Abstract
In plants and invertebrates, viral-derived siRNAs processed by the RNaseIII Dicer guide Argonaute (AGO) proteins as part of antiviral RNA-induced silencing complexes (RISC). As a counterdefense, viruses produce suppressor proteins (VSRs) that inhibit the host silencing machinery, but their mechanisms of action and cellular targets remain largely unknown. Here, we show that the Turnip crinckle virus (TCV) capsid, the P38 protein, acts as a homodimer, or multiples thereof, to mimic host-encoded glycine/tryptophane (GW)-containing proteins normally required for RISC assembly/function in diverse organisms. The P38 GW residues bind directly and specifically to Arabidopsis AGO1, which, in addition to its role in endogenous microRNA-mediated silencing, is identified as a major effector of TCV-derived siRNAs. Point mutations in the P38 GW residues are sufficient to abolish TCV virulence, which is restored in Arabidopsis ago1 hypomorphic mutants, uncovering both physical and genetic interactions between the two proteins. We further show how AGO1 quenching by P38 profoundly impacts the cellular availability of the four Arabidopsis Dicers, uncovering an AGO1-dependent, homeostatic network that functionally connects these factors together. The likely widespread occurrence and expected consequences of GW protein mimicry on host silencing pathways are discussed in the context of innate and adaptive immunity in plants and metazoans.
Keywords: Argonaute, GW motif, TCV, viral suppressor
In the model plant Arabidopsis, four paralogs of the RNaseIII Dicer are at the core of multiple RNA silencing pathways with specialized functions (Baulcombe 2004). Dicer-like 1(DCL1) mainly contributes to produce 19- to 24-nucleotide (nt)-long microRNAs (miRNAs) from noncoding, imperfect stem–loop precursor RNAs transcribed mostly from intergenic regions (Voinnet 2009). Upon unwinding, one miRNA strand incorporates into an RNA-induced silencing complex (RISC) that contains Argonaute 1 (AGO1), one of 10 AGO proteins that effect RNA silencing in Arabidopsis. The miRNA-loaded AGO1 then guides post-transcriptional gene silencing of complementary or partially complementary mRNA by inhibiting their stability and/or translation (Voinnet 2009). miRNA targets include transcription factor mRNAs required for plant development, as well as transcripts encoding proteins involved in metabolic and hormonal pathways. Unlike miRNAs, the 24-nt-long siRNA products of DCL3 act mostly in cis upon their incorporation into AGO4 or its surrogate, AGO6, to direct cytosine methylation and chromatin modifications at endogenous loci, including transposons, DNA repeats, and complexly rearranged gene arrays (Matzke et al. 2009). DCL4 and its surrogate, DCL2, guide AGO1-dependent post-transcriptional gene silencing of endogenous transcripts, including those involved in phase transitions, via the production of trans-acting RNAs (tasiRNAs) (Vaucheret 2005). Unlike DCL1, DCL2, DCL3, and DCL4 display higher affinity for perfect or near-perfect dsRNA molecules produced by sense/antisense transcription, converging transcription, folding of inverted repeat (IR) transcripts, or the action of endogenous RNA-dependent RNA polymerases.
The affinity of DCL2, DCL3, and DCL4 for endogenous, long dsRNA molecules is reflected in the action of these proteins in the context of viral infection. Based on several studies (for review, see Ruiz-Ferrer and Voinnet 2009), a consensus model of antiviral RNA silencing has emerged: Replication intermediates or intramolecular folds within the genomes of ssRNA viruses (the vast majority of phytoviruses) are accessed primarily by DCL4, the major antiviral DCL, which produces diagnostic, 21-nt-long siRNAs. Both DCL2 and DCL3 compete with DCL4 for processing viral dsRNA, such that 22-nt-long siRNAs (DCL2 products) and 24-nt-long siRNAs (DCL3 products) also accumulate coincidently in infected tissues, although these siRNA species are usually more easily detected if DCL4 function is genetically inactivated or suppressed (Ruiz-Ferrer and Voinnet 2009). The dsRNA processing step or “dicing” constitutes the first layer of virus restriction; the second layer entails the subsequent recruitment of one or several species of virus-derived siRNAs by endogenous AGO proteins to direct post-transcriptional silencing of viral RNA as part of antiviral RISCs. Although the exact function of several Arabidopsis AGOs remains to be established (for review, see Vaucheret 2008), AGO1 and AGO7 are good candidates as antiviral RISC effectors, because both direct post-transcriptional silencing of endogenous and exogenous RNA (Morel et al. 2002; Montgomery et al. 2008). Moreover, hypomorphic ago1 and null ago7 mutants are hypersusceptible to viruses (Morel et al. 2002; Qu et al. 2008). In contrast, AGO4 and AGO6 are guided by the DCL3-dependent 24-nt siRNAs to mediate DNA methylation and, therefore, are unlikely to silence viruses with RNA genomes, agreeing with the lack of effects of ago4 or ago6-null mutations against these pathogens (for review, see Vaucheret 2008).
In order to multiply and invade their host systemically, viruses have evolved a vast array of proteins called suppressors of RNA silencing (VSR), which target many stages of antiviral silencing, including the processing (i.e., DCL level) and effector (i.e., AGO level) steps (for review, see Ding and Voinnet 2007). Owing to the availability of silencing suppression assays based on delivery of individual viral proteins, the discovery of VSRs has thrived over the past 10 years (for review, see Li and Ding 2006). However, their molecular mechanisms of action and exact significance in the context of authentic infections have been comparatively poorly characterized, yet this aspect remains key to the understanding of the host/parasite molecular arms race. The main difficulty lies in the fact that VSRs are usually multifunctional proteins essential for primary virus replication, so that their alteration in viral contexts is often lethal to the pathogen. Moreover, VSR expression often masks the effects of mutations in DCLs or AGOs such that clear genetic interactions linking VSRs to specific host silencing pathway components are difficult to establish (Ding and Voinnet 2007).
Studies of wild-type and recombinant Turnip crinckle RNA virus (TCV) have been somewhat instrumental in uncovering several important interactions linking specific Arabidopsis DCL and AGO to RNA virus infection (Deleris et al. 2006; Qu et al. 2008). The TCV-encoded VSR, the P38 protein, also serves as a viral capsid required for genome packaging, but this second function can be uncoupled from silencing suppression by single missense mutations within the P38 ORF (Deleris et al. 2006). In wild-type Arabidopsis, TCV-derived siRNAs are mostly 22 nt in size, indicating that DCL2—as opposed to the primary antiviral enzyme DCL4—processes viral dsRNA. However, DCL4-dependent 21-nt siRNAs accumulate in wild-type Arabidopsis infected with P38-deficient TCV (TCVΔP38) (Deleris et al. 2006). Based on this P38-dependent shift in DCL usage, we proposed that P38 somehow interferes with DCL4 function in normal infections, such that DCL2, the DLC4 surrogate, takes over antiviral defense (Deleris et al. 2006; Qu et al. 2008). Nonetheless, dsRNA processing by DCL3 also competes with DCL2 action, as large amounts of 24-nt siRNAs accumulate in dcl2-dcl4 mutant leaves infected with TCVΔP38. Only in dcl2–dcl4 double or dcl2–dcl3–dcl4 triple mutants is the lack of systemic infection by TCVΔP38 in wild-type plants rescued (Deleris et al. 2006; Qu et al. 2008). This observation indicates that the 22-nt siRNA that accumulate during normal infection have intrinsic antiviral function and, therefore, must engage into one or several AGO proteins. Therefore, to explain the genetic rescue data, we proposed previously that, in addition to its anti-DCL4 effects, P38 must also suppress the action of the 22-nt siRNA-loaded AGO protein (Deleris et al. 2006). The present study was aimed at uncovering the nature of the anti-TCV AGO and the mechanisms of its suppression by P38. The results from this study shed light on both of these issues, and uncover a novel VSR strategy that relies on viral mimicry of host-encoded glycine/tryptophane (GW)-containing proteins normally required for RISC assembly/function in diverse organisms. This finding may have relevance to silencing suppression by viruses and other pathogens, not only in plants, but also in metazoans. Our study simultaneously unravels how AGO suppression by P38 profoundly modifies a previously unsuspected homeostatic interaction network linking the four Arabidopsis DCLs. These changes incurred by P38 not only explain the shift in DCL usage observed during TCV infections, but also have broad implications for our understanding of host–parasite interactions, and of the interconnectivity of RNA silencing pathways in plants.
