Abstract
Many proteins are regulated by ubiquitin-dependent proteolysis. Substrate ubiquitylation can be stimulated by additional post-translational modifications, including small ubiquitin-like modifier (SUMO) conjugation. The recently discovered SUMO-targeted ubiquitin ligases (STUbLs) mediate the latter effect; however, no endogenous substrates of STUbLs that are degraded under normal conditions are known. From a targeted genomic screen, we now identify the yeast STUbL Slx5–Slx8, a heterodimeric RING protein complex, as a key ligase mediating degradation of the MATα2 (α2) repressor. The ubiquitin-conjugating enzyme Ubc4 was found in the same screen. Surprisingly, mutants with severe defects in SUMO–protein conjugation were not impaired for α2 turnover. Unmodified α2 also bound to and was ubiquitylated efficiently by Slx5–Slx8. Nevertheless, when we inactivated four SUMO-interacting motifs (SIMs) in Slx5 that together account for its noncovalent SUMO binding, both in vitro Slx5–Slx8-dependent ubiquitylation and in vivo degradation of α2 were inhibited. These data identify α2 as the first native substrate of the conserved STUbLs, and demonstrate that its STUbL-mediated ubiquitylation does not require SUMO. We suggest that α2, and presumably other proteins, have surface features that mimic SUMO, and therefore can directly recruit STUbLs without prior SUMO conjugation.
Keywords: Ubiquitin, SUMO, STUbL, proteasome, transcription factor
Ubiquitin and the small ubiquitin-like modifier (SUMO) proteins are highly conserved eukaryotic proteins that regulate an enormous array of target proteins through covalent modification (Pickart 2001; Kerscher et al. 2006). These modifier proteins are the most widely deployed of the ubiquitin-like proteins (Ubls). The C terminus of ubiquitin or SUMO is attached to the target protein, usually to a lysine side chain, and each is ligated to substrates by the action of a similar series of enzymes. Ubiquitin and SUMO are first activated by a dedicated activating enzyme (E1) that forms a thioester with the modifier C terminus through a cysteine side chain of the E1. The ubiquitin or SUMO is then transferred to the active-site cysteine of a modifier-specific E2-conjugating enzyme. A third enzyme, an E3 ligase, catalyzes the final step of conjugation to target proteins. For ubiquitylation, E3s are key mediators of substrate specificity, and organisms can have many hundreds of different E3s. In the case of SUMO, a much more limited set of E3 enzymes has been found despite the large number of sumoylated proteins (Johnson 2004). The most common class of E3 ligases for ubiquitin and other Ubls have a zinc-coordinating RING domain, which is involved in binding and activating E2s (Joazeiro and Weissman 2000).
A major function of protein ubiquitylation is targeting of the protein to the proteasome for degradation (Ravid and Hochstrasser 2008). Proteins destined for proteolysis by the ubiquitin–proteasome system are usually modified by a ubiquitin polymer, which is recognized by specific proteasomal receptors. Sumoylation is not known to target substrates directly to the proteasome, but an intriguing link has been revealed recently between SUMO and the ubiquitin–proteasome system (for review, see Perry et al. 2008; Geoffroy and Hay 2009). SUMO modification of certain substrates, sometimes as a poly-SUMO chain, renders them into preferred substrates for a distinct class of ubiquitin E3 ligases called SUMO-targeted ubiquitin ligases (STUbLs) (Prudden et al. 2007; Sun et al. 2007; Uzunova et al. 2007; Xie et al. 2007; Lallemand-Breitenbach et al. 2008; Mullen and Brill 2008; Tatham et al. 2008). The STUbLs are a functionally related set of RING proteins found from yeast to humans. In the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, the known STUbLs are heterodimeric species, whereas a single polypeptide comprises the human STUbL, RNF4. Other less fully studied STUbLs that are distinct in sequence have also been suggested (Geoffroy and Hay 2009).
Slx5 (also called Hex3) and Slx8 comprise the best-characterized S. cerevisiae STUbL. These proteins were identified initially from a genetic screen in which slx5 or slx8 mutations were found to be lethal in cells lacking Sgs1, a RecQ-related DNA helicase (Mullen et al. 2001). Slx5–Slx8 is required for resistance to DNA replication stress and DNA damage, as well as for wild-type rates of growth under normal conditions (Zhang et al. 2006). Large poly-SUMO chains, either free or substrate-linked, accumulate in slx5Δ or slx8Δ mutants; these species bind preferentially to Slx5–Slx8, and are degraded in a proteasome-dependent manner (Uzunova et al. 2007).
There is substantial in vitro evidence that Slx5–Slx8 is a ubiquitin E3 ligase that is stimulated by substrate attachment to SUMO (encoded by the SMT3 gene in S. cerevisiae), but no definitive in vivo evidence for this has been published. For example, the Rad52 recombination protein is ubiquitylated by Slx5–Slx8 in vitro, and this activity is stimulated by SUMO attachment to Rad52 (Xie et al. 2007). However, no detectable change in the half-life of bulk Rad52 was observed in slx5Δ or slx8Δ mutants. Recently, a mutant form of the yeast Mot1 transcriptional regulator, Mot1-301, was found to be sumoylated preferentially in vivo, and it is degraded twofold more slowly in cells lacking Slx5–Slx8 (Wang and Prelich 2009). Neither in vitro nor in vivo STUbL-dependent ubiquitylation of Mot1-301 was reported, however. Perhaps the best current evidence for a protein that is subject to (poly-)SUMO-dependent ubiquitylation by a STUbL in vivo is for the human PML protein (Lallemand-Breitenbach et al. 2008; Tatham et al. 2008). The RNF4 STUbL displays SUMO chain-dependent ubiquitylation of PML in vitro, and RNF4 is required specifically for arsenic-stimulated degradation of PML in vivo. Arsenic trioxide is used to treat patients with acute promyelocytic leukemia (APL) due to its ability to stimulate the degradation of an oncogenic PML–retinoic acid receptor-α fusion protein expressed in ∼95% of APL patients.
As part of a long-standing effort to understand regulatory protein degradation in eukaryotes, we have been studying the rapid turnover of the S. cerevisiae MATα2 (α2) cell type regulator (Hochstrasser and Varshavsky 1990; Hochstrasser et al. 1991; Chen et al. 1993; Johnson et al. 1998; Swanson et al. 2001). This model eukaryote has three distinct cell types: two haploid variants (a and α) that can mate with each other to form the third type, a nonmating a/α diploid. Mating phenotype is determined by the genetic information present at the mating type (MAT) locus, which encodes a group of transcriptional regulators that control mating and differentiation (Herskowitz 1985). In most strains found in the wild, cells of one haploid mating type can rapidly differentiate into the opposite cell type. This is accomplished by a gene conversion process in which the genetic information at the MAT locus is replaced with DNA sequences for the other mating type allele copied from one of two transcriptionally silent chromosomal sites.
For these rapid phenotypic changes to occur, the transcription factors that defined the previous cell state must be eliminated (Laney and Hochstrasser 2003; Laney et al. 2006). The ubiquitin–proteasome system is responsible for the destruction of these proteins (Hochstrasser et al. 1991; Chen et al. 1993; Chen and Hochstrasser 1995; Johnson et al. 1998). Recent work suggests that ubiquitin-dependent removal of the α2 protein from chromatin is also important (Wilcox and Laney 2009). Many other transcription factors in a variety of organisms have now also been found to be metabolically unstable ubiquitin pathway substrates (Muratani and Tansey 2003). The α2 repressor has an in vivo half-life of ∼5 min, and is targeted for proteasomal degradation by two major ubiquitylation pathways. The first pathway involves two E2s, Ubc6 and Ubc7, as well as an integral membrane E3, Doa10 (Chen et al. 1993; Swanson et al. 2001). Doa10 moves to the inner nuclear membrane, where it targets a degron, Deg1, in the N-terminal 67 amino acids of α2 (Deng and Hochstrasser 2006). The second major α2 ubiquitylation pathway is defined by the Ubc4 E2 (Chen et al. 1993). Neither the E3 nor the degron in α2 that is recognized by this Ubc4-dependent pathway was known previously. The physiological rationale for having multiple α2 ubiquitylation mechanisms is unknown; this might allow continued α2 degradation (and cell type switching) if one or the other pathway was inhibited under certain environmental conditions, or differential localization of the relevant E2s and E3s might favor complete clearance of α2 throughout the cell.
