Abstract
VASA is an evolutionarily conserved RNA helicase essential for germ cell development. The mouse PIWI family proteins MILI and MIWI2 are involved in production of Piwi-interacting RNAs (piRNAs) in fetal male germ cells through a ping-pong amplification cycle. Expression of retrotransposons is elevated in MILI- and MIWI2-deficient male germ cells due to defective de novo DNA methylation, which is presumably caused by impaired piRNA expression. Here, we report that essentially the same abnormalities are observed in MVH (mouse VASA homolog)-deficient mice. Comprehensive analysis of piRNAs in MVH-deficient fetal male germ cells showed that MVH plays crucial roles in the early phase of the ping-pong amplification cycle.
Keywords: Spermatogenesis, MVH, small RNA, piRNA, retrotransposon, epigenetic regulation
There are many evolutionarily conserved proteins that play essential roles in germ cell development. Some proteins—such as VASA, PIWI, and NANOS in Drosophila, and their mammalian homologs—are involved in the processing and binding of RNA, suggesting the importance of RNA processing in germ cell development and differentiation (Saga 2008; Siomi and Kuramochi-Miyagawa 2009). VASA, a germ cell-specific DEAD-box RNA helicase of Drosophila, is essential for germ cell development and oogenesis through regulation of target mRNA such as Nanos (Noce et al. 2001; Becalska and Gavis 2009). Expression of its murine homolog, MVH (mouse vasa homolog), is also restricted to germ cell lineage (Toyooka et al. 2000). Primordial germ cells (PGCs) develop in MVH-deficient mice, but spermatogenesis is blocked at the first meiotic cell division (Tanaka et al. 2000). In round spermatids of the adult testis, MVH is localized in the chromatoid body, a unique cloud-like structure of male germ cells that contains mRNA, microRNA (miRNA), and various proteins, including MVH (Kotaja and Sassone-Corsi 2007). As the chromatoid body would be an intracellular focal domain necessary for RNA processing, MVH is likely to have some pivotal role(s) in RNA processing in male germ cells. However, its molecular role has not been elucidated.
Piwi family genes also show germ cell-specific expression and are essential for germ cell maintenance and spermatogenesis in Drosophila and mammals, respectively (Lin 2007; Peters and Meister 2007; Siomi and Kuramochi-Miyagawa 2009). Piwi was originally identified as a gene essential for germ stem cell maintenance in Drosophila, and its gene family is found in a wide range of organisms from Arabidopsis to mammals (Cox et al. 1998). The three mouse Piwi homologs Miwi, Mili, and Miwi2 are all essential for spermatogenesis (Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007). The phenotypes of Mili and Miwi2 gene targeted mice were essentially the same and showed male sterility due to apoptosis of the germ cells at early pachytene phase (Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007). In addition, both mouse mutants showed enhanced retrotransposon expression in the male germ cells due to defective de novo DNA methylation of the genes (Kuramochi-Miyagawa et al. 2008).
PIWI proteins are bound to a novel class of germ cell-specific small RNAs called Piwi-interacting RNAs (piRNAs) (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). MILI, which is expressed from PGCs at embryonic day 12.5 (E12.5) to round spermatids, binds with 26- to 27-nucleotide (nt) piRNAs (Kuramochi-Miyagawa et al. 2001; Aravin et al. 2006). On the other hand, MIWI2, which is expressed in fetal gonocytes from E15.5 until soon after birth, binds to 28- to 29-nt piRNAs (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). Previously, we showed that most piRNAs at the fetal stage were derived from repetitive retrotransposon genes, and that the production of piRNA was markedly impaired in MILI- and MIWI2-deficient mice (Kuramochi-Miyagawa et al. 2008). These data suggest that MILI and MIWI2 are involved in piRNA production in the fetal male gonads, and that the piRNA would play some important role(s) in gene silencing of retrotransposons via DNA methylation.