Results
TCV-derived 22-nt siRNAs are loaded preferentially into AGO1
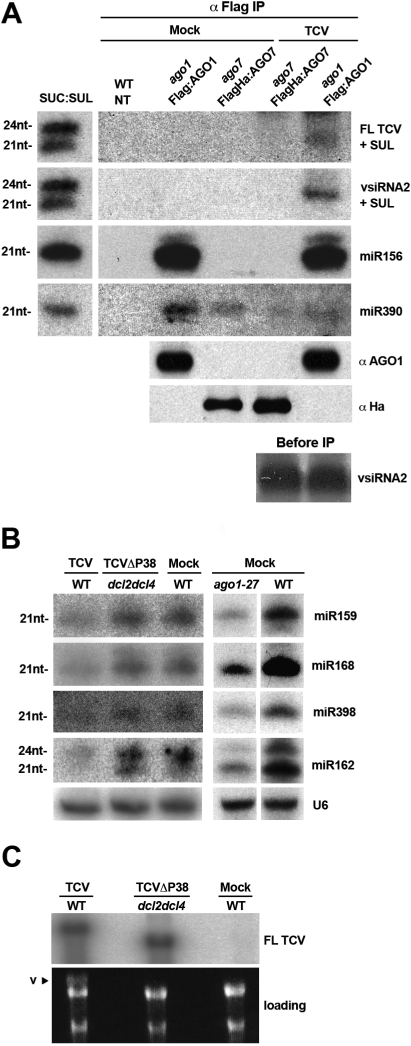
Arabidopsis ago1 and ago7 mutants have been found previously to be hypersusceptible to wild-type and recombinant TCV, respectively (Qu et al. 2008). Thus, both AGO1 and AGO7 are plausible effectors of the 22-nt siRNAs produced during TCV infections of wild-type plants. To address this issue, we used ago1 and ago7 mutant plants expressing Flag-tagged versions of AGO1 or AGO7, respectively, under their own promoter. These constructs rescue the molecular and developmental defects of the corresponding mutants (Supplemental Fig. S1A; Baumberger and Baulcombe 2005). As a likely negative control, we used ago4 mutant plants expressing a Myc-tagged AGO4 construct under its own promoter, which restores AGO4 functions in the DNA methylation pathway (Li et al. 2006). Wild-type, ago1/Flag:AGO1, ago7/Flag:AGO7, or ago4/Myc:AGO4 plants were infected with TCV. Twenty-one days post-infection (dpi), inflorescences were harvested and immunoprecipitations were carried out with anti-Flag (AGO1 or AGO7) or anti-Myc (AGO4) antibodies. Northern analyses of RNA extracted from immunoprecipitates revealed that, of AGO1 and AGO7, only the former associates with viral 22-nt-long siRNAs, as assessed with a full-length TCV probe (FL) (Fig. 1A), or with an oligonucleotide probe detecting a single, highly abundant, viral siRNA (vsRNA2) (Fig. 1A). As expected, there was no TCV siRNA signal in immunoprecipitates from Myc-tagged AGO4 (Supplemental Fig. S1B). Therefore, among the three AGOs tested, AGO1 is loaded preferentially with TCV-derived 22-nt siRNAs, consistent with AGO1 being a major effector of siRNAs that guide post-transcriptional silencing in Arabidopsis (Vaucheret 2008).
Figure 1.
Preferential loading of TCV 22-nt siRNAs into AGO1 and effects of P38 on accumulation of AGO1-dependent miRNAs in infected plants. (A) Small RNA fraction associated with Flag-AGO1 and Flag-HA-AGO7 immunoprecipitates (IP anti-Flag) in mock- or TCV-inoculated crude inflorescence extracts. RNAs and proteins were extracted directly from α-Flag immunoprecipitates (αFlag IP). sRNA blots were hybridized with either DNA oligonucleotide probes complementary to the indicated miRNAs or TCV-derived siRNA2 or a full-length virus probe (FL TCV). The panel on the left contains RNA from the SUC:SUL transgenic line that expresses an RNAi hairpin targeting the SUL endogenous mRNA (Dunoyer et al. 2007). The resulting dsRNA is processed into 21-nt and 24-nt siRNAs, used here as size references. The SUL RNA signals were detected on the same blots upon stripping. miR156 and miR390 are used as qualitative controls for their established loading preferences into AGO1 and AGO7, respectively. AGO1 was detected using an anti-AGO1 antibody, and AGO7 was detected using an anti-HA antibody. (B, left panel) miRNA accumulation in inflorescences of mock-, TCVΔP38-, or TCV-inoculated plants with a dcl2dcl4 or wild-type background. (Right panel) miRNA levels in noninfected ago1-27 versus wild-type inflorescences. The U6 RNA was used as a loading control. (C) Compared levels of TCV and TCVΔP38 in wild-type or dcl2dcl4 mutant plants. The virus genomic RNA (v) can be visualized by ethidium bromide staining; rRNA serves as a loading control.
The P38 effects on cellular miRNA are genetically equivalent to those of an hypomorphic ago1 mutation
Optimal accumulation of P38-deficient TCV (TCVΔP38) is observed only in dcl2–dcl4 double mutants, as opposed to dcl4 single mutants (Deleris et al. 2006). Thus, besides its primary DCL4-antagonizing function, P38 must also suppress the antiviral activity of the DCL2-dependent siRNAs that normally accumulate in TCV-infected, wild-type Arabidopsis. Having established that TCV-derived 22-nt siRNAs are loaded into AGO1, we thus investigated if P38 suppresses AGO1 function. We measured accumulation of several endogenous miRNAs whose activity and stability also require loading into AGO1. Northern analyses indeed revealed a significant reduction in miRNA levels in TCV-infected, but not healthy, Arabidopsis, an effect strikingly similar to that observed in noninfected ago1-27 hypomorphic mutants (Fig. 1B; Morel et al. 2002). Moreover, this effect was P38-dependent because it was not observed in dcl2–dcl4 double mutants infected by TCVΔP38 (Fig. 1B), which accumulated the TCV RNA to high levels similar to TCV-infected wild-type plants (Fig. 1C).