From a targeted genomic screen, we identified the Slx5–Slx8 heterodimer as the cognate ubiquitin E3 ligase in the Ubc4 pathway of α2 degradation. We show that this E3 works strictly in the Ubc4 pathway and not in the Ubc6/Ubc7/Doa10 pathway of α2 degradation. Previous evidence, from our group and others, suggested that Slx5–Slx8 and related STUbLs exert their diverse physiological functions through targeting of sumoylated proteins in the cell (Heideker et al. 2009). Therefore, we were surprised to find that yeast mutants with little or no ability to conjugate SUMO to proteins degrade α2 normally. Furthermore, the Slx5 subunit of the Slx5–Slx8 complex binds nonsumoylated α2, and the purified recombinant E3 supports efficient α2 ubiquitylation in vitro. We also identified four SUMO-interacting motifs (SIMs) in Slx5 that together are responsible for noncovalent Slx5–SUMO interactions. A yeast strain expressing a version of Slx5 with point mutations in all four of these SIMs (which is expressed at wild-type levels) is hypersensitive to DNA-damaging agents and, surprisingly, is defective for α2 degradation. Slx5–Slx8-dependent ubiquitylation of α2 in vitro was also strongly inhibited by inactivation of the Slx5 SIMs. These unanticipated data suggest that certain proteins have structural features that mimic SUMO and thereby expand the function of STUbLs beyond SUMO-conjugated substrates.
Results
Identification of an E3 ligase active in α2 degradation
As noted, the yeast α2 protein is degraded by two ubiquitin-conjugation pathways (Fig. 1A). The first pathway uses two E2s, Ubc6 and Ubc7, as well as an integral membrane E3, Doa10. A second ubiquitylation mechanism, which uses Ubc4 as the major E2 (Chen et al. 1993), is much less well characterized.
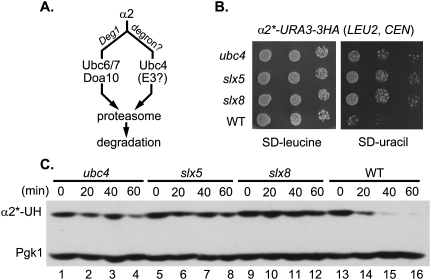
Figure 1.
SLX5 and SLX8 are required for degradation of the α2*-Ura3-3HA reporter protein. (A) Schematic of the two major ubiquitin conjugation pathways that regulate α2 degradation. (B) Growth assay for α2*-Ura3-3HA metabolic stabilization. The slx5Δ and slx8Δ mutants were transformed with a centromeric plasmid carrying the α2*-URA3-3HA allele, grown in liquid medium lacking leucine (to maintain the LEU2 plasmid), and then spotted on both SD-leucine and SD-uracil plates in 10-fold serial dilutions. The α2* protein carries the I4T and L10S substitutions, which inhibit degradation by the Ubc6/7–Doa10 pathway. Pictures were taken after 1 d (SD-leucine) or 3 d (SD-uracil) at 30°C. (C) Cycloheximide-chase/immunoblot analysis of α2*-Ura3-3HA degradation. Cell extracts from the indicated strains were harvested at the indicated times after addition of cycloheximide, and were resolved by 8% SDS-PAGE followed by anti-HA immunoblotting. Anti-Pgk1immunoblotting allowed comparison of protein loading between samples. (α2*-UH) α2*-Ura3-3HA.
To determine what E3 ubiquitin ligase targets α2 for Ubc4-dependent degradation, a targeted genomic screen was performed. We designed a reporter construct in which the Ura3 protein (which is required for uracil biosynthesis) and a triplicated hemagglutinin (3HA) epitope tag were fused to the C terminus of full-length α2. Point mutations (I4T and L10S) in the Deg1 degron of α2 were known to impede α2 degradation by the Doa10 pathway (J Laney and M Hochstrasser, unpubl.), so these mutations were introduced into the reporter to yield the α2*-Ura3-3HA reporter. The α2*-Ura3-3HA fusion protein was rapidly degraded in yeast (see Fig. 1C, lanes 13–16), and the I4T/L10S mutations biased its degradation toward the Ubc4 pathway (Supplemental Fig. 1).
A LEU2 plasmid encoding α2*-Ura3-3HA was transformed into a targeted ubiquitin system (TUS) yeast gene deletion library. The TUS library contains a set of chromosomal gene deletion strains that lack proteins implicated in the ubiquitin system by previous experimental data or by sequence motifs (such as RING domains) (Supplemental Table 2). The TUS strains could be fit onto a single 96-well microtiter plate, which greatly streamlined the analysis. Transformants were initially selected on medium lacking leucine and were subsequently tested for growth on plates lacking uracil. Because the α2*-Ura3-3HA fusion provides the only source of Ura3 in these cells, inhibition of α2*-Ura3-3HA degradation should allow cells to grow more rapidly on the latter plates. Of the 81 mutants tested, deletion of three genes (SLX5, SLX8, and UBC4) led to detectably enhanced growth on medium lacking uracil (Fig. 1B).
Ubc4 is the major E2 for the Doa10-independent pathway of α2 degradation (Fig. 1A), so isolation of ubc4Δ in the screen served as a positive control. To confirm a role for Slx5 and Slx8 in α2*-Ura3-3HA degradation, we used a cycloheximide-chase/immunoblot assay (Fig. 1C). After protein synthesis was blocked, α2*-Ura3-3HA was depleted rapidly in wild-type cells. In contrast, deletion of UBC4, SLX5, or SLX8 strongly stabilized the fusion protein, consistent with a role in α2 degradation. Slx5 and Slx8 are both RING proteins; they form a nuclear-localized heterodimeric complex that plays a critical role in maintaining genome stability (Mullen et al. 2001; Yang et al. 2006; Zhang et al. 2006). Identification of deletions of both SLX5 and SLX8 is consistent with their concerted involvement in α2 degradation.
Slx5–Slx8 targets endogenous α2 as part of the Ubc4 pathway
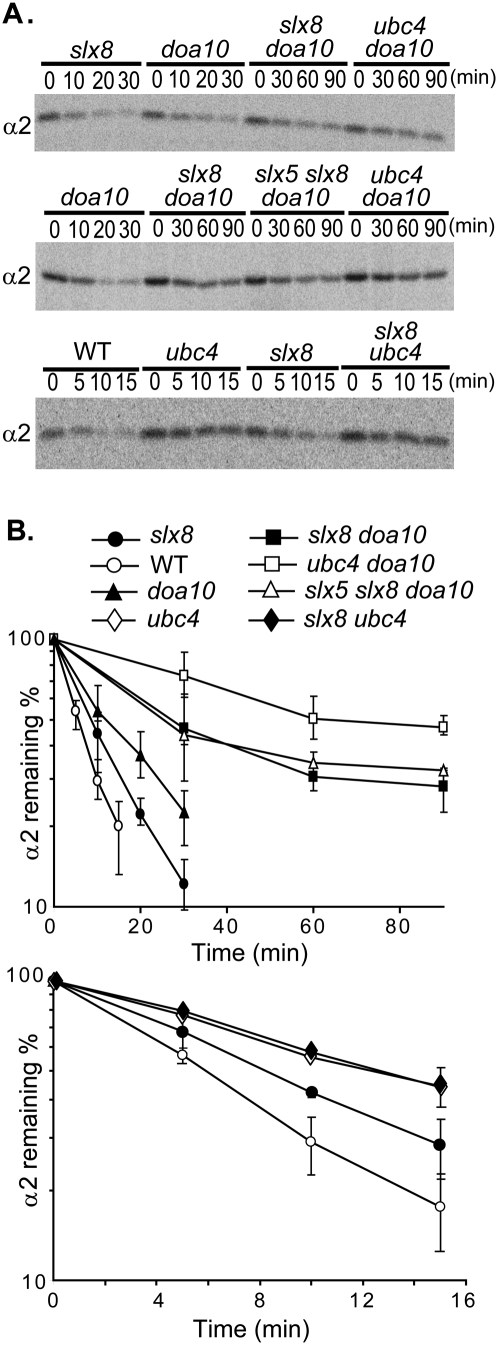
To test the role of Slx5–Slx8 in degrading endogenous α2, pulse-chase analyses were performed (Fig. 2). An ∼1.5-fold stabilization of α2 was observed in slx8Δ compared with wild-type cells. By comparison, α2 was stabilized approximately twofold to threefold in doa10Δ cells, which lack the E3 gene for the other major α2 ubiquitylation pathway (Fig. 1A). When slx8Δ was combined with doa10Δ, greater stabilization of α2 was seen than in either single mutant. These data indicate that Slx8 functions in a pathway distinct from the Doa10 pathway. Mutant slx5Δ cells exhibited kinetics of α2 degradation similar to those observed in slx8Δ (data not shown), and α2 degradation in a slx5Δ slx8Δ doa10Δ triple mutant was indistinguishable from that in slx8Δ doa10Δ cells (Fig. 2). From this epistasis analysis, we conclude that Slx5 and Slx8 function together in a Doa10-independent pathway that targets the endogenous α2 repressor.