Many proteins are involved in piRNA production in Drosophila (Malone et al. 2009). A feed-forward loop to mediate the generation of piRNAs was originally postulated for Drosophila piRNA production (Brennecke et al. 2007; Gunawardane et al. 2007). This ping-pong amplification cycle is mediated by two Drosophila PIWI family proteins, AUB and AGO3, which bind primarily to antisense primary piRNA and secondary sense piRNAs, respectively. Based on the observation that MIWI2 binds preferentially to secondary antisense piRNAs compared with MILI, a similar ping-pong cycle would presumably involve MILI and MIWI2 in the mouse fetal testes instead of AUB and AGO3 in Drosophila (Aravin et al. 2008). It is conceivable that the ping-pong cycle cannot proceed by the actions of MILI and MIWI2 alone, and we attempted to identify other molecules essential for the ping-pong cycle.
MVH is expressed in the male germ cells from E10.5 to around spermatid (Toyooka et al. 2000), which covers the period of de novo DNA methylation of retrotransposons. In addition, we reported previously that the defective spermatogenesis and impairment of gene expression in MILI-deficient mice were similar to those of MVH-deficient mice (Kuramochi-Miyagawa et al. 2004). We also found that both MILI and MIWI bound to MVH. Therefore, we postulated that MVH may play some role(s) in piRNA production and subsequent DNA methylation of retrotransposons. Here, we showed that MVH plays essential roles in de novo DNA methylation of retrotransposons, presumably due to the defective piRNA production, and that MVH is an essential factor in the piRNA processing pathway.
Results and Discussion
Expression and DNA methylation of intracisternal A particle (IAP) and Line-1 retrotransposons in MVH-deficient testes
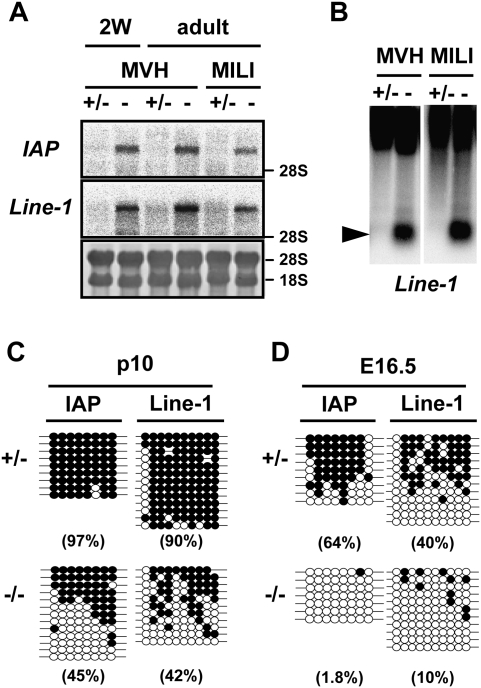
First, we analyzed the expression of IAP and Line-1 retrotransposons, both of which were derepressed in the MILI- and MIWI2-deficient testes. As shown in Figure 1A, expression levels of the 5.4-kb transcript IΔI-type IAP and the Line-1 transcripts were elevated in the testes of MVH-deficient mice. As heavy methylation of the 5′ long terminal repeat (LTR) of IAP and the promoter region of Line-1 was the major cause of transcriptional silencing of these genes, the methylation status of the regulatory regions of these retrotransposons was examined. Methylation-sensitive Southern blotting of Line-1 (Fig. 1B) and bisulfite sequencing analyses of IAP and Line-1 (Fig. 1C) in male germ cells clearly showed that DNA methylation was significantly impaired in MVH-deficient male germ cells at the postnatal stage.
Figure 1.
Expression of the IAP and Line-1 retrotransposons and DNA methylation of their regulatory regions in the MVH-deficient testes. (A) Northern blotting analysis of IAP and Line-1 retrotransposons in the testes of 2-wk-old and 2-mo-old MVH- and MILI-deficient mice. The 3′ noncoding region of IAP and the 5′ noncoding regions of Line-1 were used as probes. (B) Methylation-sensitive Southern blotting analysis of the Line-1 promoter region. DNA was extracted from the testes of 2-wk-old heterozygous and homozygous mice. The probe was the same as that used in A. (C,D) Bisulfite sequencing of IAP and Line-1. Germ cells were collected from p10 testis (C) and fetal testis (D) using EGFP as a marker. The LTR region from the IΔI-type IAP in chromosomes 3qD and the 5′ noncoding regions of Line-1 were analyzed. Filled and open circles represent methylated and unmethylated CpGs, respectively. The percentages of methylated CpGs are shown in parentheses.
