we generated two Coop alleles (hereafter referred to as coop) that encode truncated proteins (Supplemental Fig. S11). coop mutant flies seemed normal, but showed enhanced Wg signaling in a sensitized background: Loss of Coop enhanced a rough eye phenotype caused by ectopic Wg signaling (sev-Wg) (Supplemental Fig. S10C–E). In coop mutant clones, we also observed moderately enhanced expression of Dll in the wing disc (Fig. 4C). This is consistent with our in vivo RNAi results, indicating that Coop is a repressor of Wg target genes. Although coop clones had a weaker effect than RNAi, this difference might be due to the perdurance of Coop protein. The RNAi was induced earlier than the clones of coop, thus probably eliminating Coop more thoroughly (cf. Supplemental Fig. S9D,E).
Figure 4.
Loss of Coop enhances expression of Dll in vivo. (A) Expression of Dll in wild-type control. (B) Expression of dsRNA targeting Coop mRNA by enGal4-enhanced Dll expression. (C) Expression of Dll was enhanced in coop clones induced by hs-flp; FRT42 ubi-GFP Minute/FRT42 coopR62. Bar, 100 μm.
There are 15 sequence homologs of Coop in Drosophila, and it is possible that one or several of them also had similar roles in Wg signaling. We examined some Coop family members, and found overexpression of CG6854 and Adf1 also repressed Wg signaling in Drosophila cultured cells (data not shown). As a next step, we tested the possibility that CG6854 or Adf1 acts like Coop in vivo. Overexpression of CG6854 strongly repressed expression of Dll and sens (Supplemental Fig. S12A–E). However, in contrast to Coop, it also repressed expression of wg and targets of other pathways (Supplemental Fig. S12F,G; data not shown), suggesting the repressive effect of CG6854 may be less specific than that of Coop. Adf1 behaved like CG6854 (data not shown). Taking these results together, we propose that, unlike Coop, Adf1 and CG6854 are not specific repressors of the Wg pathway.
Coop competes with Arm for binding to Pan
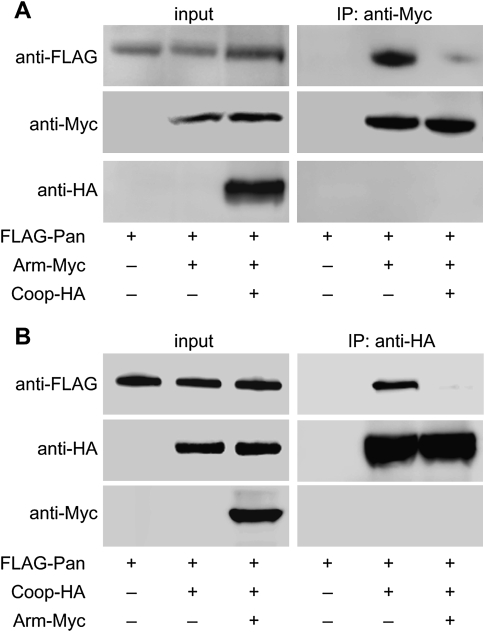
Having established that Coop has a defined role in Wg signaling, in contrast to CG6854 and Adf1, we then investigated the mechanism by which Coop represses Wg target genes. As the interaction between Arm and Pan is essential for activation of Wg target genes, we tested whether Coop functions by preventing this process. In cultured Drosophila Kc cells, we overexpressed Pan and Arm in the absence or presence of overexpressed Coop, and coimmunoprecipitated Pan via Arm. The presence of Coop greatly reduced the amount of Pan coimmunoprecipitated (Fig. 5A). Similarly, when we coimmunoprecipitated Pan via Coop, the presence of overexpressed Arm also prevented this interaction (Fig. 5B). These results suggest that the binding of Coop to Pan and the binding of Arm to Pan are mutually exclusive.
Figure 5.
Coop and Arm compete for Pan binding. (Left panels) Presence of transfected Pan, Arm, and Coop are shown by Western blotting. (A, right panels) Coop reduced the amount of Pan coimmunoprecipitated along with Arm. (B, right panels) Arm reduced the amount of Pan coimmunoprecipitated along with Coop.