Evidence for a higher-order complex between P38 and AGO1
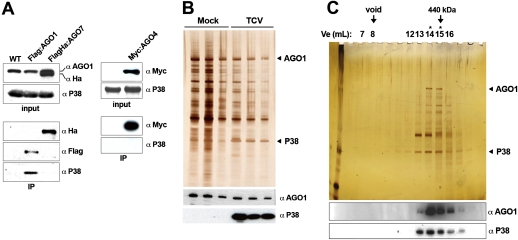
To test if these effects entail physical association of P38 with AGO1, we used the Flag and Myc immunoprecipitates employed in Figure 1A and Supplemental Figure S1B and assayed the presence of P38 by Western analysis. P38 was indeed detected in Flag:AGO1 immunoprecipitates; this association was specific because it was not found in either Flag:AGO7 or Myc:AGO4 immunoprecipitate fractions (Fig. 2A). Silver nitrate staining of Flag:AGO1 immunoprecipitates further confirmed these findings, and also showed that the P38–AGO1 interaction was unaltered by treatments of up to 800 mM KCl (Fig. 2B). In addition, gel filtration of Flag:AGO1 immunoprecipitates showed that the AGO1 and P38 elution peaks overlap perfectly (Fig. 2C). Therefore, P38 associates strongly and specifically with AGO1, the effector of the 22-nt-long siRNAs produced in TCV-infected tissues (Fig. 1A). An analysis of coeluted fractions revealed that the intensity of P38 silver staining was at least as strong as that of AGO1 (Fig. 2C), despite the highly dissimilar molecular mass of the two proteins (38 kDa and 120 kDa, respectively). Thus, AGO1 and P38 likely form a complex that contains substoichiometric amounts of AGO1. This observation was of particular interest in light of two P38 missense mutations previously isolated by us (Deleris et al. 2006). These lesions (m1: R74 → W; m2: E122 → K) individually abolish P38 VSR function without altering its stability (Fig. 3A; Deleris et al. 2006). Using X-ray crystallographic data (Qu and Morris 1997), we found that m1 and m2 are sterically juxtaposed at the interface of C:C P38 homodimers (Fig. 3B,C). TCV virions are indeed arranged in T = 3 icosahedral symmetry, and consist of 30 copies of hexameric capsomers assembled via A:B, A:C, B:C, and C:C interactions between P38 monomers (Fig. 3B). This structural information makes it likely, therefore, that P38–P38 interactions involving at least C:C dimers are required for its VSR activity, presumably by binding and further sequestering AGO1.
Figure 2.
AGO1 is a physical target of P38. (A) Coimmunoprecipitation of P38 with AGO proteins in flower extracts from the indicated transgenic lines. Anti-Flag or anti-Myc immunoprecipitates were analyzed by immunodetection using anti-P38 serum. AGO7 and AGO4 were detected using anti-HA- and anti-Myc antibodies, respectively, and AGO1 was detected with anti-AGO1 antibodies. All immunoprecipitates were washed with high salt concentrations (see the Materials and Methods). (B) Polypeptides elution profile of AGO1 immunoprecipitates from inflorescence extracts of mock- or TCV-inoculated Flag-AGO1 plants. Flag-AGO1 immunoprecipitate (IP anti-Flag) products were released through three stepwise elutions by competition (Flag peptide), and were analyzed by either silver nitrate staining or immunodetection for AGO1 and P38. AGO1 and P38 positions in the stained gel are indicated with arrows. (C) Separation, by gel filtration, of Flag-AGO1 immunoprecipitate products (IP anti-Flag) from TCV-infected inflorescences under high stringency conditions. Fractions eluted from Superose 6 column were analyzed by either silver nitrate staining or protein immunodetection for AGO1 and P38. Void volume and Ferritine (440 kDa) are marked with arrows. The elution peak of the AGO1 complex is centered on 14 mL (asterisks).
Figure 3.
P38 VSR function likely requires C:C homodimerization. (A) Schematic of P38 structural domains with the position of the amino acids mutated in the m1 and m2 alleles impaired for VSR function (in yellow and pink, respectively). The same colors are used throughout the figure. The m1 and m2 proteins are stable, as visualized by immunodetection using total extracts from transgenic lines expressing mutant and wild-type P38. (B) Representation of a P38 hexamer made of two A–B–C trimers that interact through C:C dimerization at the twofold axis, as viewed from the inside of the capsid. (C) A close-up view of B in which the C backbone and all side chains are represented with simple lines. This allows visualization of m2 (E122) embedded within the β barrels that form the S domain, and unravels the sterical proximity of m1 and m2. The C backbone is represented by a green line, and the two amino acids of interest are represented by space-filling.
P38 is a virus-encoded GW repeat, AGO anchor protein
Which P38 feature(s) could account for its association with AGO1? In plants and metazoans, assembly and activity of RNA silencing effector complexes require proteins with specialized AGO-binding platforms. These evolutionarily conserved platforms are composed of reiterated tryptophane/glycine (WG) or GW residues first revealed in the C terminus of the plant-specific RNA Polymerase V subunit NRPE1 (El-Shami et al. 2007), and in the TAS3 component of the RITS complex in Schizosaccharomyces pombe (Till et al. 2007). Both proteins bind to AGO4 and SpAGO1, respectively, in a tryptophan-dependent manner, and are key components of the RNA-induced heterochromatin formation pathway. WG/GW motifs are also implicated in the recruitment of AGO proteins to effect miRNA functions through binding to the human GW182 protein and its homologs in fly and worm (for review, see Eulalio et al. 2009). While the number of GW motifs may vary greatly among AGO-binding platforms (from two to up to 40), computer analyses have uncovered consensus features surrounding GW domains (El-Shami et al. 2007). These include a higher representation of G, W, S, and T residues, and an enrichment in charged amino acids (prominently E, D, and K). There is also a notable deficit in F, Y, C, H, and M residues.