As reported previously, de novo DNA methylation of retrotransposons was reduced in MILI- and MIWI2-deficient male germ cells (Kuramochi-Miyagawa et al. 2008). We carried out bisulfite sequencing of the regulatory regions of the Line-1 and IAP retrotransposons in MVH-deficient fetal germ cells at E16.5, when de novo DNA methylation occurred (Fig. 1D). Compared with the control male germ cells, DNA methylation of the MVH-deficient male germ cells was severely impaired. Two paternal imprinted genes, H19 and Dlk-Gtl2, were methylated normally in the MVH-deficient male germ cells, suggesting that de novo DNA methyltransferases function normally even without Mvh (Supplemental Fig. 1). These observations indicated that MVH plays essential roles in de novo DNA methylation and subsequent silencing of the retrotransposons.
Expression and binding of piRNAs in MVH-deficient fetal testes
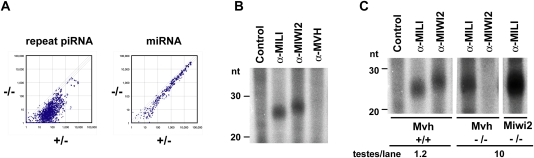
The abnormalities of spermatogenesis and the regulation of retrotransposon gene expression in MVH-deficient mice were quite similar to those of MILI- and MIWI2-deficient mice. As piRNA expression in MILI- and MIWI2-deficient fetal male germ cells was significantly reduced (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008), we analyzed the expression of piRNA in fetal male germ cells of MVH-deficient mice by microarray analysis (Fig. 2A). Although the expression of miRNA was not altered, the expression of piRNA of repetitive sequences was significantly impaired under MVH-deficient conditions. General reduction of piRNA expression in MILI-deficient male germ cells was significantly more severe than that of MIWI2-deficient cells (Kuramochi-Miyagawa et al. 2008). Gross comparison of the piRNA microarray analyses also showed that expression of piRNA in MVH-deficient fetal male germ cells was intermediate between those of MILI- and MIWI2-deficent cells. Reduced expression of arbitrary chosen piRNAs was confirmed by Northern blot analysis (Supplemental Fig. 2).
Figure 2.
piRNA expression in the MVH-deficient fetal testis. (A) Microarray analysis of the repeat-associated piRNAs and miRNAs. A microarray containing 672 types of repeat-associated piRNA and 150 types of miRNA, both of which were expressed in fetal testes, was produced. Testis RNA samples isolated from E16.5 MVH-deficient and heterozygous mice were used. The values normalized with control probes are plotted, and diagonal lines indicate a 1.5-fold difference in expression. (B) Binding of piRNA with MILI, MIWI2, and MVH. The immunoprecipitated RNAs from E16.5 wild-type testicular lysates were 32P-end-labeled and separated in 15% denaturing urea–polyacrylamide gels. (C) MILI- and MIWI2-bound piRNAs in the mutant embryonic testes. The immunoprecipitated RNAs from E16.5 wild-type and mutant testicular lysates were 32P-end-labeled and separated in 15% denaturing urea–polyacrylamide gels. The numbers of testes used for the immunoprecipitation assays are indicated at the bottom.