As Coop can interact with a domain in Pan that is conserved in other TCFs, we were therefore interested in examining whether Coop could also affect Wnt signaling. Interestingly, activation of a Wnt signaling reporter was repressed by ectopic expression of Coop in HEK293T cells (Supplemental Fig. S3I). It is likely that Coop achieved this by interfering with the conserved interaction between β-catenin and TCF. These results indicate a way in which Wnt signaling could be additionally regulated by a functional homolog of Coop. Since BESS domain proteins apparently do not exist in mammals, we postulate that a Coop-like repressor function is carried out by a TCF-interacting protein that is not necessarily structurally related to Coop. Proteomic analysis of TCF interaction partners may help to identify functional homologs of Coop in mammals.
It has been shown in vitro that β-catenin and TLE1 compete for binding to Lef1 (Daniels and Weis 2005). By competing for an overlapping binding site adjacent to the DNA-binding domain of Lef1, TLE1 prevents the recruitment of β-catenin. Here we show that Coop and Arm also compete to bind Pan. Thus, the levels of nuclear β-catenin/Arm, determined by the levels of Wnt/Wg signaling, decide the transcriptional activity of TCF/Lef/Pan. This dosage-dependent, reversible mechanism helps to shape Wnt/Wg gradient-induced expression of downstream targets.
Ectopic Wnt signaling, transduced via interaction between β-catenin and TCF, is often detected in human cancers. Several β-catenin-binding proteins, including ICAT and Chibby, can interfere with the interaction of these two proteins (Tago et al. 2000; Takemaru et al. 2003). Here we report the identification of Coop as another potential blocker of the β-catenin–TCF interaction. As β-catenin has divergent functions in more than Wnt signaling, TCF-binding proteins may help to specifically decrease the transcriptional outputs of ectopic Wnt signaling. Given the specific effect of Coop in the Wg pathway, we believe Coop may function as a specific inhibitor of Arm–Pan interaction in Drosophila. Further studies to map the Coop–Pan interaction may uncover novel ways to prevent the interaction between β-catenin and TCF.
Materials and methods
Antibodies and immunochemistry
The following antibodies were used: mouse anti-Dll (1:500; a gift from Ian Duncan), mouse anti-Wg (4D4, 1:500; Hybridoma Bank), guinea pig anti-Sens (1:800; a gift from Hugo Bellen), mouse anti-Cut (2B10, 1:20; Hybridoma Bank), rabbit anti-HA (1:1000 for Western blot, and 1:200 for immunostaining; ICL), mouse anti-HA (1:1000; Covance Research Products), rabbit anti-Lgs (1:500), rabbit anti-β-galactosidase (1:2000; Cappel), mouse anti-Flag (1:5000 for Western blot, and 1:1000 for immunostaining; Sigma), mouse anti-Myc (9E10, 1:1000; Hybridoma Bank), Alexa-fluor-conjugated secondary antibodies (Molecular Probes), HRP-conjugated secondary antibodies (1:2000; Jackson Laboratories). Anti-Coop antibody was generated using purified Coop (1–160). For immunoprecipitation, mouse anti-HA-conjugated or rabbit anti-myc-conjugated agarose beads (Sigma) were used. X-Gal staining was performed as described (Song et al. 2004).
Fly stocks and plasmid constructs
The following fly lines were used for overexpression: apGal4/SM5a^TM6b, enGal4/cyo, act>CD2>Gal4/TM6b, salEGal4/TM6b, dppGal4/cyo, UAS-GFP/cyo, UAS-Coop, UAS-CG6854, UAS-Adf1. Coop RNAi lines were from VDRC. The coopR62 and coopR56 alleles contain nonsense mutations at W89 and Q182, respectively. The PCR fragments corresponding to Coop 1–358, 1–160 (CoopN), and 161–358 (CoopC); Pan 1–719, 1–160, and 247–362; and Arm 1–853 were cloned into expression vectors and used for interaction studies.