Figure 2.
Slx5 and Slx8 contribute to degradation of endogenous α2 as part of the Ubc4-dependent pathway. (A) Representative pulse-chase analysis of α2 degradation in wild-type (WT) and mutant yeast strains. (B) Quantitation of α2 degradation rates in the indicated strains (error bars depict standard deviations; n = 3).
The Ubc4 pathway is known to target α2 independently of the Ubc6/Ubc7/Doa10 pathway (Chen et al. 1993), so Slx5–Slx8 might function with the Ubc4 E2. Epistasis analysis supported this hypothesis: α2 was stabilized to the same degree in the slx8Δ ubc4Δ double mutant as in ubc4Δ alone, with a half-life of ∼12 min (Fig. 2). We also observed that α2 was more stable in ubc4Δ cells compared with slx8Δ cells, and was more stable in doa10Δ ubc4Δ than doa10Δ slx8Δ cells (Fig. 2). These data show that Slx8 functions in the Ubc4-dependent α2 degradation pathway, possibly as part of an E3 ligase; they also suggest that another as-yet-unknown E3 might have a minor role in targeting nascent α2 for Ubc4-dependent degradation (see the Discussion).
Ubiquitylation of α2 by the Ubc4/Slx5–Slx8 machinery
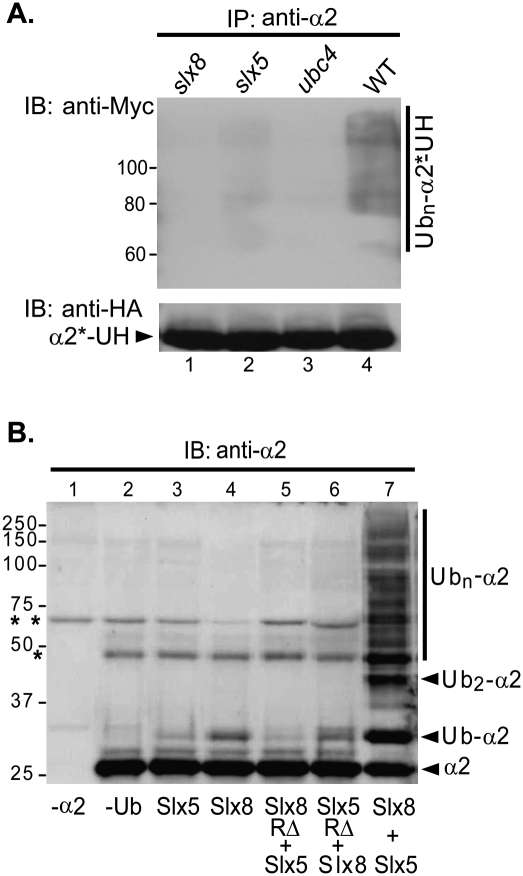
To test if the Slx5–Slx8 E3 uses Ubc4 as its cognate E2 in catalyzing α2 ubiquitylation, we carried out in vivo and in vitro ubiquitylation assays. For the in vivo analysis, the α2*-Ura3-3HA fusion was immunoprecipitated from yeast cells that also expressed Myc epitope-tagged ubiquitin. Ubiquitin conjugation was visualized by anti-Myc immunoblotting (Fig. 3B). A broad array of ubiquitylated α2*-Ura3-3HA species was detected in wild-type cells (Fig. 3A, lane 4). Deletion of SLX8, SLX5, or UBC4 severely compromised this modification (Fig. 3A, lanes 1–3).
Figure 3.
Slx5–Slx8-dependent ubiquitylation of α2 in vivo and in vitro. (A) Ubiquitylation of α2*-Ura3-3HA in yeast cells. A plasmid expressing α2*-Ura3-3HA was transformed along with YEp105, a plasmid encoding Myc epitope-tagged ubiquitin, into wild-type (WT) (MHY501), ubc4Δ (MHY498), slx5Δ (MHY3712), and slx8Δ cells (MHY3716). The α2*-Ura3-3HA protein was precipitated with anti-α2 antibodies bound to Protein A-agarose, and the precipitated species were analyzed by immunoblotting. (α2*-UH) α2*-Ura3-3HA. (B) Ubiquitin conjugation to α2 with recombinant proteins. (Lanes 3–7) Purified α2 substrate (1 μM; from Escherichia coli) was incubated for 2 h at 30°C with Uba1 (E1) (0.1 μM), Ubc4 (E2) (0.37 μM), and 1 μM each the indicated recombinant proteins. Control reactions lacked α2 (lane 1) or ubiquitin (lane 2). Reaction samples were resolved by 10% SDS-PAGE and visualized by anti-α2 immunoblotting. (RΔ) Recombinant Slx5 or Slx8 protein, with the respective RING domain deleted (Xie et al. 2007); (**) nonspecific contaminating band from the purified Slx5 and Slx8 preparations; (*) nonspecific cross-reacting species that copurified with α2.
To determine if the Ubc4/Slx5–Slx8 machinery can directly modify α2, we performed in vitro ubiquitylation assays with recombinant α2, Ubc4, Slx5, and Slx8 proteins purified from bacteria (as well as ubiquitin, E1, and ATP). In the absence of Slx5, purified Slx8 weakly stimulated monoubiquitylation of α2 (Fig. 3B, lane 4); Slx5 by itself did not have a reproducible stimulatory effect. However, addition of Slx5 to Slx8 greatly increased E3 activity (Fig. 3B, lane 7). A RING domain deletion in either Slx5 or Slx8 strongly compromised α2 ubiquitylation (Fig. 3B, lanes 5,6), consistent with the essential role of the RING domains for Slx5–Slx8 activity in vivo (Zhang et al. 2006; Xie et al. 2007). The same assay was performed using α2 purified directly from yeast as a substrate. By this protocol, α2 was ubiquitylated at levels above background only when both Slx5 and Slx8 were present (Supplemental Fig. 2). Our data suggest that Slx8 has an extremely weak basal E3 activity toward α2, and that Slx5 greatly stimulates this activity, possibly by enhancing substrate binding.
Role of SUMO pathway enzymes in Slx5–Slx8-mediated α2 degradation
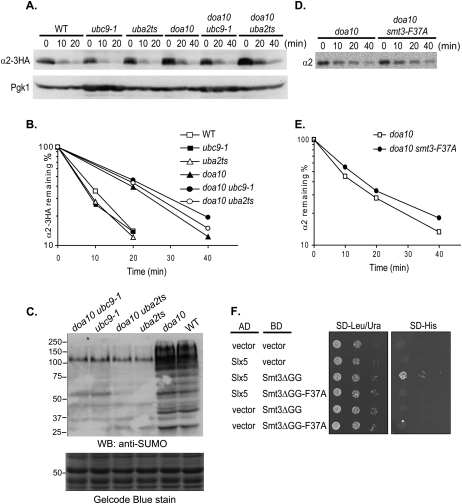
Slx5–Slx8 was originally identified as a STUbL that can ubiquitylate SUMO-modified substrates (for review, see Heideker et al. 2009). However, most of the evidence for the designation as a STUbL has come from in vitro studies with model substrates (e.g., Rad52 [Xie et al. 2007], GST-SUMO [Sun et al. 2007], and Siz2 [Mullen and Brill 2008]). In SUMO–protein conjugation, there is a single E1, the Aos1–Uba2 heterodimer, and one E2, Ubc9; both enzymes are essential for viability (Johnson 2004). Mutants with a temperature-sensitive allele of either UBA2 (uba2ts) or UBC9 (ubc9-1) exhibit a drastic reduction of SUMO conjugates in vivo (see Fig. 4C). Using a functional α2 derivative tagged with a triple HA epitope (α2-3HA) (Wilcox and Laney 2009), we measured degradation of the steady-state pool of α2 by cycloheximide chase/immunoblotting in uba2ts and ubc9-1 cells; α2-3HA was degraded at wild-type rates in both mutants (Fig. 4A,B). In uba2ts doa10Δ and ubc9-1 doa10Δ double mutants, α2-3HA was degraded with kinetics similar to those in the doa10Δ single mutant. To exclude the possibility that the internal 3HA tag might have perturbed degradation, turnover of untagged α2 was studied by the same cycloheximide-chase assay but with anti-α2 immunoblotting. Again, no significant increase in α2 half-life was observed in ubc9-1 doa10Δ cells or uba2ts doa10Δ cells compared with doa10Δ cells (data not shown).