We examined whether the piRNA was bound to MVH. Extracts of E16.5 wild-type fetal male germ cells were immunoprecipitated with the appropriate antibodies, and the bound small RNAs were radiolabeled and subjected to gel electrophoresis. As shown in Figure 2B, 26- to 27-nt-long and 28- to 29-nt-long piRNAs were bound to MILI and MIWI2, respectively. However, the band of piRNA was undetectable in the complex immunoprecipitated with the anti-MVH antibody.
piRNA expression was reduced in the MVH-deficient testes, but small amounts of piRNA remained. We analyzed the binding of residual piRNAs to MILI and MIWI2 in E16.5 MVH-deficient testes. As shown in Figure 2C, MILI-bound piRNA was detected in the MVH-deficient testes, but MIWI2-bound piRNA was not. These observations were not due to the disappearance of MIWI2 caused by defective MVH expression, because MIWI2 was expressed in MVH-defective mice (Supplemental Fig. 3). Taken together, these results indicate that, although piRNA did not bind to MVH, piRNA expression was significantly decreased in the MVH-deficient fetal testes. In addition, MILI-bound piRNA was present in the MVH-deficient fetal testes, whereas MIWI2-bound piRNA was not.
Characterization of small RNAs in MVH-deficient fetal testes by deep sequencing
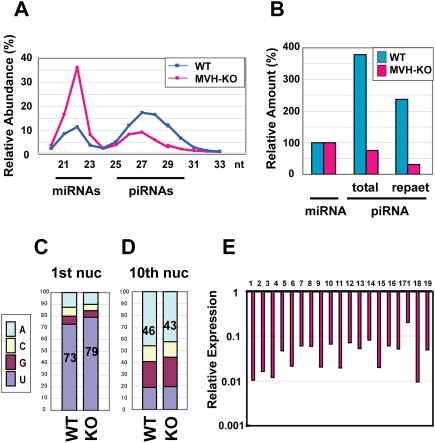
To characterize the piRNAs of the MVH-deficient E16.5 male germ cells in more detail, we sequenced a large number of small RNAs from these cells. In comparison with the controls, the amount of piRNAs was reduced in the MVH-deficient fetal male germ cells (Fig. 3A; Supplemental Table 1). Especially, the number of 28- to 29-nt piRNAs, which were MIWI2-bound piRNAs, was markedly reduced. These observations were consistent with those of microarray (Fig. 2A) and binding (Fig. 2C) analyses. The sequences of small RNAs of control and MVH-deficient E16.5 fetal male germ cells were classified according to the corresponding genes (Supplemental Table 1), and their relative abundances compared with the numbers of miRNAs were calculated (Fig. 3B). The relative amount of piRNAs in the mutant male germ cells was about one-fifth that of the control. piRNAs corresponding to repetitive sequences were markedly reduced in the MVH-deficient fetal male germ cells.
Figure 3.
piRNA expression analysis in the MVH-deficient fetal testis by deep sequencing. (A) The size distribution of the total small RNAs in E16.5 testes. Total numbers of small RNAs sequenced from wild-type and MVH-deficient testis libraries were 2,090,035 and 2,812,945, respectively. (B) Ratio of total and repeat-associated piRNA to miRNA. (C,D) Ratio of the first (C) and 10th (D) nucleotides of the repeat-associated piRNAs. (E) Expression of piRNA cluster-derived piRNAs in the MVH-deficient testes. The data of piRNAs derived from the large 19 piRNA clusters are shown. The ratios of piRNAs in the MVH-deficient testes per those in wild-type testes are shown. (Supplemental Table 2) Number 1 represents the data of the largest piRNA cluster, and the following are ordered according to the sizes of the clusters.
piRNA production can be divided into primary and secondary piRNA processing pathways (Kim et al. 2009). Primary processing is from the precursor RNA, presumably ssRNA, to the MILI-bound sense primary piRNA in mice (Aravin et al. 2008). The secondary processing pathway is the so-called ping-pong cycle, in which MILI-bound piRNAs and MIWI2-bound piRNAs are produced in sequence. piRNAs corresponding to repetitive sequences are believed to be produced by amplification through the ping-pong cycle. The absence of MIWI2-bound piRNAs in the MVH-deficient male germ cells indicated clearly that the ping-pong cycle with MILI and MIWI2 did not progress in the mutant mice. Meanwhile, the first nucleotide of the repeat piRNAs was uridine in >79% of cases (Fig. 3C), indicating that the primary processing pathway functions even in the absence of MVH.