Protein interaction studies
For GST pull-down, [35S]methionine-labeled proteins were diluted into binding buffer (20 mM HEPES at pH 7.9, 100 mM NaCl, 2 mM EDTA, 10% glycerol, 0.2% NP-40, 0.5 mM DTT, protease inhibitor cocktail) and incubated with glutathione beads bearing GST or GST fusion proteins. For coimmunoprecipitation, transfected cells were lysed in the buffer containing 20 mM phosphate (pH 7.4), 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, 0.5 mM DTT, and protease inhibitor cocktail. After extensive washing, bound proteins were eluted and resolved by Nu-PAGE, and analyzed by autoradiography or Western blotting.
Kc cell culture
RNAi was performed as described (Worby et al. 2001). The dsRNA-treated cells were incubated for 4 d, and a second round of dsRNA treatment was performed. Total RNA was isolated after another 3 d. Quantitative RT–PCR was performed and analyzed as described previously (Mosimann et al. 2006). The mRNA levels of Actin5C, α-tubulin, and TBP were used as internal control for normalization. Two independent sets of dsRNA were used to target Coop mRNA and gave similar results.
EMSA
We previously identified a 200-base-pair (bp) Wg-responsive element ∼8 kb upstream of the wf transcription starting site (M Kuster and K Basler, unpubl.). We used a 61-bp oligonucleotide from this element containing a single putative Pan-binding site (PBS) and flanking sequence for our assay (forward strand, tttCTCTGCCGACGTCGCTGCC GCTGCCGCTGCTCTGCAGCTGCCGGCTGAACACTTTGATGTC; reverse strand, tttGACATCAAAGTGTTCAGCCGGCAGCTGCAGAGCAGCGGCAGCGGCAGCGACGTCGGCAGAG). The control oligonucleotide has the same sequence, with a mutated Pan-binding site (PBS*). The oligonucleotides were end-labeled by 32P, and were incubated with purified Pan (247–362) and/or GST-Coop in binding buffer (20 mM HEPES at pH 7.9, 50 mM KCl, 6 mM MgCl2, 0.1 mM EDTA, 0.1 mg/mL BSA, 2 mM DTT, 50 μg/mL d[I-C], 5% glycerol). The bound and free DNA were resolved in 6% DNA retardation gel (Invitrogen) followed by autoradiography.
Reverse-phase LC-MS analysis
Proteins isolated via immunoprecipitation were reduced with 5 mM TCEP and treated with 10 mM iodoacetamide to modify cystein residues. Tryptic digestion was carried out overnight. Samples were purified by reverse-phase C-18 chromatography (Sep-PacK, Waters). For mass spectrometry analysis samples were resuspended in buffer A (5% acetonitrile, 0.2% formic acid). ESI-based LC-MS/MS analyses were performed on an LTQ linear ion trap (Thermo Fisher Scientific). MS/MS spectra were searched using the Mascot software version 2.2 (Matrix Science).
Acknowledgments
We thank G. Hausmann for critical reading of the manuscript, and I. Duncan and H. Bellen for the antibodies. This work was supported by the Swiss National Science Foundation and the Kanton of Zürich. Additional funds were provided by ta Marie Heim-Voegtlin fellowship (PMPDP3_122836) to S.G., the SystemsX.ch initiative within the framework of the WingX Project, and the University Research Priority Program Systems Biology/Functional Genomics of the University of Zurich.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.561310.