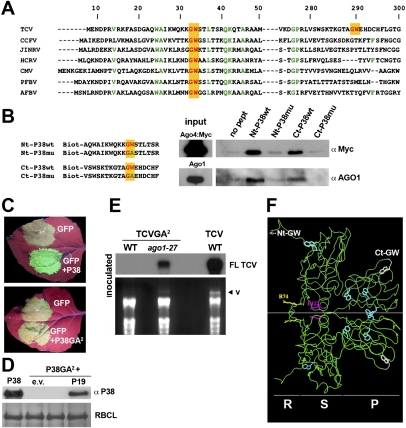
Two discrete GW motifs were detected in the entire P38 ORF, in the N and C termini, respectively (Fig. 4A); strikingly, both motifs have favorable AGO-binding amino acid contexts, according to the above criteria (Supplemental Fig. S2A). Moreover, the N-terminal GW motif is found within an otherwise poorly conserved region in the P38 ORFs of nearly all members of the Carmovirus genus, indicating strong positive selection (Fig. 4A). The C-terminal GW motif, in contrast, was found to be unique to the TCV P38, although the favorable amino acid environment was still detected in the P38 ORFs from other Carmoviridae (Fig. 4A). To test if P38 interacts directly with AGO via the GW motifs, we first attempted to express the protein in Escherichia coli. However, and as expected from its aggregative nature, P38 formed insoluble inclusion bodies that precluded further investigation. Previous work reported the use of short and sometimes concatemerized portions of GW repeat proteins for in vitro AGO-binding assays (El-Shami et al. 2007; Till et al. 2007). Accordingly, we used chemically synthesized, 18-amino-acid-long biotinylated peptides encompassing the N-terminal and C-terminal GW motifs, within their favorable AGO-binding context (Fig. 4B). As a control, we used GW → GA mutated forms of both peptides, predicted to lose any putative interaction with AGO (El-Shami et al. 2007). Wild-type and mutant peptides were then bound onto streptavidin beads for affinity chromatography using inflorescence extracts from Myc:AGO4 and Flag:AGO1 transgenic Arabidopsis. Upon washing, eluted fractions were subjected to Western analysis using anti-Myc or anti-AGO1 antibodies. Both N-terminal and C-terminal peptides, but not their corresponding GW → GA mutated forms, could indeed bind to AGO1 (Fig. 4B). Similar results were obtained with Myc:AGO4 (Fig. 3B), indicating that the AGO-binding specificity of P38 in planta (Fig. 2A) is not recapitulated in vitro, consistent with previously demonstrated in vitro interactions involving truncated AGO and GW proteins from unrelated organisms and functioning in distinct silencing pathways (El-Shami et al. 2007). The specificity of AGO binding might require full-length P38 and/or in vivo parameters such as subcellular localization or cell type-specific expression. Collectively, these and additional data pertaining to the AGO protein domain that is likely involved in the interaction (Supplemental Fig. S2B) strongly suggest that P38 is a bona fide GW protein that requires both N-terminal and C-terminal GW motifs to act as an AGO-binding platform.
Figure 4.
The physical interaction between P38 and AGO1 is mediated by two GW motifs that are essential for VSR activity. (A) Amino acid alignment of the P38 proteins from various Carmoviruses. The N-terminal GW residues (in red and yellow) are highly conserved in an otherwise poorly conserved domain (other conserved residues are shown in green). (B) Equimolar amounts of biotin GW peptides (P38wt) or biotin mutated peptides (P38mu) were coupled to streptavidin beads and subjected to inflorescence extracts from Myc-AGO4 or Flag-AGO1 transgenic plants. Bound proteins were detected by immunoblotting using anti-Myc or anti-AGO1 antibodies. (C) Effect of dual GW mutations on the VSR activity of P38, as assayed by cotransient expression of a reporter GFP transgene together with P38 or P38GA2 in N. benthamiana leaves. Photographs were taken 5 d post-infiltration (5 dpi) under ultraviolet light to show green fluorescence from GFP and red fluorescence from chlorophyll. (D) Accumulation of P38 and P38GA2 in the leaves depicted in C, as assayed by immunoblotting using the P38 serum. (E) RNA blot hybridizations showing accumulation levels of TCVGA2 and wild-type TCV in inoculated leaves of wild-type or ago1-27 plants. The virus genomic RNA (v) can be visualized by ethidium bromide staining; rRNA serves as a loading control. (F) GW motifs are exposed to the surface and are positioned away from the C:C dimerization interface. R, S, and P domains of P38 C/C homodimer viewed normal to the dyad. In capsid context, the center of the particle (S) would lie on the left of the model and the outside of the particle (P), at the right. The R domain and the fold-back region of the arm, where R74 lies, point to the interior of the capsid. The N-terminal and C-terminal GW motifs are indicated in white, although the former cannot be modeled in the structure presented. The other W residues that normally facilitate AGO binding are shown in light blue, alongside the m1 and m2 mutations, as depicted in Figure 3.
AGO1-binding GW residues are essential to the VSR function of P38 and for systemic TCV infection
To address if AGO binding by P38 is required for its VSR function in vivo, we engineered point mutation alleles of P38 carrying GW → GA lesions in each or both of the N-terminal and C-terminal motifs. Wild-type P38 and its modified versions were then used in a classical Agrobacterium-mediated transient gene silencing assay in Nicotiana benthamiana leaves (Voinnet et al. 1998). In this assay, expression of a GFP reporter transgene is turned off at 5 dpi owing to intrinsic T-DNA-triggered, AGO1-dependent post-transcriptional gene silencing (Dunoyer et al. 2006). In contrast, transient coexpression of GFP with a second, P38-producing T-DNA maintains high levels of green fluorescence at 5 dpi and beyond, due to P38-mediated silencing suppression (Fig. 4C, top panel; Voinnet et al. 1998). The P38 GW → GA double mutant (referred to here as P38GA2) was unable to sustain GFP expression in the silencing assay (Fig. 4C, bottom panel) and accumulated below the detection limit of Western analysis (Fig. 4D). This was not due to protein instability incurred by the mutations, as P38GA2 accumulation was restored if it was transiently codelivered with a T-DNA expressing the Tomato bushy stunt virus P19 protein (Fig. 4D), a distinct VSR that sequesters siRNA duplexes (Vargason et al. 2003). Therefore, the failure of P38GA2 to sustain GFP expression in the transient assay results from an inability to suppress intrinsic silencing triggered by T-DNAs, including by the P38GA2-encoding T-DNA. Similar results were obtained with single GW → GA point mutations in either the N-terminal or C-terminal GW motif of P38 (data not shown), consistent with the loss-of-function effects of each lesion in the in vitro AGO-binding assays (Fig. 4B). Thus, mutations in either of the two P38 GW motifs that prevent AGO binding in vitro also compromise the P38 VSR function in vivo. Further proof that AGO1 binding contributes significantly to the VSR function of P38 came from studies of TCVGA2, in which the GA2 allele replaces the wild-type allele of P38 in recombinant TCV. Hence, TCVGA2 accumulation was below the detection levels of Northern analysis in inoculated leaves of wild-type Arabidopsis, but it was restored to high levels in ago1-27 mutant plants (Fig. 4E). Therefore, the loss of the AGO1-binding GW residues in P38 is sufficient to abolish TCV infection in an AGO1-dependent manner.
Possible structural and molecular bases of P38-mediated inhibition of AGO1 function
Using partial structural data, the C-terminal GW motif was mapped onto the surface of the P domain of the C:C P38 dimer (Figs. 3A, 4F), away from the internal homodimerization domain that is likely disrupted by the m1 and m2 mutations (Fig. 3C). Thus, while it compromises P38 VSR functions, the C-terminal GW → GA mutation is not predicted to affect P38 dimerization, at least under C:C forms. We also noticed that the additional tryptophane residues in the vicinity of the C-terminal GW motif that are known to provide a favorable AGO-binding environment (Fig. 4F; Supplemental Fig. S2A; El-Shami et al. 2007) are also exposed to the surface, consistent with an interaction of P38 with a ligand. Although similar information could not be obtained for the N-terminal GW motif, these structural data are consistent with the idea that C-terminal GW motifs of C:C P38 homodimers interact with AGO at the surface of P38 P domains. What are the molecular consequences of the binding of AGO1 by P38? We rule out an effect on AGO1 stability, which is unchanged in TCV-infected, as compared with noninfected, tissues (Fig. 2A; Supplemental Fig. S3D,E). Comparing the vsRNA2 input signal to that found in AGO1 immunoprecipitates , however, showed that only a modest fraction of vsRNA2 is associated with AGO1 in TCV-infected Arabidopsis (Fig. 1A). In addition, the levels of cellular miRNAs are reduced in infected plants (Fig. 1B), possibly due to their enhanced turnover as a result of defective incorporation into AGO1, as has been proposed in previous analyses of the ago1-27 mutant protein (Vaucheret 2008). P38 might thus compromise small RNA loading into AGO1, either by preventing their incorporation into nonloaded RISC, or by disrupting the integrity of preloaded RISC. Of the two possibilities, we favor the former, because transgenic coexpression of P38 and an IR construct—used as a surrogate to viral infection—caused a dramatic reduction in the loading of IR-derived siRNAs into AGO1 (Supplemental Fig. S3C,D). In contrast, there was little or no effect if P38 was delivered virally onto the IR transgenic plants in which AGO1 had been preloaded with IR-derived siRNAs (Supplemental Fig. S3B,E).