Figure 4.
Mutations in the SUMO pathway do not impair degradation of cellular α2. (A) A plasmid expressing α2-3HA was transformed into the indicated congenic strains. Cells were grown at 30°C and shifted for 30 min to 37°C before the addition of cycloheximide. Whole-cell extract was prepared at various time points after cycloheximide addition, and the disappearance of α2-3HA was followed by anti-HA immunoblot analysis. (B) Plot of α2-3HA degradation rates for the experiment in A. (C) SUMO conjugation profiles of yeast strains shown in A. Extracts from logarithmically growing cells at 30°C were analyzed by anti-SUMO immunoblotting. A portion of the Gelcode Blue-stained membrane is shown to verify comparable sample loading. (D) A SUMO mutant defective for SIM binding does not impair α2 degradation. Representative pulse-chase analyses of endogenous α2 degradation at 30°C in doa10Δ cells or doa10Δ smt3Δ cells harboring the smt3-F37A allele on a centromeric plasmid. (E) Plot of α2 degradation for the experiment in D. (F) Yeast two-hybrid analysis of interaction of Slx5 (pGAD-Slx51–443) (Hannich et al. 2005) with Smt3ΔGG (pGBD-UC1-Smt3ΔGG) or Smt3ΔGG-F37A (pGBD-Smt3ΔGG-F37A). PJ69-4a cells were spotted in sixfold serial dilutions. Two-hybrid interaction was determined by analyzing growth on medium lacking histidine. The F37A mutation did not affect SM3ΔGG protein levels (not shown). (AD) Gal4 transcriptional activation domain; (BD) Gal4 DNA-binding domain.
Pulse-chase analysis confirmed that endogenous untagged α2 was still degraded at wild-type rates in both uba2ts and ubc9-1 cells (Supplemental Fig. 3). When these mutations were combined with doa10Δ, the degradation kinetics were the same as for doa10Δ cells. All of these results are consistent with our inability to detect significant α2 sumoylation in vivo (O Kerscher, Y Xie, and M Hochstrasser, unpubl.). Taken together, these data strongly suggest that Slx5–Slx8-mediated α2 degradation in vivo is not affected by a deficiency in SUMO conjugation.
Noncovalent SUMO interaction and Slx5–Slx8-mediated α2 degradation
Although the data presented above indicated that covalent protein modification by SUMO is not necessary for α2 degradation, it remained possible that noncovalent SUMO–protein interactions nevertheless contribute in some way. Interaction between Slx5–Slx8 and the SUMO moieties of SUMO-linked proteins is mediated by noncovalent binding of SUMO to SIMs in Slx5 (Uzunova et al. 2007; Xie et al. 2007). Mutating the SUMO surface that binds to SIMs (Song et al. 2004), thereby inhibiting the interaction, would allow us to test if there is a function for noncovalent SUMO–protein binding at any stage of α2 degradation.
The SMT3 gene encoding endogenous SUMO was replaced with a mutant version, smt3-F37A; Smt3-Phe37 corresponds to human SUMO1 Phe36, which is essential for SIM binding (Song et al. 2004). Yeast two-hybrid analysis confirmed that the F37A mutation in yeast Smt3 abolished Slx5 binding (Fig. 4F). For these assays, we used a form of SUMO that lacked the C-terminal diglycine motif (Smt3ΔGG). This prevented conjugation of the BD-Smt3 fusion to endogenous wild-type Smt3, which otherwise would lead to an Slx5 two-hybrid interaction regardless of whether the F37A mutation was present (data not shown). Yeast expressing Smt3-F37A as the only source of Smt3 exhibited a weak growth defect at 30°C and were sensitive to ultraviolet (UV) radiation (Supplemental Fig. 4), indicating that Smt3-Phe37 was required for its normal function. However, when α2 degradation was measured in smt3-F37A doa10Δ cells, little or no stabilization was observed relative to doa10Δ cells (Fig. 4D,E). Since much stronger stabilization of α2 was observed in doa10Δ slx8Δ compared with doa10Δ cells (Fig. 2), we conclude that SUMO has little if any role in regulating α2 degradation by the Slx5–Slx8 E3.
Physical interaction between Slx5 and α2
In vitro ubiquitylation of α2 with recombinant proteins purified from bacteria implied that the Slx5–Slx8 complex is able to interact directly with α2 (Fig. 3A). Our observation that α2 proteolysis in vivo is SUMO-independent (Fig. 4; Supplemental Fig. 3) is also consistent with a direct E3–α2 association. We were unable to verify direct E3–α2 interaction in vitro by reciprocal immunoprecipitation due to high background binding of recombinant α2 and Slx5 to the affinity resin. To bypass this technical limitation, an in vivo binding assay between α2 and Slx5–Slx8 was performed. All three proteins were expressed from genes driven by the strong galactose-inducible GAL1 promoter.
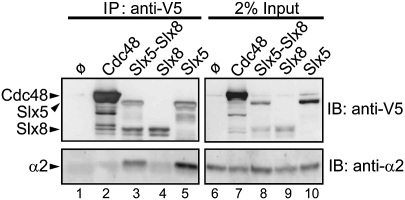
Using V5 epitope-tagged Slx5 or Slx8, we observed that α2 was coimmunoprecipitated by tagged Slx5 but not Slx8 (Fig. 5, lanes 4,5). V5-tagged Cdc48 provided a negative control for interaction with α2 (Fig. 5, lane 2). To test if the Slx5–Slx8 complex binds α2 more efficiently than Slx5 alone, yeast cells that separately overexpressed V5-tagged Slx5 and Slx8 were mixed, lysed, and immunoprecipitated (Fig. 5, lane 3). Since mixing bacterially expressed Slx5 and Slx8 in vitro resulted in robust ubiquitylation of α2, the two proteins should be able to associate appropriately when derived from separate yeast strains. We made extracts from the same total number of cells as was used for the extracts from single subunit-expressing cells—one-half derived from cells expressing Slx5-V5, and the other derived from Slx8-V5. Levels of coprecipitated α2 did not increase when Slx5 and Slx8 were combined, but rather were reduced proportionately to the amount of Slx5 pulled down (Fig. 5, lane 3 vs. lane 5). Overall, these data indicate that, in the Slx5–Slx8 complex, Slx5 provides the α2-binding activity, independent of Slx8.
Figure 5.
Slx5, but not Slx8, interacts physically with α2. Yeast ubc4Δ ubc6Δ matα2Δ cells (MHY3765) cells were cotransformed with pRS425-GAL1-α2 and a pYES2.1 (2 μm, GAL1) plasmid expressing V5-His6-tagged Slx5, Slx8, or Cdc48 (negative control). After induction with galactose, cells were lysed under native conditions, and V5-tagged proteins were immunoprecipitated with anti-V5-agarose. Slx5, Slx8, and Cdc48 proteins were detected by anti-V5 (Invitrogen), and coprecipitated α2 by anti-α2 immunoblotting. For lane 3, half of each of the number of cells used for lanes 4 and 5 were mixed and lysed together, so the amount of input Slx8 and Slx5 was half of that used for each of the proteins in lanes 4 and 5. (Lane 1) A control immunoprecipitation with cells that overexpressed α2 but lacked any V5-tagged protein.