Supplemental material is available at http://www.genesdev.org.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R 1997. β-Catenin is a target for the ubiquitin–proteasome pathway. EMBO J 16: 3797–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H 2001. The chromatin remodelling factor Brg-1 interacts with β-catenin to promote target gene activation. EMBO J 20: 4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V, Courey AJ 2002. The MADF–BESS domain factor Dip3 potentiates synergistic activation by Dorsal and Twist. Gene 299: 173–184 [DOI] [PubMed] [Google Scholar]
- Brannon M, Brown JD, Bates R, Kimelman D, Moon RT 1999. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126: 3159–3170 [DOI] [PubMed] [Google Scholar]
- Brook WJ, Cohen SM 1996. Antagonistic interactions between wingless and decapentaplegic responsible for dorsal–ventral pattern in the Drosophila Leg. Science 273: 1373–1377 [DOI] [PubMed] [Google Scholar]
- Carrera I, Janody F, Leeds N, Duveau F, Treisman JE 2008. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc Natl Acad Sci 105: 6644–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A 1998. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395: 604–608 [DOI] [PubMed] [Google Scholar]
- Chang JL, Chang MV, Barolo S, Cadigan KM 2008. Regulation of the feedback antagonist naked cuticle by Wingless signaling. Dev Biol 321: 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ 1999. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes & Dev 13: 2218–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H 2006. Wnt/β-catenin signaling in development and disease. Cell 127: 469–480 [DOI] [PubMed] [Google Scholar]
- Cutler G, Perry KM, Tjian R 1998. Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol Cell Biol 18: 2252–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI 2005. β-Catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 12: 364–371 [DOI] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM 2006. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J 25: 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M, Sanchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Muller J, Evans P, Bienz M 2008. Decoding of methylated histone H3 tail by the Pygo–BCL9 Wnt signaling complex. Mol Cell 30: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz O, Basler K 2002. Wingful, an extracellular feedback inhibitor of Wingless. Genes & Dev 16: 1055–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Copley RR, Cohen SM 2002. HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev Cell 2: 667–676 [DOI] [PubMed] [Google Scholar]
- Gu B, Sun P, Yuan Y, Moraes RC, Li A, Teng A, Agrawal A, Rheaume C, Bilanchone V, Veltmaat JM, et al. 2009. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol 185: 811–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of β-catenin in vertebrates. EMBO J 19: 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R, Hausmann G, Basler K 2009. The PHD domain is required to link Drosophila Pygopus to Legless/β-catenin and not to histone H3. Mech Dev 126: 752–759 [DOI] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Zullig S, Basler K 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear β-catenin–TCF complex. Cell 109: 47–60 [DOI] [PubMed] [Google Scholar]
- Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM 2007. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J 26: 2284–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810 [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X 2009. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev Cell 17: 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K 2006. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with β-catenin/Armadillo. Cell 125: 327–341 [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K 2009. β-Catenin hits chromatin: Regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10: 276–286 [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102: 349–362 [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M 1997. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development 124: 2255–2266 [DOI] [PubMed] [Google Scholar]
- Prestwich TC, Macdougald OA 2007. Wnt/β-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol 19: 612–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS 2007. Repression by Groucho/TLE/Grg proteins: Genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 28: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar EB, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA 2006. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes & Dev 20: 586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Hasson P, Paroush Z, Courey AJ 2004. Groucho oligomerization is required for repression in vivo. Mol Cell Biol 24: 4341–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Spichiger-Haeusermann C, Basler K 2009. The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway. EMBO Rep 10: 1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, et al. 2000. Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes & Dev 14: 1741–1749 [PMC free article] [PubMed] [Google Scholar]
- Takemaru KI, Moon RT 2000. The transcriptional coactivator CBP interacts with β-catenin to activate gene expression. J Cell Biol 149: 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT 2003. Chibby, a nuclear β-catenin-associated antagonist of the Wnt/Wingless pathway. Nature 422: 905–909 [DOI] [PubMed] [Google Scholar]
- Thompson B, Townsley F, Rosin-Arbesfeld R, Musisi H, Bienz M 2002. A new nuclear component of the Wnt signalling pathway. Nat Cell Biol 4: 367–373 [DOI] [PubMed] [Google Scholar]
- Worby CA, Simonson-Leff N, Dixon JE 2001. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci STKE 2001: PL1. doi: 10.1126/stke.200195.pl1 [DOI] [PubMed] [Google Scholar]
- Wright KJ, Tjian R 2009. Wnt signaling targets ETO coactivation domain of TAF4/TFIID in vivo. Proc Natl Acad Sci 106: 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Wharton KA Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP 2000. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature 403: 789–795 [DOI] [PubMed] [Google Scholar]