Rescue of systemic TCVGA2 infection in Arabidopsis dcl2–dcl4 double mutants
While this study establishes both genetic and physical interactions between viral P38 and host AGO1, a genetic link also exists between P38, the primary antiviral enzyme DCL4, and its functional surrogate, DCL2 (Fig. 1C; Deleris et al. 2006). Therefore, we tested if loss of the DCL4 and DCL2 antiviral pathways could rescue the inability of TCVP38GA2 to accumulate in wild-type Arabidopsis. As shown in Figure 5A (top panel), TCVP38GA2 accumulation was restored in inoculated leaves of dcl2–dcl4 double mutants. Detection of P38GA2 by Western analyses in dcl2–dcl4 inoculated leaves confirmed that the mutant protein is not intrinsically unstable (Fig. 4A, top panel), in agreement with the P19 coinfiltration results in N. benthamiana leaves (Fig. 4C,D). Analyses of systemically infected inflorescences at 21 dpi further indicated that TCVP38GA2 and P38GA2 accumulation was as high in dcl2–dcl4 double-mutant plants as was accumulation of wild-type TCV and P38 in wild-type inflorescences (Fig. 5A, bottom panel; Supplemental Fig. S4A). Therefore, the combined loss of DCL4 and DCL2 functions fully rescues the systemic infection of TCVP38GA2, which differs from wild-type TCV in its ability to bind to AGO1 via the P38 GW residues.
Figure 5.
Restoration of TCVGA2 virus level in dcl2dcl4 mutants and global changes in DCL levels caused by P38 interference with AGO1. (A) RNA blot hybridizations showing accumulation of TCVGA2 and wild-type TCV in inoculated leaves (top panel) and systemic inflorescences (bottom panel) of wild-type or dcl2–dcl4 plants. The virus genomic RNA (v) can be visualized by ethidium bromide staining of rRNA. Coomassie staining was used as a protein loading control. (B) DCL1 levels were evaluated by immunodetection using an anti-DCL1 antibody (see the Materials and Methods) in total inflorescence extracts. Samples from mock-inoculated plants (wild type [WT] or dcl2–dcl4), TCVΔP38-infected dcl2–dcl4; TCVGA2-infected plants (wild type [WT] or dcl2–dcl4), and TCV-infected wild-type plants were analyzed. (C). Expression of GFP under the control of the DCL1 promoter (see the Materials and Methods) was monitored by quantitative RT–PCR analysis of three biological replicates involving three TCV infections each. Systemic leaf tissue was harvested at 21 dpi, and cDNA inputs were normalized using Actin2 abundances; expression ratios are given relative to levels in mock-inoculated plants (set to 1). Data are displayed as averages ± standard deviation (three PCR replicates). (D, left and middle panels) The tissues from wild-type Arabidopsis inflorescences used in B were employed for immunodetection of DCL4 and DCL3, respectively (see the Materials and Methods). Coomassie staining was used as a protein loading control. (Right panel) DCL2 accumulation was analyzed by semiquantitative RT–PCR on total RNA from the inflorescence tissues used in B. Ubiquitin (Ub) was used as a loading control.
P38-mediated inhibition of AGO1 modifies the cellular availability of the four Arabidopsis DCLs
The above findings are consistent with AGO1 being the main effector of TCV-derived DCL4-dependent 21-nt siRNAs and DCL2-dependent 22-nt siRNAs. They are also consistent with P38 suppressing the function of both viral siRNA species through its interaction with AGO1. However, the results fail to explain why DCL2—as opposed to the main antiviral enzyme, DCL4—contributes prominently to anti-TCV silencing in wild-type Arabidopsis (Fig. 1A; Deleris et al. 2006). In a previous analysis, Qu et al. (2008) showed that Arabidopsis dcl1 hypomorphic mutants accumulate higher levels of the DCL4 and DCL3 transcripts, suggesting that DCL1, the main miRNA-processing enzyme, normally regulates negatively DCL4 and DCL3; accordingly, TCVΔP38 levels are lower in hypomorphic dcl1 Arabidopsis mutants than in wild-type plants (Qu et al. 2008). Both DCL4 and DCL3 compete with DCL2 for access and processing of TCV dsRNA molecules; in particular, we showed that high levels of viral-derived DCL3-dependent, 24-nt-long siRNAs accumulate in TCVΔP38-infected plants with a dcl2–dcl4 background (Deleris et al. 2006). Moreover, TCVΔP38 accumulates higher in Arabidopsis with dcl2–dcl3 and dcl3–dcl4 backgrounds than in plants with single dcl2 or dcl4 backgrounds (Qu et al. 2008), consistent with the immunoprecipitation of TCV-derived 24-nt-long siRNAs in infected ago4/Myc:AGO4 transgenic plants (Supplemental Fig. S1B). Strikingly, the DCL1 levels are themselves negatively controlled by the AGO1-loaded miR162 (Xie et al. 2003), and miR162 accumulation is strongly reduced in a P38-dependent manner in TCV-infected plants (Fig. 1B). Using an antibody raised against the endogenous protein (Supplemental Fig. S4B), we found that DCL1 levels were dramatically enhanced in TCV-infected inflorescences of wild-type plants (Fig. 5B, left panel). This effect was clearly P38-dependent, because it was not observed in inflorescences of dcl2–dcl4 double mutants infected with TCVΔP38, which accumulated the TCV RNA to similar levels as TCV-infected wild-type tissues (Figs. 1C, 5B [left panel]). Furthermore, the effect was also lost in dcl2–dcl4 mutant inflorescences infected with TCVP38GA2 (Fig. 5A,B, right panel), strongly suggesting that DCL1 up-regulation depends on the AGO1-binding property of P38. TCV infection had only a modest effect on DCL1 transcriptional activity, as assessed in three independent experiments involving a transgenic line in which a 2.5-kb fragment that likely encompasses the DCL1 promoter is fused to GFP (Fig. 5C). Therefore, the observed post-transcriptional enhancement of DCL1 levels is most likely caused by a P38-dependent reduction of the activity of AGO1-dependent miR162. As expected from the proposed DCL1-dependent suppression of DCL4 and DCL3 (Qu et al. 2008), and from the dramatically enhanced DCL1 levels uncovered in this study (Fig. 4B), both DCL4 and DCL3 accumulated at lower levels in TCV-infected inflorescences, an effect that was particularly pronounced for DCL3 (Fig. 4D, left and middle panels). Due to a lack of a reliable antibody, the DCL2 protein levels could not be determined, but semiquantitative RT–PCR revealed only modest changes in DCL2 transcript accumulation in TCV-infected inflorescences (Fig. 5D, right panel). Collectively, these findings strongly suggest that the prominent contribution of DCL2 to anti-TCV silencing in wild-type plants results, at least partly, from a deficit in DCL4 and DCL3 caused by a P38-dependent enhancement of DCL1 levels.