The Slx5 SIMs are required for SUMO binding and α2 degradation
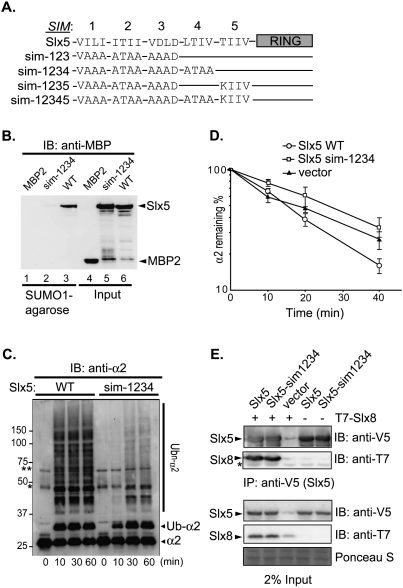
In its function as a STUbL, the Slx5–Slx8 E3 is believed to function primarily through its interaction with SUMO or poly-SUMO molecules attached to substrates. These interactions are mediated by the SIMs on the Slx5 subunit; several SIM-like elements have been identified in Slx5, each featuring a characteristic hydrophobic core usually flanked by acidic residues (Uzunova et al. 2007; Xie et al. 2007). In these earlier two studies, site-specific mutagenesis of SIM-1 and SIM-2 together or SIM-1 and SIM-3 together (Fig. 6A) impaired Slx5–SUMO interaction in vivo, based on yeast two-hybrid analysis. However, these SIM mutations did not cause major cellular defects, nor did they abolish Slx5–SUMO association in vitro (Xie et al. 2007; our unpublished results). These findings suggested that additional SIMs might exist in Slx5. Based on the SIM consensus sequence (Hecker et al. 2006; Kerscher 2007), we identified two additional potential SIMs in Slx5, here named SIM-4 and SIM-5 (Fig. 6A).
Figure 6.
The SIMs of Slx5 are required for SUMO binding and α2 degradation. (A) Schematic of Slx5 (wild type and mutants) with its SIMs and RING domain highlighted. Four residues that constitute the hydrophobic core of each SIM are indicated, with the altered sequences indicated for each mutant. The SIM core sequences that were tested were SIM-1, 24VILI27; SIM-2, 93ITII96; SIM-3, 116VDLD119; SIM-4, 155LTIV158; and SIM-5, 476TIIV479. (B) Protein binding to SUMO1-agarose. (Lanes 1–3) Purified recombinant MBP-Slx5 (wild type and sim-1234) or MBP2 proteins were incubated with SUMO1-agarose, and proteins that bound to the resin were eluted, resolved by SDS-PAGE, and detected by anti-MBP immunoblotting. Input proteins (20%) are shown in lanes 4–6. (C) Slx5–Slx8-mediated ubiquitylation of α2 in vitro (conditions as in Fig. 3). Reactions were stopped at the indicated times by addition of 2× SDS gel loading buffer. Proteins were resolved by SDS-PAGE, and were detected by anti-α2 immunoblotting. Asterisks denote the same two cross-reacting proteins noted in Figure 3. (D) Quantitation of α2 degradation in doa10Δ slx5Δ strains transformed with plasmid-borne SLX5 alleles measured by pulse-chase analysis at 30°C. (E) Coimmunoprecipitation of T7-tagged Slx8 with variants of Slx5. Analysis was done as in Figure 5. Slx5 variants were detected by anti-V5 immunoblotting, and coprecipitated T7-Slx8 was detected by anti-T7 immunoblotting. Protein staining of a portion of the membrane shows similar loading.
To test the contribution of these five potential SIMs to Slx5 function in vivo, residues within the hydrophobic cores of the respective SIMs were mutagenized. Cells with SIM-mutated Slx5 were tested for sensitivity to DNA-damaging concentrations of hydroxyurea (HU) as an indication of Slx5 function in the maintenance of genome stability (Zhang et al. 2006). Simultaneous mutation of SIM-1, SIM-2, and SIM-3 or of SIM-1, SIM-2, SIM-3, and SIM-5 had little detectable effect on cell growth or HU sensitivity (Supplemental Fig. 5A,B). In contrast, mutating SIM-1, SIM-2, SIM-3, and SIM-4 (sim-1234 allele) impaired cellular growth on rich medium and rendered cells more sensitive to HU, although growth was still slightly better than with an slx5-null allele (Supplemental Fig. 5B; data not shown). Mutating SIM-4 alone did not impede cellular growth in either condition. Thus, Slx5 depends on the composite function of multiple SIMs.
We hypothesized that the HU sensitivity of the slx5-sim1234 mutant was due to weakened interaction between Slx5 and SUMO. To address this, wild-type and sim-1234 mutant Slx5 proteins were bacterially expressed, purified, and tested for direct SUMO binding. Whereas wild-type Slx5 was precipitated efficiently by SUMO-agarose, very little Slx5-sim1234 bound to the matrix (Fig. 6B), strongly suggesting that Slx5 uses a redundant set of SIMs (SIM-1, SIM-2, SIM-3, and SIM-4) for SUMO association.
Inasmuch as SUMO was dispensable for Slx5–Slx8-dependent α2 degradation in vivo (Fig. 4; Supplemental Fig. 3) and for Slx5–Slx8-mediated α2 ubiquitylation in vitro (Fig. 3B), we assumed that the Slx5-sim1234 mutant protein would still be active for α2 ubiquitylation and degradation. Surprisingly, Slx5-sim1234 supported only very weak α2 ubiquitylation in vitro (Fig. 6C). The solubility and expression of the recombinant mutant protein were similar to wild-type Slx5 (Fig. 6B), suggesting that the SIM mutations were not causing major misfolding.
Consistent with our in vitro observations, a mutant slx5-sim1234 yeast strain was defective for Slx5–Slx8-dependent α2 degradation (Fig. 6D). Expression of mutant Slx5-sim1234 protein in yeast was similar to that of wild-type Slx5 (Supplemental Fig. 5C). We note that the SIMs are embedded in regions predicted to be largely intrinsically disordered (http://dis.embl.de), so mutations in the SIMs were not likely to induce misfolding. Moreover, V5-tagged wild-type Slx5 or Slx5-sim1234 coprecipitated Slx8 with similar efficiencies in vivo (Fig. 6E). The SIMs are thus not required for E3 heterodimerization, and their mutation does not grossly perturb Slx5 protein structure in cells. These data indicate that, even though Slx5–Slx8-mediated α2 degradation is SUMO-independent, the four SIMs of Slx5, which together account for its SUMO-binding activity, nevertheless contribute to α2 ubiquitylation by the Slx5–Slx8 STUbL. Using the in vivo coimmunoprecipitation assay (Fig. 5), we tested whether α2 binding to Slx5 was affected by the sim1234 mutations in Slx5. The E3 subunit coprecipitated α2 similarly, regardless of SIM integrity (Supplemental Fig. 5D). Although these results indicate that, under conditions of strong protein overexpression, Slx5–α2 binding is not abolished, binding may be impaired at physiological concentrations. These results have interesting implications for substrate recognition by the STUbLs (see the Discussion).
Discussion
Our targeted genomic screen for a ubiquitin E3 ligase that functions in the Ubc4 pathway of α2 degradation identified the Slx5–Slx8 heterodimer. These two RING proteins were shown previously, by our laboratory and others, to form a STUbL. Unexpectedly, we found that SUMO conjugation plays little if any role in the degradation of α2. Instead, the Slx5 subunit of the STUbL associates with nonsumoylated α2, and the recombinant heterodimer functions with Ubc4 in vitro to direct efficient α2 ubiquitylation. In parallel to this analysis, we defined, using point mutagenesis of Slx5, the SIMs that together account for noncovalent Slx5–SUMO binding. Surprisingly, yeast cells expressing a SUMO-binding-defective Slx5 protein are also defective for α2 degradation. Ubiquitylation of α2 in vitro was also strongly inhibited by inactivation of the Slx5 SIMs. These results lead to the proposal that some proteins, including α2, have structural features that allow their direct recognition and ubiquitylation by STUbLs without prior SUMO modification. This allows the selective uncoupling of STUbL-mediated ubiquitylation of such proteins from regulation by the SUMO system.
