Abstract
Cancer stem cells (CSCs) display plasticity and self-renewal properties reminiscent of normal tissue stem cells, but the events responsible for their emergence remain obscure. We recently identified CSCs in Ewing sarcoma family tumors (ESFTs) and showed that they retain mesenchymal stem cell (MSC) plasticity. In the present study, we addressed the mechanisms that underlie ESFT CSC development. We show that the EWS-FLI-1 fusion gene, associated with 85%–90% of ESFTs and believed to initiate their pathogenesis, induces expression of the embryonic stem cell (ESC) genes OCT4, SOX2, and NANOG in human pediatric MSCs (hpMSCs) but not in their adult counterparts. Moreover, under appropriate culture conditions, hpMSCs expressing EWS-FLI-1 generate a cell subpopulation displaying ESFT CSC features in vitro. We further demonstrate that induction of the ESFT CSC phenotype is the result of the combined effect of EWS-FLI-1 on its target gene expression and repression of microRNA-145 (miRNA145) promoter activity. Finally, we provide evidence that EWS-FLI-1 and miRNA-145 function in a mutually repressive feedback loop and identify their common target gene, SOX2, in addition to miRNA145 itself, as key players in ESFT cell differentiation and tumorigenicity. Our observations provide insight for the first time into the mechanisms whereby a single oncogene can reprogram primary cells to display a CSC phenotype.
Keywords: Ewing sarcoma, SOX2, cancer stem cells, mesenchymal stem cells, miRNA145, reprogramming
Hematopoietic malignancies as well as a growing number of solid tumors have been shown to contain a subpopulation of cells possessing tumor-initiating capability associated with stem cell properties that include expression of embryonic stem cell (ESC) genes and asymmetric division (Clarke et al. 2006). These cells, termed cancer stem cells (CSCs), are generally believed to represent the tumor repopulating force, preserving their own numbers through self-renewal, and generating more differentiated progeny that compose the bulk of the tumor (Clarke et al. 2006). Such cells should, in principle, be able to arise by at least two mechanisms: transformation of resident stem cells within a tissue that allows them to maintain their “stemness” in the transformed state, or genetic reprogramming of differentiated somatic cells during transformation that endows them with stem cell properties. Recent work has identified a small number of transcription factors normally expressed in ESCs—including OCT4, SOX2, NANOG, C-MYC, KLF4, and LIN28—that can reprogram fibroblasts and other differentiated cells to acquire pluripotency and become what have been termed human induced pluripotent stem cells (hiPSs) (Jaenisch and Young 2008). Thus, it is conceivable that transformation of primary differentiated or lineage-committed cells may incur similar reprogramming, and that some degree of reprogramming may even be inherent to transformation itself. In support of these views, loss of the retinoblastoma (RB) gene that occurs in a wide variety of malignancies has been shown to cause primary mouse embryonic fibroblasts (MEFs) to undergo reprogramming and acquire CSC properties (Liu et al. 2009). In addition, recent observations suggest that two major tumor suppressor genes, INK4A and TP53, impair efficient iPS generation (Banito et al. 2009; Kawamura et al. 2009; Marion et al. 2009), consistent with the existence of a relationship between genetic reprogramming and transformation.
Some types of cancer, including a subset of sarcomas and hematopoietic malignancies, can, at least in their early phases, display only a single detectable oncogenic event, usually in the form of a nonrandom reciprocal chromosomal translocation. Chromosomal translocations that constitute a phenotypic signature of defined tumor types commonly generate a functional fusion gene believed to initiate tumor development (Helman and Meltzer 2003). For transformation and subsequent tumor initiation to occur, the corresponding fusion protein should be able to generate CSCs by either exploiting a primary cellular microenvironment that is permissive for expression of its oncogenic properties, or early reprogramming of primary target cells to render them permissive for its subsequent transforming activity.