A model for silencing suppression by TCV in Arabidopsis
The results presented in this and previous studies (Deleris et al. 2006; Qu et al. 2008) can now be integrated into a mechanistic model in which the GW-dependent, AGO1-binding property of P38 is potentially entirely accountable for most of the complex changes incurred by TCV to the Arabidopsis RNA silencing machinery (Fig. 6). We propose that, during initial phases of the infection, the TCV dsRNA is processed mainly by DCL4 into 21-nt-long siRNAs, which incorporate into AGO1 to effect primary antiviral silencing. AGO1 also mediates the activity of cellular miRNAs, including that of miR162, which normally dampens DCL1 accumulation (Fig. 6, panel 1). In a second phase of the infection, the build-up in virus replication would promote accumulation of P38, which exerts its VSR function presumably as a C:C homodimer or multiples thereof. P38 would bind to AGO1 through both its GW motifs, resulting in a deficit of viral siRNA-loaded and cellular miRNA-loaded AGO1. In particular, reduced levels of miR162-loaded AGO1 would enhance DCL1 accumulation. This would in turn promote a decrease in DCL4 and DCL3 levels through as-yet-unidentified mechanisms (Fig. 6, panel 2). Because both DCL4 and DCL3 compete with DCL2 for processing the TCV dsRNA, and because the DCL2 levels are only modestly affected by TCV, DCL2 would then take over the antiviral function upon loading of its 22-nt-long siRNA products into AGO1. The continued action of P38 would explain why only a small fraction of DCL2 products would incorporate into AGO1, resulting in the steady-state infection status normally observed in TCV-infected Arabidopsis tissues (Fig. 6, panel 3).
Figure 6.
A three-step model for the molecular effects of TCV infection on Arabidopsis RNA silencing pathways. See the text for details.
Discussion
The present study provides two fundamentally novel insights into the mechanisms and molecular consequences of antiviral gene silencing and its suppression in plants, and perhaps in metazoans. First, we uncovered how a viral protein has evolved as a specific anti-AGO1 device through positive selection of GW residues within optimal amino acid contexts that had so far been found exclusively in cellular, AGO-interacting proteins. This finding provides yet another original example of how parasites mimic and usurp existing host regulatory pathways. All known GW repeat proteins characterized so far exert positive effects on transcriptional and post-transcriptional gene silencing pathways by facilitating the assembly and/or action of silencing effector complexes in a variety of organisms (Eulalio et al. 2009). Unlike its host-encoded counterparts, however, TCV P38 prevents RNA silencing through its tight interaction with a cellular AGO protein that promotes both antiviral and miRNA functions in Arabidopsis. The most straightforward interpretation of P38's effects on the two silencing pathways is that it sequesters AGO1 away from cognate siRNA and miRNA effector complexes, preventing their assembly and, consequently, loading. A nonmutually exclusive interpretation entails that P38 acts as a dominant negative of an as-yet-unidentified endogenous, GW-rich protein required for miRNA and siRNA RISC assembly/function in plants. This, in turn, would cause the silencing complexes to fall apart, resulting in enhanced small RNA turnover. Whatever the precise mechanism involved, the deployment of GW-rich proteins by viral and nonviral pathogens might well represent a widespread strategy to counteract the effects of both siRNA- and miRNA-directed innate immune pathways, not only in plants, but also in animals. Hence, our own unpublished bioinformatics survey has identified additional putative proteins with clear signatures of GW repeat in both plant and vertebrate viruses, with some of these factors corresponding to known virulence or pathogenicity determinants. Given that the vertebrate miRNA pathway has been implicated in both innate and adaptive immunity against various parasites that include RNA viruses (Lu and Liston 2009), it will be highly interesting to test if, and how, such factors might compromise the integrity/function of mammalian miRNA effector complexes. We also showed that the Arabidopsis miRNA pathway contributes, via AGO1, to basal defense against Pseudomonas syringae; as a counterdefensive strategy, the bacterium secrete proteins into host cells, some of which have effects that are genetically equivalent to those of hypomorphic ago1 mutations, as shown here with P38 (Navarro et al. 2008). These bacterial suppressors of RNA silencing (BSR) thus constitute plausible candidates as GW motif-containing proteins from nonviral pathogens. Intriguingly, the mammalian bacterium Listeria monocytogenes produces several surface proteins directly exposed to the host cytosol that contain extensive GW repeats (Cabanes et al. 2002), which may contribute to the suspected virulence function of these factors by interacting with AGO proteins that effect mammalian miRNAs.
The second important insight from this study is the unsuspected degree of interconnectivity between siRNA and miRNA pathways in Arabidopsis. Notably, we uncovered an AGO1-dependent regulatory network that completely reconfigures the respective intracellular levels of plant DCLs upon TCV and, presumably, other virus infections. While the DCL1 hyperinduction is most likely contributed through effects involving miR162 (Xie et al. 2003), the basis for the DCL1-dependent negative impact on DCL4 and DCL3 levels remains to be established, although preliminary promoter fusion analyses suggest that the dampening of DCL3 accumulation in TCV-infected plants is largely transcriptional (O Voinnet and S Ohnesorge, unpubl.). This newly discovered, global homeostatic control of DCLs not only explains the complex molecular phenotype of the TCV infection, but, more importantly, it also suggests that alterations to AGO1 integrity or function are likely to have much more profound consequences on silencing-based regulation of plant gene expression than originally anticipated. In particular, the dramatic reduction in DCL3 levels incurred by the P38–AGO1 interaction is predicted to have major consequences on RNA-directed DNA methylation (RdDM) and ensuing transcriptional silencing of many endogenous loci, including transposons and repeats (Matzke et al. 2009). This might incidentally explain why viruses reactivate transcription of retroposons in certain plant species (Grandbastien et al. 1997). It can be further inferred that other nonviral pathogens that target AGO1 functions in the miRNA or siRNA pathways, including P. syringae (Navarro et al. 2008), probably will have similar effects on DCL3 and RdDM. Generally, perturbations of AGO1 function/integrity might thus constitute a generic sensor to parasite aggressions that signals the global dampening of the RdDM pathway, consistent with gene hypomethylation reported previously in tobacco upon a pathogen's attacks (Wada et al. 2004). Implicit to this idea is the notion that the RdDM pathway might contribute dynamically to dampen basal resistance to a wide variety of pathogens in healthy plants. We proposed that, in the absence of infection, such a process might help reduce the fitness costs incurred by constitutive expression of defense-related genes, including classical disease resistance (R) genes whose complex genomic arrays are prone to undergo RdDM (Wada et al. 2004; Voinnet 2008; Yi and Richards 2009). This proposed AGO1-dependent “sentinel” effect and its expected impact and seemingly unrelated endogenous silencing pathways might well explain why, unlike in Drosophila, the Arabidopsis DCL4-/DCL2-dependent siRNA pathway and the DCL1-dependent miRNA pathway have not been completely separated and share a common effector, AGO1.