The first step of the ping-pong cycle involves the slicer activity of MILI, which cleaves the 10th nucleotide of the complimentary transcript annealed to the primary piRNA. This cleavage gives rise to adenine at the 10th nucleotide from the 5′ end, which corresponds to the first uridine of the primary piRNA (Brennecke et al. 2007; Gunawardane et al. 2007). As shown in Figure 3D, the percentage of piRNAs from MVH-deficient germ cells with adenine at the 10th nucleotide was almost the same as that of control cells. Therefore, although the ping-pong cycle with MILI and MIWI2 did not proceed, it is possible that the primary piRNA production and the initial part of the cycle—namely, the cleavage of the mRNA corresponding to the primary piRNA—took place without MVH. Meanwhile, we cannot exclude the possibility that MILI alone can proceed the ping-pong cycle in a very inefficient manner. As no MIWI2-bound piRNA was observed in the MVH-deficient cells, MVH appears indispensable between the first step of the secondary piRNA production and the loading of piRNA to MIWI2.
A significant proportion of piRNAs are derived from clusters scattered on the genome. The relative amounts of piRNAs derived from large piRNA clusters are shown in Figure 3E. Compared with the reduction of total piRNAs (∼20% of the control, as shown in Fig. 3B; Supplemental Table 1), the relative abundance of cluster-derived piRNAs was decreased more markedly (∼2.5% of the control, as shown in Fig. 3E; Supplemental Table 2). RNA transcripts from the clusters were postulated to play significant roles in the ping-pong cycle. Thus, the marked reduction of cluster-derived piRNAs is consistent with the defective ping-pong cycle with MILI and MIWI2.
Alterations in the subcellular localizations of MVH, MILI, and MIWI2 in deficient mice
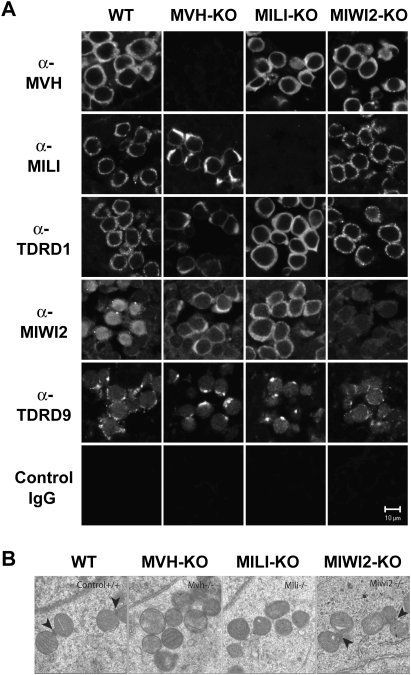
Nuage or germ cell granules are cytoplasmic ribonucleoprotein (RNP) aggregates with amorphous ultrastructural granules localized specifically in the male germ cells (Eddy 1975; Chuma et al. 2009). It has been reported that germ granules have a close relationship with piRNA production and subsequent retrotransposon silencing in the germline. Two groups, including our group, have reported recently that MILI and MIWI2 are components of two distinct subcellular compartments in fetal post-spermatogonia/gonocytes in which de novo DNA methylation is established (Aravin et al. 2009; Shoji et al. 2009). MILI is localized to the intermitochondrial cement, a form of germ granule, and MIWI2 is a component of processing bodies (P-bodies; an mRNP assembly), which are reported to be involved in RNA degradation/translational control, including miRNA- and siRNA-mediated pathways (Aravin et al. 2009; Shoji et al. 2009). Aravin et al. (2009) proposed naming the former and the latter pi-body and piP-body, respectively. In addition, two tudor domain-containing proteins, TDRD1 and TDRD9, both of which are important for piRNA production and gene silencing, belong to the intermitochondrial cement and processing bodies, respectively (Aravin et al. 2009; Shoji et al. 2009). We next analyzed the subcellular localizations of MVH, MILI, MIWI2, TDRD1, and TDRD9 in the mutant gonocytes (Fig. 4A).
Figure 4.