We were able to identify mutants defective for Ubc4-dependent α2 degradation through generation of a sensitive reporter construct that did not require knowledge of the cognate degron in α2. Because the Deg1 sequence was mutated, proteolysis of the α2*-Ura3-3HA reporter was relatively insensitive to loss of the Doa10-dependent α2 degradation pathway, a necessary condition for the screen to work. By collecting all known or suspected E3 and E2 gene knockout strains on a single 96-well microtiter plate (the TUS library), standard procedures for yeast transformation could be used to create the strains needed for the assay. Although this approach has the potential disadvantage, compared with screening the entire ∼5800 gene knockout collection, of missing additional novel factors that contribute to α2 degradation, it allowed us to detect subtle colony growth differences between strains in the initial replica plating; in all likelihood, these would have been missed in a whole-genome screen. The value of the TUS library has also been shown recently by our identification of an E3 involved in an E2 autoubiquitylation-mediated degradation pathway (Ravid and Hochstrasser 2007), and by the identification of two E3s that participate in cytosolic protein quality control (Heck et al. 2010).
The evidence that Slx5–Slx8 functions as the major E3 for Ubc4-dependent degradation of α2 is extensive. Epistasis analysis places Slx5 and Slx8 in the same pathway as Ubc4. We also see a loss of ubiquitylation of the α2*-Ura3-3HA test substrate in slx5Δ and slx8Δ mutant cells. Biochemical analysis with purified proteins shows a striking dependence of α2 ubiquitylation on both Slx5 and Slx8. Only very weak monoubiquitylation of α2 in the absence of Slx5 is observed (Fig. 3B), consistent with previous data suggesting that Slx8 is the catalytic subunit of the Slx5–Slx8 RING protein complex (Xie et al. 2007). Binding assays indicate that Slx5, but not Slx8, binds to α2 (Fig. 5), suggesting that the major role of the Slx5 subunit is substrate recognition and presentation to the Slx8 RING protein (and/or Ubc4).
The degree to which α2 is stabilized by simultaneous loss of both the Doa10 and Slx5–Slx8 pathways is significantly different when measured by pulse-chase versus cycloheximide-chase/immunoblot analyses. Pulse-chase measurements yield substantially shorter half-lives (Fig. 2; Laney and Hochstrasser 2003; data not shown). Polypeptides radiolabeled during a brief pulse period represent the pool of nascent α2 proteins, of which a sizeable fraction may reside in the cytoplasm and be in the process of folding and assembly. Cycloheximide-chase/immunoblotting, on the other hand, measures loss of the steady-state pool of α2, which will be dominated by mature substrate concentrated in the nucleus and on chromatin. We suggest that the substantial residual degradation observed by pulse-chase analysis in doa10Δ slx8Δ (or ubc4Δ ubc6Δ) cells represents proteolysis of nonnative or unassembled nascent α2, and may occur primarily in the cytoplasm. At late times in the chase (e.g., Fig. 2), a plateau in α2 protein loss is seen, which is in line with this hypothesis. Conversely, we regard Doa10 and Slx5–Slx8 as the E3s responsible for the vast majority of ubiquitin-dependent degradation of mature α2 in the nucleus. Slx5–Slx8 is in the nucleus and is at least partly chromatin-associated (Yang et al. 2006). Doa10 is known to target α2 in the nucleus (Deng and Hochstrasser 2006).
In the studies identifying the yeast Slx5–Slx8 complex as a STUbL, all of the specific SUMO-dependent substrates tested were characterized only in vitro. More recently, the mutant Mot1-301 protein was reported to be degraded in an Slx5–Slx8-dependent manner in vivo (Wang and Prelich 2009). However, it remains to be determined whether Slx5–Slx8 specifically targets sumoylated Mot1-301. Intriguingly, α2 degradation by the Slx5–Slx8 E3 is affected minimally, if at all, by SUMO. Deficiencies in components of the SUMO pathway have little impact on α2 turnover (Fig. 4; Supplemental Fig. 3). Moreover, Slx5–Slx8 efficiently ubiquitylates α2 in vitro in the absence of SUMO (Fig. 3B). Therefore, our work has established that a STUbL can ubiquitylate a physiological substrate independent of substrate modification by SUMO.
The Slx5 subunit of the Slx5–Slx8 heterodimer associates physically with unmodified α2 substrate and strongly enhances Slx8-initiated α2 ubiquitylation in vitro. Previously, the SIMs in Slx5 had been implicated in SUMO–substrate interaction, although no Slx5 point mutant had been described that fully blocked SUMO binding (Uzunova et al. 2007; Xie et al. 2007). Here we explored the molecular basis for E3–α2 substrate recognition and its relationship to E3–SUMO interaction. We identified four SIMs in Slx5 (SIM-1, SIM-2, SIM-3, and SIM-4) that act redundantly in SUMO binding. Introducing point mutations into all four blocks Slx5–SUMO binding in vitro and causes substantial cellular defects. This is consistent with an analysis of the human RNF4 STUbL, in which point mutations were made in four SIMs; the results indicated these SIMs functioned redundantly (although not identically) in poly-SUMO binding (Tatham et al. 2008). Our assays for SUMO binding used a SUMO-conjugated agarose resin; the high concentration of SUMO on the beads might mimic effects of SUMO polymerization into chains. Multiple SIMs might increase the avidity of Slx5 binding to the locally concentrated SUMO moieties. Interestingly, in two-hybrid assays, Slx5 interacts much more strongly with wild-type Smt3 (which can form poly-SUMO chains) than Smt3ΔGG (which cannot be conjugated to substrates or extant SUMO chains) (Hannich et al. 2005; data not shown). This probably reflects a preference of Slx5 for poly- or multisumoylated proteins over monosumoylated proteins or free SUMO. On the other hand, we found previously that a Rad52–SUMO fusion with only a single SUMO element also bound to Slx5 in vitro (Xie et al. 2007). Therefore, yeast Slx5 may have a higher affinity for monomeric SUMO than does human RNF4.
Surprisingly, even though α2 degradation is SUMO-independent, the SIMs in Slx5 are nevertheless required for efficient α2 turnover (Fig. 6D; data not shown). The defect of the Slx5-sim1234 mutant is unlikely to be due to gross protein misfolding induced by the point mutations in the SIMs, since mutant and wild-type Slx5 proteins are expressed at similar levels and can bind Slx8 similarly. The continued binding to Slx8 is consistent with data on the S. pombe orthologs of these proteins, which show that the RING domains are both necessary and sufficient for heterodimerization (Prudden et al. 2007; Sun et al. 2007). Notably, the four SIMs are all within the first 200-residue stretch of Slx5, of which >80% is predicted to be intrinsically disordered.
What, then, is the function of the SIMs in α2 ubiquitylation? Coimmunoprecipitation experiments suggest that the SIMs of Slx5 are not absolutely required for interaction with α2. A limitation of this assay is that these proteins are expressed at very high (nonphysiological) levels; this technique may not be sufficiently sensitive to allow detection of more subtly reduced binding. Alternatively, interactions of the SIM elements of Slx5 with α2, while not essential for binding, may be required for proper substrate orientation with respect to the Slx8 catalytic subunit or E2. In SUMO–SIM interactions, the hydrophobic SIM core adopts an extended conformation that fits into a hydrophobic groove between the α helix and β2 strand of the SUMO molecule, extending the SUMO β sheet (Song et al. 2004, 2005; Hecker et al. 2006). Available sequence and structural information do not reveal any obvious similarity to SUMO in the α2 protein. Thus, α2 is not directly analogous to Rad60 (S. pombe) and Esc2 (S. cerevisiae), related proteins that both contain easily recognizable SUMO-like domains (SLDs) and can also bind the Slx5–Slx8 E3, presumably through SLD–SIM interactions (Prudden et al. 2009; our unpublished data). Preliminary data suggest that SIM1 and SIM2 of Slx5 may have a dominant role in the in vivo ubiquitylation of α2 (Y Xie and M Hochstrasser, unpubl.), consistent with overlapping but not identical SUMO and α2 binding to Slx5. It is unlikely that Slx5–Slx8 requires SIM-mediated interaction with other proteins (such as those with SLDs) for targeting α2, since a ubiquitylation defect associated with the SIM mutations is apparent in a fully purified in vitro system.