Materials and methods
Plant material
Ararabidopsis thaliana dcl1-9, dcl2–dcl4, ago1-27, and ago7-1 mutants and the ago1-36/Flag:AGO1, ago4/Myc:AGO4, and SUC:SUL lines were described previously (Jacobsen et al. 1999; Morel et al. 2002; Baumberger and Baulcombe 2005; Deleris et al. 2006; Garcia et al. 2006; Li et al. 2006). The pCAMBIA1200-pAGO7-FlagHA-AGO7, pBIN61-P38, and DCL1-GFP constructs (see below) were mobilized into Agrobacterium strain GV3101 and transformed into ago7-1 and SUC:SUL plants, respectively, by the floral dip method (Bechtold and Pelletier 1998). Selection of transgenic plants was on medium containing 25 mg/mL hygromycine or 50 mg/mL kanamycin, respectively. All Arabidopsis plants were grown in standard greenhouse conditions for 3 wk, then in short-day conditions for 2–3 wk before infection assays.
DNA constructs
Individual GW → GA substitutions in the P38 ORF were carried out by PCR-based site-directed mutagenesis using mismatched primers. The pBin61-P38 cDNA template was used for PCR using High-fidelity Mastermix Phusion Taq polymerase (Finnzyme). Oligonucleotide sequences are available on request. Wild-type and GW mutated cDNAs were cloned into pENTR/D vectors and then transferred to the pK7WG2D destination vectors by recombination using the LR Clonase enzyme (Invitrogen). The GA2 virus fragment was reintroduced into pBin61-TCV (Thomas et al. 2003) digested with BamHI and XmaI. The first GW mutation creates a new restriction site (HhaI), which was used to monitor the integrity of viral progenies in systemic infection assays employing TCVGA2. Wild-type and mutated P38 constructs were mobilized into Agrobacterium strain GV3101 and used for transient expression in N. benthamiana for silencing suppression assays, or for virus infection in Arabidopsis. The AGO7 promoter (a 2.1-kb fragment upstream of At1g69440) and downstream genomic sequence were cloned in a custom-made pCAMBIA-1200 vector containing a sequence encoding a double Flag–double HA tag introduced into the NcoI site (vector and MCS sequence can be provided on request). The PCR fragment was inserted between the SalI–SacI sites. Primers used for amplification were as follows: AGO7-SalI, 5′-GTCGACCAACTTTACCTGTCTCTTCTTCTG-3′; and AGO7-SmaI, 5′-CCCGGGTCAGCAGTAAAACATGAGATTC-3′. The presumptive DCL1 promoter sequence (Xie et al. 2003) was PCR-amplified as a 2.5-kB fragment upstream of At1g01040 and cloned in place of the 35S promoter into pBin61-GFP (Voinnet et al. 2003), creating DCL1-GFP. Primers used for amplification were as follows: Frwd-DCL1PRO, 5′-GAGGTCCATGTCCACAATAAGAAAT-3′; and Rev-DCL1PRO, 5′-TCTCTTCTGGTCTGTTTTCCACTTG-3′.
Agro infiltration assay
The transient silencing suppression assay was described previously (Voinnet et al. 1998). The various Agrobacterium tumefaciens clones (strain GV3101) were grown to OD600 = 1 and mixed in equal volumes with strain pBin61-GFP prior to coinfiltration into leaves of 4-wk-old nontransgenic N. benthamiana. GFP expression was monitored under ultraviolet (UV) light as described (Voinnet et al. 1998).
Virus infection assays
Infection of TCVΔP38 on dcl2–dcl4 mutant plants was with in vitro transcripts, as described previously (Deleris et al. 2006; Qu et al. 2008). FlagAGO1, MycAGO4, FlagHA-AGO7, and their respective reference ecotypes (Col0 and La-er) were infected by TCV using sap inoculation as described in Deleris et al. (2006). Infected inflorescences were collected 21 dpi. Derivatives of pBin61-TCV (TCV and TCVGA2) were propagated in A. tumefaciens strain GV3101, grown to OD600 = 1, and infiltrated into 3-wk-old Arabidopsis leaves. Four independent plants were infected (four leaves per plant) for both dcl2–dcl4 and ago1-27 mutants as well as wild-type controls (Col0). RNA and protein analyses in inoculated leaves were carried out at 8 dpi, by pooling the infected tissues. Mock-inoculated (10 mM MES/10 mM MgCl2) leaves were collected at the same time. Systemically infected leaves were collected at 17 dpi, and inflorescences were collected at 21 dpi.
Antibodies
Peptide antibodies were prepared in rabbits according to the DoubleX program of Eurogentec. Peptides used for DCL1 antibodies were H2N-VME DEP REA TIK PSY C-CONH2 and H2N-CNN YRR DGR DRE VRG Y-CONH2. The sera were affinity-purified on the first peptide by Eurogentec. Rabbit antisera were raised against a peptide designed in DCL3 (QTMSDPPSRNEQLPPC), and affinity-purified by Eugentec (Eurogentec SA). Antibodies for AGO1, DCL3, and DCL4 were described previously (Qi et al. 2005; Li et al. 2006; Nakazawa et al. 2007) Affinity-purified anti-HA and anti-MYC antibodies coupled to HRP (Sigma) were used for detection of AGO7 and AGO4, respectively, in transgenic tagged lines.
RNA blot analysis
Total RNA was extracted from A. thaliana tissues or immunoprecipitation products with Tri-Reagent (Sigma) according to the manufacturer's instructions. RNA gel blot analysis of high- and low-molecular-weight RNA was conducted on 1–5 μg and on 10–25 μg of total RNA, respectively, as described previously (Blevins et al. 2006). Ethidium bromide staining of total RNA before transfer was used to check loading. Radiolabeled probes for the SUL or full-length TCV RNA were made by random priming reactions in the presence of α-32P-dCTP (Amersham). The template used was either a 400-base-pair (bp) (SUL) amplified from A. thaliana cDNA, or full-length PCR product for TCV amplified from pBin61-TCV (Thomas et al. 2003). DNA oligonucleotides complementary to miRNAs, tasiRNAs (tasi255 and TAS3), and the TCV P28-derived siRNA2 were end-labeled with γ-32P-ATP using T4 PNK (New England Biolabs). The sequence of the vs-siRNA2 antisense oligonucleotide used as a probe was 5′-ACTACCTACCGCCTCCACCAA-3′.
Real-time RT–PCR analyses of DCL1-GFP
Total RNA was extracted using Rneasy Plant Mini kit (Qiagen). RNA samples were reverse-transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen). The cDNA was quantified using a SYBR Green qPCR kit (Eurogentec) and eGFP gene-specific primers. The primer sequences were as follows: eGFP Forward, 5′-TATATCATGGCCGACAAGCA-3′; and Reverse, 5′-GAACTCCAGCAGGACCATGT-3′. PCR was performed in 96-well optical reaction plates heated for 10 min at 95°C, followed by 45 cycles of denaturation for 15 sec at 95°C, annealing for 20 sec at 60°C, and elongation for 40 sec at 72°C. A melting curve was performed at the end of the amplification by steps of 1°C (from 95°C to 50°C). Transcript levels were normalized to that of Actin2 (At3g18780): Forward, 5′-GCACCCTGTTCTTCTTACCG-3′; and Reverse, 5′-AACCCTCGTAGATTGGCACA-3′.