Localization of MILI and MIWI2 in the MVH-deficient fetal germ cells. (A) Immunofluorescence staining of wild-type and mutant testes. Sections of E16.5 testes were stained with anti-MVH, anti-MILI, anti-TDRD1, anti-MIWI2, and anti-TDRD9 antibodies, control IgG. (B) Electron microscopic analyses of MVH-, MILI-, and MIWI2-deficient and control E17.5 testes. Arrowheads indicate intermitochondrial cement.
Immunostaining for MVH, MILI, and TDRD1 in wild-type E16.5 male germ cells showed staining with many cytoplasmic granules, which corresponded to the intermitochondrial cement (Fig. 4A). On the other hand, immunostaining with anti-MIWI2 and anti-TDRD9 antibodies showed a few larger granules, presumably corresponding to the processing bodies, and MIWI2 was detected in the nucleus (Aravin et al. 2009). In MVH-deficient mice, nuclear localization and large granular staining of MIWI2 was not detectable, but that of TDRD9 remained, which suggested that the processing bodies would exist, but MIWI2 was mislocalized. As the subcellular localization of MIWI2 has been suggested to be dependent on piRNA expression (Aravin et al. 2008), it is possible that the mislocalization of MIWI2 is due to the decreased piRNA levels.
Granular staining of both MILI and TDRD1 was abolished in the MVH-deficient germ cells. Examination by electron microscopy indicated that this alteration was not due to the mislocalization of the proteins, but was attributable to the loss of intermitochondrial cement (Fig. 4B). In the MILI-deficient germ cells, intermitochondrial cement was not detected and the numbers of the MVH-stained granules were reduced (Supplemental Fig. 4), suggesting the functional loss of the intermitochondrial cement. The disappearance of intermitochondrial cement was not caused solely by the loss of piRNAs, as the intermitochondrial cement was essentially normal in MIWI2-deficient male germ cells in which piRNA production was significantly reduced. Therefore, it is possible that the loss of intermitochondrial cement was due to the loss of MVH per se or by some molecular sequence downstream from MVH other than piRNA production. Taken together, these observations indicated that MVH is essential for the construction, and presumably also the function, of the intermitochondrial cement. It is unclear whether the molecular function of MVH as an RNA helicase on RNA per se, the role in the formation of intermitochondrial cement, or both is essential for normal piRNA production. We are carrying out the experiments to clarify this point by producing the transgenic mouse bearing the mutant MVH without helicase activity (Sengoku et al. 2006). However, MVH and nuage are essential for the ping-pong amplification in which MVH is involved.
Essential proteins in ping-pong amplification cycle and intermitochondrial cement formation
As shown in Figure 4B, the intermitochondrial cement was missing in the MVH-deficient fetal male germ cells. Intermitochondrial cement was also absent in the TDRD1-deficient fetal male germ cells (Chuma et al. 2006). Recently, several groups have reported that TDRD1 binds physically to MILI, and that the protein is crucial for the production of piRNA (Chen et al. 2009; Kirino et al. 2009; Kojima et al. 2009; Reuter et al. 2009; Vagin et al. 2009). TDRD1-deficient fetal male germ cells showed decreases in levels of MIWI2-bound piRNAs, antisense piRNAs, and 10th A piRNAs, suggesting impairment of the ping-pong cycle (Vagin et al. 2009). Taken together, the similarities in impairment of piRNA production in TDRD1-deficient and MVH-deficient fetal male germ cells support the suggestion that appropriate intermitochondrial cement formation is essential for the progression of ping-pong cycle amplification.
The ping-pong amplification cycle cannot proceed with only MILI and MIWI2. TDRD1 and TDRD9 also play essential roles, presumably in the intermitochondrial cement and processing bodies, respectively (Aravin et al. 2009; Shoji et al. 2009). These two RNP complexes often show neighboring, suggesting that they cooperate and have interdependent functions in piRNA biogenesis. Therefore, it is possible that piRNAs are transferred from one domain to the other during the cycle. The findings of the present study indicating the existence of MILI-bound primary piRNA and the involvement of MVH in the construction and/or function of intermitochondrial cement suggested that MVH is essential for transfer of piRNA from the intermitochondrial cement to the processing bodies.