A crystal structure of the α2 homeodomain and its upstream linker sequence in complex with the Mcm1 corepressor and DNA operator site revealed an interesting structural dimorphism in the α2 linker region (Tan and Richmond 1998), which might provide a clue to α2–Slx5 SIM binding. The linker adopts a β hairpin in one α2 monomer, but in the other monomer, the downstream sequence that had formed the second strand of the hairpin adopts an α-helical conformation instead. When this “chameleon sequence” forms an α helix, it may allow the largely hydrophobic upstream β strand to bind to the hydrophobic Slx5 SIM core. Preliminary pulse-chase data suggest that this hydrophobic β strand (residues 113–120) is indeed important for α2 degradation in vivo (our unpublished data). The downstream chameleon sequence may allow regulation of α2 ubiquitylation by factors that bias the sequence toward a particular conformation.
As noted, Slx5–Slx8 plays a critical role in the maintenance of genomic stability (Mullen et al. 2001; Zhang et al. 2006), which is unlikely to be linked to α2 function. To understand the role of Slx5–Slx8 in this capacity, more STUbL substrates must be characterized. Our data on α2 argue that Slx5–Slx8 and other STUbLs do not act only on sumoylated proteins. It will be interesting to determine whether the DNA damage control exerted by Slx5–Slx8 is strictly SUMO-dependent, or is regulated through targeting of both SUMO-dependent and SUMO-independent protein substrates.
Materials and methods
Yeast and bacterial methods
Yeast rich (yeast extract/peptone/dextrose, YPD) and minimal (SD) media were prepared as described previously, and standard methods were used for genetic manipulation of yeast (Guthrie and Fink 1991). Standard techniques were used for recombinant DNA work in Escherichia coli (Ausubel et al. 1989).
Yeast strain constructions
S. cerevisiae strains used in this study are listed in Supplemental Table 1. To create null alleles of SLX5 and SLX8 in the MHY501 background, deletion cassettes were PCR-amplified from slx5Δ∷kanMX4 and slx8Δ∷kanMX4 strains (Open Biosystems) with primer pairs corresponding to regions 200 base pairs (bp) upstream of and downstream from each ORF. The resulting fragments were then transformed into wild-type (MHY501), doa10Δ (MHY1631), and ubc4Δ (MHY498) cells to generate slx5Δ (MHY3712), slx8Δ (MHY3716), doa10Δ slx8Δ (MHY3718), and ubc4Δ slx8Δ (MHY3747) deletion strains. The slx5Δ slx8Δ double-deletion strain (MHY3861) was generated from a cross between the corresponding single mutants. The ubc9-1 doa10Δ (MHY3986) strain was generated from a cross between ubc9-1 (MHY1620) and doa10Δ (MHY1631). Congenic uba2ts (MHY4664) and uba2ts doa10Δ (MHY4659) strains were created by sporulation of a doa10Δ∷HIS3/+ uba2Δ∷kanMX6/+ doubly heterozygous diploid (MHY1684) containing the pIS-uba2ts plasmid (Schwienhorst et al. 2000). To make the doa10Δ smt3-F37A strain, smt3Δ cells (MHY1283) carrying plasmid-borne SMT3 (URA3) first had the DOA10 gene replaced by natMX4 (Goldstein and McCusker 1999). The resulting strain was transformed with pRS315-SMT3-F37A (LEU2) and cured of wild-type SMT3 plasmid by FOA selection.
Mutant and reporter gene generation
To create the α2-URA3-3HA fusion, the URA3-3HA sequence (along with a His6 tag, which for brevity is not noted explicitly in subsequent constructs) was PCR-amplified from a Deg1-URA3-3HA-His6-bearing plasmid (Ravid et al. 2006) with primers containing 45-nucleotide (nt) sequences that correspond to the sequences immediately upstream of and downstream from, respectively, the α2 stop codon (TGA). The URA3-3HA DNA fragment was then inserted in frame downstream from α2 by cotransforming the URA3-3HA DNA with an α2-containing plasmid (pJM130; CEN/LEU2) (Mead et al. 1996) digested with XhoI restriction enzyme. After gap repair in yeast, the resulting pJM130-α2-URA3-3HA plasmid was recovered in E. coli, and the α2-URA3-3HA fusion was verified by DNA sequencing. The Ile4 → Thr and Leu10 → Ser mutations within the Deg1 region of α2 were introduced by QuikChange mutagenesis (Stratagene). Unless indicated otherwise, these two mutations were included in the α2-Ura3-3HA protein for all experiments reported in this study; the altered protein is denoted as α2*-Ura3-3HA.
The pRS316-α2-3HA plasmid was made using homologous recombination in yeast between a PCR-amplified α2-3HA-containing sequence (from pRS306-pGAL-α2x3HA, a gift from J.D. Laney, Brown University) cotransformed with AgeI-digested pRS316-α2 plasmid, which carries a 4.3-kb HindIII MATα fragment (subcloned from pAV115-α2) (Johnson et al. 1998). The plasmid was recovered in E. coli, and the α2-3HA segment was sequenced. The resulting construct fuses the 3HA tag after Glu103 and deletes Leu104 and Thr105 from the α2 ORF. Site-directed mutagenesis of SMT3 and SLX5 (SIM mutations) was performed using QuikChange.
For protein overproduction in yeast, the corresponding genes were placed under the control of the galactose-inducible GAL1 promoter. To overproduce Slx5, Slx8, and Cdc48, the corresponding ORFs (without stop codons) were amplified by Taq polymerase from genomic DNA, and were inserted into the pYES2.1/V5-His-TOPO vector upstream of the V5 tag using the TOPO TA cloning protocol (Invitrogen). pRS425-GAL1-α2 was constructed by insertion of a PCR-amplified α2 ORF from pRS316-α2 into a pRS425-GAL1 plasmid after BamHI/SalI restriction digestion of both DNAs.
Genetic screen for genes required for α2*-Ura3-3HA degradation
A TUS yeast gene deletion library (Supplemental Table 2) was assembled in a single 96-well microtiter plate to screen for functional E3(s) in the Ubc4 pathway of α2 degradation. For this, 81 yeast strains were selected from the full deletion library containing ∼5800 strains (BY4741 background; Open Biosystems). Each strain carried a deletion of a nonessential gene implicated in the ubiquitin pathway. Proteins encoded by the indicated genes include 40 RING domain proteins, four HECT ubiquitin ligases, 20 proteins with other domains characteristic of ubiquitin ligases or domains thought to characterize such enzymes (e.g., U-box, PHD, and RWD proteins), seven Ubx proteins, and 10 ubiquitin-conjugating enzymes (E2s). The pJM130-α2*-URA3-3HA plasmid was transformed into the TUS library in a 96-well plate, and the transformants were spotted onto selective SD-leucine medium. Transformants were replica-plated from the SD-leucine plate onto SD-uracil medium. After 3 d of incubation at 30°C, the size and density of colonies on SD-uracil were determined. The fastest growing transformants were retested by serial dilution onto fresh SD-uracil plates; degradation rates of α2*-Ura3-3HA in the remaining candidates were estimated by cycloheximide-chase/anti-HA immunoblotting.
Recombinant protein expression and purification
Slx5, Slx8, and their derivatives were expressed and purified from E. coli according to Xie et al. (2007). Recombinant Ubc4 (human) was purified from bacteria as in Bays et al. (2001). Recombinant α2 was expressed with a His6 tag from the pJM163 plasmid in BL21 (DE3)/pLysS E. coli cells (Promega) according to Mead et al. (1996), except that EDTA was omitted from the lysis buffer and TALON Superflow metal affinity resin (Clontech) was used for protein binding.