Semiquantitative RT–PCR analyses
Total RNA was extracted using Rneasy Plant Mini kit (Qiagen). To control equal cDNA amounts in each reaction, a PCR was performed with primers corresponding to ubiquitin. The sequence of primers were as follows: UBQ: Forward, 5′-CGAAGATCCAAGACAAGGAAG-3′, and Reverse, 5′-GATCAAAATGACTCGCCATGA-3′; and DCL2: Forward, 5′-GGTTGGAGTTCCTCGGTGAC-3′, and Reverse, 5′-GGAGATGATGAGAGGCGTACAG-3′.
Total protein extraction and analyses
Total plant protein extracts from A. thaliana inflorescences were obtained following the Tanaka method (Hurkman and Tanaka 1986), and were resolved on SDS-PAGE. After electroblotting proteins on Immobilon-P membrane (Millipore), protein blot analysis was performed using antiserum to DCL1 at a dilution of 1/3000, to DCL3 at 1/1000, to DCL4 at 1/5000 (Nakazawa et al. 2007), and to TCV CP (1/50,000 to 1/100,000 for N. benthamiana leaf infiltrations; 1/500,000 to 1/10−9 for Arabidopsis infections).
Immunoprecipitation of Arabidopsis extracts
Inflorescence tissue from wild-type and transgenic Arabidopsis (ago1/FlagAGO1, ago4/mycAGO4, and ago7/FlagHA:AGO7) were homogenized in BC100 or BC300 (50 mM Tris-HCl at pH 7.5, 100 or 300 mM KCl, 5 mM MgCl2, 0.1% v/v NP40, 10% glycerol) containing the EDTA-free protease inhibitor cocktail (Roche). Cell debris were removed by centrifugation at 18,000g for 30 min at 4°C. The clarified lysate was incubated for 1 h at 4°C at 5 rpm, with anti-Flag M2 agarose beads or anti-MYC agarose beads (Sigma). Beads were packed by gravity into a polyprep column (Bio-Rad) and washed with 50 column volumes (cv) of BC500 or BC800, followed by 10 cv of dPBS (GIBCO). Immunoprecipitates were eluted three times with 1 cv of 3XFlag peptide (Sigma) at 250 μg/mL in BC100, twice with 1 cv of dPBS, and finally with 2 cv of glycine 0.1N (pH 2.5). Aliquots of these fractions and from inputs were subjected to either protein analysis or RNA analysis. Proteins were analyzed by Western blotting or silver nitrate staining with ProteoSilver staining kit (Sigma) according to the suppliers' protocols. For AGO-associated sRNA analysis, immunoprecipitates were subjected to Tri-Reagent extraction (Sigma).
Peptide pull-down assays
Eighteen-amino-acid-long peptides corresponding to the N or C terminus part of P38 containing a GW motif or a mutated GA version were synthesized as biotinylated N-terminal molecules (Eurogentec). Twenty micrograms of each peptide was incubated with 20 μL of washed Dynabeads MyOne Streptavidin T1 beads (Invitrogen) in PBS for 30 min, rotating at room temperature. After three washes in 750 μL of PBS, the beads were then incubated for 2 h at 4°C, with 350 μL of extracts from myc-AGO4 inflorescences, prepared in immunoprecipitation buffer containing 150 mM NaCl and 0.1% NP40. After two washes, the beads were resuspended in 80 μL of SDS-PAGE loading buffer and incubated 5 min at 95°C. The samples were then analyzed by SDS-PAGE. Western blots were performed using polyclonal anti-cmyc (TEBU) and anti-AGO1 antibodies at 1/4000 and 1/1000 dilutions, respectively.
Gel filtration chromatography
For size exclusion chromatography of TCV-infected Flag:AGO1 immunoprecipitation products, peptide eluted fractions (washed with BC800: 50 mM Tris-HCl at pH 7.5, 800 mM KCl, 5 mM MgCl2, 0.1% v/v NP40, 10% glycerol, protease inhibitor cocktail EDTA-free [from Roche]) were loaded onto a Superose 6 GL 10/30 column equilibrated with BC800 buffer. The flow rate was fixed at 0.35 mL/min, and 0.5-mL fractions were collected at 4°C. Aliquots of fractions were analyzed by immunoblotting and silver nitrate staining.
P38 modeling
The crystallographic coordinates of the TCV capsid monomer were kindly provided by Stephen Harrisson. The residues of interest (R74 and E122) were replaced (W and K, respectively) using O (Jones et al. 1991), and the adjacent monomer was generated by building the whole capsid with VIPERdb's oligomer generator (http://viperdb.scripps.edu). Representations of TCV P38 were drawn using PyMOL (http://www.pymol.org).
Acknowledgments
This work was supported by a grant from the Agence Nationale de la Recherche, project ANR-08-BLAN-0206-01 “Ago-hook” attributed to O.V., T.L., and M.A.H. O.V. also acknowledges financial support from a starting grant of the European Research Council “Frontiers of RNAi” ERC 210890 and a prize from the Bettencourt Foundation for Life Science.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1908710.
Supplemental material is available at http://www.genesdev.org.
References
- Baulcombe D 2004. RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC 2005. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F Jr, Hohn T, et al. 2006. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34: 6233–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol 10: 238–245 [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O 2007. Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O 2006. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat Genet 38: 258–263 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O 2007. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet 39: 848–856 [DOI] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T 2007. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Dev 21: 2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E 2009. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA 15: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA 2006. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Grandbastien MA, Lucas H, Morel JB, Mhiri C, Vernhettes S, Casacuberta JM 1997. The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica 100: 241–252 [PubMed] [Google Scholar]
- Hurkman WJ, Tanaka CK 1986. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol 81: 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM 1999. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Li F, Ding SW 2006. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 60: 503–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE 2006. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Lu LF, Liston A 2009. MicroRNA in the immune system, microRNA as an immune system. Immunology 127: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ 2009. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC 2008. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Morel J-B, Gordon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T 2007. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol Biol 63: 777–785 [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O 2008. Suppression of the microRNA pathway by bacterial effector proteins. Science 321: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Qu F, Morris TJ 1997. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J Virol 71: 1428–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris JT 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O 2009. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60: 485–510 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Leh V, Lederer C, Maule AJ 2003. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306: 33–41 [DOI] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG 2007. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol 14: 897–903 [DOI] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811 [DOI] [PubMed] [Google Scholar]
- Vaucheret H 2005. MicroRNA-dependent trans-acting siRNA production. Sci STKE 2005: pe43. doi: 10.1126/stke.3002005pe43 [DOI] [PubMed] [Google Scholar]
- Vaucheret H 2008. Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Voinnet O 2008. Post-transcriptional RNA silencing in plant-microbe interactions: A touch of robustness and versatility. Curr Opin Plant Biol 11: 464–470 [DOI] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Vain P, Angell S, Baulcombe DC 1998. Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell 95: 177–187 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wada Y, Miyamoto K, Kusano T, Sano H 2004. Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol Genet Genomics 271: 658–666 [DOI] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC 2003. Negative feedback regulation of Dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 13: 784–789 [DOI] [PubMed] [Google Scholar]
- Yi H, Richards EJ 2009. Gene duplication and hypermutation of the pathogen Resistance gene SNC1 in the Arabidopsis bal variant. Genetics 183: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]