Materials and methods
RNA extraction, Northern blotting, and microarray analysis
Total RNA was prepared from 2-wk and 2-mo testes and E16.5–E17.5 embryonic testes using Isogen (Nippon Gene). Northern blotting analysis was performed as described in a previous study (Kuramochi-Miyagawa et al. 2008). The probes of IAP and Line-1 were as follows: the 3′ noncoding region of IAP (nucleotides 6486–6793 of GenBank accession no. M17551), and the 5′ noncoding region of Line-1 (nucleotides 2621–2625 of D84391).
Microarray analysis
Total RNA from E16.5–E17.5 testes was purified using Isogen. The target sequences for the microarray were the same as described previously (Kuramochi-Miyagawa et al. 2008). Microarray analysis was performed by a service provider (LC Sciences).
Antibodies
The antibody for MVH was ab13840 (Abcam). The anti-MILI antibodies were PM044 (MBL) for immunoprecipitation and Western blotting, and ab36763 (Abcam) for immunostaining. The anti-MIWI2 antibody for immunoprecipitation was described previously (Kuramochi-Miyagawa et al. 2008), and that for immunostaining was a kind gift from Javier Martines (Aravin et al. 2008).
Small RNA cloning, sequencing, and annotation
To isolate small RNAs, the total RNAs from E16.5 testes were gel-fractionated, and those 16–40 nt in length were purified. The libraries were constructed using the Digital Gene Expression for Small RNA Sample prep kit (Illumina). Sequencing of the small RNA library was performed using a Solexa Genome Sequencer (Illumina). Small RNA annotation was performed as described previously (Kuramochi-Miyagawa et al. 2008). The accession number is GSE20327.
Detailed information is available in the Supplemental Material
Acknowledgments
We thank Dr. D. Pezic and J. Martinez for providing the anti-MIWI2 antibody, and Dr. Takashi Tanaka for supporting the electron microscopy. We also thank Ms. A. Mizokami for secretarial work. This work was supported in part by grants from the Japan Society for the Promotion of Science, the Ministry of Education, Science, Sports and Culture. S.K.-M. is a Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology researcher.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1902110.
Supplemental material is available at http://www.genesdev.org.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A 2009. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. doi: 10.1371/journal.pgen.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska AN, Gavis ER 2009. Lighting up mRNA localization in Drosophila oogenesis. Development 136: 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, et al. 2009. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci 106: 20336–20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N 2006. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci 103: 15894–15899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N 2009. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol 306: 17–23 [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- Eddy EM 1975. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol 43: 229–280 [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev 20: 1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Kuramochi-Miyagawa S, Chuma S, Tanaka T, Nakatsuji N, Kimura T, Nakano T 2009. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells 14: 1155–1165 [DOI] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P 2007. The chromatoid body: A germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol 8: 85–90 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T 2001. Two mouse piwi-related genes: miwi and mili. Mech Dev 108: 121–133 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Lin H 2007. piRNAs in the germ line. Science 316: 397. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noce T, Okamoto-Ito S, Tsunekawa N 2001. Vasa homolog genes in mammalian germ cell development. Cell Struct Funct 26: 131–136 [DOI] [PubMed] [Google Scholar]
- Peters L, Meister G 2007. Argonaute proteins: Mediators of RNA silencing. Mol Cell 26: 611–623 [DOI] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Saga Y 2008. Mouse germ cell development during embryogenesis. Curr Opin Genet Dev 18: 337–341 [DOI] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125: 287–300 [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. 2009. The TDRD9–MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Kuramochi-Miyagawa S 2009. RNA silencing in germlines—Exquisite collaboration of Argonaute proteins with small RNAs for germline survival. Curr Opin Cell Biol 21: 426–434 [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T 2000. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes & Dev 14: 841–853 [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T 2000. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev 93: 139–149 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA 2009. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes & Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev 20: 1732–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]