Coimmunoprecipitation and pull-down assays
To study the interaction between the Slx5–Slx8 E3 and α2 in yeast, a pYES2.1-GAL1-V5 vector containing the SLX5, SLX8, or CDC48 ORF (the last as a negative control) was cotransformed into ubc4Δ ubc6Δ matαΔ cells (MHY3765) together with pRS425-GAL1-α2. Transformants were grown overnight in SD-uracil-leucine medium with 2% raffinose, diluted to OD600 0.2 in 30 mL of SD-uracil-leucine medium containing 2% raffinose and 2% galactose the next morning, and allowed to grow for another 12 h before harvesting (OD600 = 1.2). The cell pellet was washed in 5 mL of ice-cold PBS and was lysed in 400 μL of TBT buffer (100 mM potassium acetate, 50 mM HEPES at pH 7.5, 2 mM MgCl2, 1% Triton X-100, 20 μg/mL pepstatin A, 1 mM PMSF, 1× protease inhibitor cocktail [Complete Mini; Roche Diagnostics]) with 400 μL of acid-washed glass beads (435–600 μm; Sigma-Aldrich) in a FastPrep-24 bead beater (two 20-sec pulses) (MP Biomedicals). After the lysate was collected from the top of the beads, another 400 μL of TBT buffer was added to the beads. The buffer–bead mixture was vortexed again for 20 sec as above. Lysate was eluted through a hole poked into the tube bottom using a brief centrifugation, and the eluate was combined with the lysate collected earlier (total ∼800 μL). After a 10-min, 15,000-rpm microcentrifuge spin, the supernatant of the lysate was harvested and precleared with 20 μL of a 50% Protein A-agarose slurry (Repligen) for 1 h at 4°C before it was incubated with 15 μL of a 50% anti-V5-agarose suspension (Sigma-Aldrich) for 1.5 h at 4°C. Antigen-bound beads were washed in TBT buffer + 250 mM NaCl and resuspended in 50 μL of 2× SDS gel loading buffer before heating for 10 min at 100°C. V5-tagged proteins were detected with an anti-V5 antibody (1:5000 dilution; Invitrogen), and α2 was detected with an anti-α2 antibody (1:1000) (Chen et al. 1993). Interaction of V5-tagged Slx5 with T7-tagged Slx8 was performed in the same manner except that Slx8 was encoded by a 2-μm (high-copy) plasmid (pRS424-T7-Slx8) and was not under the control of the GAL1 promoter. T7-Slx8 was detected with an anti-T7 antibody (1:2500; Novagen). SUMO pull-down assays were performed as described (Xie et al. 2007). MBP-fusion proteins were detected with an anti-MBP antibody.
In vitro α2 ubiquitylation assays
Unless otherwise noted, α2 ubiquitylation assays contained 0.5 μg of yeast Uba1 (Boston Biochem), 0.2 μg of recombinant human Ubc4, 2.4 μg of recombinant human ubiquitin, and 2 mM ATP. Wild-type Slx5 and/or Slx8 (1 μM) or their respective mutants were incubated with 1 μM recombinant α2 (purified from bacteria) in a 30-μL reaction. Reaction buffer was the same as in Xie et al. (2007), except that it contained 100 mM NaCl and 5 μM ZnSO4. Ubiquitylation of α2 was detected by immunoblotting with anti-α2 antibodies (Chen et al. 1993) at 1:1000 dilution. For ubiquitylation of yeast-expressed α2, α2 was purified as a fusion protein containing His6, HA, and ZZ (from Protein A) domains immediately downstream from the α2 ORF (Gelperin et al. 2005). Expression of the α2 fusion construct was induced with galactose in W303 yeast cells; following cell lysis, the fusion protein was bound to IgG-Sepharose (GE Healthcare). By incubating with 3C protease overnight, the α2-His6-HA moiety was cleaved and eluted from the IgG resin-bound ZZ domain, as described (Gelperin et al. 2005). Ubiquitylation reactions were performed as detailed above except that 6 μL of the eluate purified from yeast was used as substrate. The eluate contained α2-His6-HA (29 kDa) (cut by 3C protease) and a small amount of full-length α2-His6-HA-ZZ (43 kDa) that had dissociated from the IgG-Sepharose. Samples were loaded on a gradient (6.5%-15%) SDS-PAGE gel and were analyzed by immunoblotting with the anti-HA 16B12 antibody (Covance).
Yeast protein degradation assays
Pulse-chase experiments were performed and radioactive α2 signal was quantified according to Chen et al. (1993). Cycloheximide-chase/immunoblot assays were based on Ravid et al. (2006). Protein degradation after immunoblotting was quantified using a G:Box system (Syngene).
Acknowledgments
We thank Jeff Laney, Oliver Kerscher, and Drew Vershon for plasmids, and Rachael Felberbaum and Dan Su for comments on the manuscript. This work was supported by grants from the NIH (GM46904 and GM53756) to M.H. E.M.R. was supported in part by an NIH Ruth Kirschstein post-doctoral fellowship.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1906510.
Supplemental material is available at http://www.genesdev.org.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K 1989. Current protocols in molecular biology Wiley, New York [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol 3: 24–29 [DOI] [PubMed] [Google Scholar]
- Chen P, Hochstrasser M 1995. Biogenesis, structure, and function of the yeast 20S proteasome. EMBO J 14: 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74: 357–369 [DOI] [PubMed] [Google Scholar]
- Deng M, Hochstrasser M 2006. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature 443: 827–831 [DOI] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes & Dev 19: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC, Hay RT 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 10: 564–568 [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR 1991. Guide to yeast genetics and molecular biology Academic Press, San Diego [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280: 4102–4110 [DOI] [PubMed] [Google Scholar]
- Heck JW, Cheung SK, Hampton RY 2010. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci 107: 1106–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I 2006. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281: 16117–16127 [DOI] [PubMed] [Google Scholar]
- Heideker J, Perry JJ, Boddy MN 2009. Genome stability roles of SUMO-targeted ubiquitin ligases. DNA Repair (Amst) 8: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I 1985. Master regulatory loci in yeast and λ. Cold Spring Harb Symp Quant Biol 50: 565–574 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Varshavsky A 1990. In vivo degradation of a transcriptional regulator: The yeast α2 repressor. Cell 61: 697–708 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Ellison MJ, Chau V, Varshavsky A 1991. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci 88: 4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM 2000. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Johnson ES 2004. Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Johnson PR, Swanson R, Rakhilina L, Hochstrasser M 1998. Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin–proteasome pathway. Cell 94: 217–227 [DOI] [PubMed] [Google Scholar]
- Kerscher O 2007. SUMO junction—What's your function? New insights through SUMO-interacting motifs. EMBO Rep 8: 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H 2008. Arsenic degrades PML or PML–RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10: 547–555 [DOI] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M 2003. Ubiquitin-dependent degradation of the yeast Matα2 repressor enables a switch in developmental state. Genes & Dev 17: 2259–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney JD, Mobley EF, Hochstrasser M 2006. The short-lived Matα2 transcriptional repressor is protected from degradation in vivo by interactions with its corepressors Tup1 and Ssn6. Mol Cell Biol 26: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead J, Zhong H, Acton TB, Vershon AK 1996. The yeast α2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins of higher eukaryotes. Mol Cell Biol 16: 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Brill SJ 2008. Activation of the Slx5–Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J Biol Chem 283: 19912–19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Tansey WP 2003. How the ubiquitin–proteasome system controls transcription. Nat Rev Mol Cell Biol 4: 192–201 [DOI] [PubMed] [Google Scholar]
- Perry JJ, Tainer JA, Boddy MN 2008. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33: 201–208 [DOI] [PubMed] [Google Scholar]
- Pickart CM 2001. Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN 2009. Molecular mimicry of SUMO promotes DNA repair. Nat Struct Mol Biol 16: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M 2007. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol 9: 422–427 [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M 2008. Degradation signal diversity in the ubiquitin–proteasome system. Nat Rev Mol Cell Biol 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M 2006. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J 25: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienhorst I, Johnson ES, Dohmen RJ 2000. SUMO conjugation and deconjugation. Mol Gen Genet 263: 771–786 [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci 101: 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang Z, Hu W, Chen Y 2005. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: A reversal of the bound orientation. J Biol Chem 280: 40122–40129 [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T 2007. Conserved function of RNF4 family proteins in eukaryotes: Targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26: 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes & Dev 15: 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Richmond TJ 1998. Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature 391: 660–666 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10: 538–546 [DOI] [PubMed] [Google Scholar]
- Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282: 34167–34175 [DOI] [PubMed] [Google Scholar]
- Wang Z, Prelich G 2009. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol 29: 1694–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AJ, Laney JD 2009. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodeling a repressor-promoter DNA complex. Nat Cell Biol 11: 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M 2007. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282: 34176–34184 [DOI] [PubMed] [Google Scholar]
- Yang L, Mullen JR, Brill SJ 2006. Purification of the yeast Slx5–Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res 34: 5541–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Roberts TM, Yang J, Desai R, Brown GW 2006. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 5: 336–346 [DOI] [PubMed] [Google Scholar]