Ewing sarcoma family tumors (ESFTs) provide a malignancy of choice to address this issue because they are associated with a single chromosomal translocation that appears to be the only detectable oncogenic event in as many as 20% of the tumors (Riggi and Stamenkovic 2007). Furthermore, they display both mesenchymal stem cell (MSC) and neural crest stem cell (NCSC) features, contain a tumor cell subpopulation with CSC properties, and are thought to arise from primary MSCs (Suva et al. 2009).
ESFTs constitute a highly aggressive bone and soft tissue malignancy of children and young adults associated with a unique chromosomal translocation that generates a fusion protein composed of EWS (or, rarely, FUS/TLS) and a member of the ets transcription factor family (Riggi et al. 2007). In 85%–90% of cases, the t(11;22)(q24;q12) translocation fuses the 5′ end of the EWS gene to the 3′ end of the FLI-1 gene, giving rise to the EWS-FLI-1 fusion protein where sequences containing the potent EWS transactivation domain are joined to sequences containing the DNA-binding domain (DBD) of FLI-1 (Delattre et al. 1992).
EWS-FLI-1 behaves as an aberrant transcription factor—with both inducer and suppressor activity—that displays distinct target gene specificity from those of its component parts (Riggi and Stamenkovic 2007). Expression of the full transcriptional potential of EWS-FLI-1 is highly cell context-dependent, as illustrated by its ability to transdifferentiate neuroblastoma and rhabdomyosarcoma cells, but to induce p53-dependent growth arrest and apoptosis in primary human embryonic fibroblasts (Lessnick et al. 2002) and MEFs (Deneen and Denny 2001). We showed previously that, among primary human cells, only MSCs display permissiveness for stable EWS-FLI-1 expression without undergoing growth arrest (Riggi et al. 2005, 2008), and that, in mouse mesenchymal progenitor cells (MPCs), expression of EWS-FLI-1 alone is sufficient to induce transformation and development of Ewing sarcoma-like tumors in vivo (Riggi et al. 2005). In adult human MSCs (hMSCs), EWS-FLI-1 was observed to induce a transcriptome reminiscent of that of Ewing sarcoma, including expression of insulin-like growth factor-1 (IGF1), on whose activity ESFT cells are dependent, and the polycomb group gene enhancer of zeste-2 (EZH2) (Riggi et al. 2008). Subsequent work showed that IGF1 is a direct target of EWS-FLI-1 in MSCs (Cironi et al. 2008), whereas EZH2, which was found to be highly expressed in primary ESFTs, plays a central role in the maintenance of Ewing sarcoma cell line tumorigenicity (Riggi et al. 2008). More recently, we identified ESFT CSCs (Suva et al. 2009) and showed that they express the CD133/Prominin-1 stem cell surface marker in addition to representing the tumor-initiating population of ESFTs. Interestingly, ESFT CSCs retain MSC plasticity in vitro, and express genes implicated in ESC maintenance and reprogramming, including SOX2, OCT4, and NANOG.
To address the mechanisms that may underlie the emergence of ESFT CSCs, we assessed the effects of EWS-FLI-1 expression in human pediatric MSCs (hpMSCs), given that ESFTs are primarily a pediatric/young adult malignancy. We show that hpMSCs constitute a far more permissive cellular environment toward EWS-FLI-1 activity than their adult counterparts, and display gene expression profile changes that more accurately recapitulate the molecular profile of ESFT. Moreover, pediatric MSCs grown in a defined serum-free (henceforth KO) culture medium undergo genetic reprogramming in response to EWS-FLI-1, reflected by strong induction of the ESC genes OCT4, NANOG, and SOX2; the appearance of a NCSC molecular phenotype; and the generation of a subpopulation of cells that displays ESFT CSC features. We further provide mechanistic evidence that the robust induction of SOX2, OCT4, and NANOG associated with the emergence of an ESFT CSC population results in part from the in vivo repression of the microRNA-145 (miRNA145) promoter by EWS-FLI-1. Finally, we uncover the existence of a negative regulatory feedback loop between EWS-FLI-1 and the miRNA145, and identify their common target, Sox-2, as a key player in determining ESFT cell differentiation and tumorigenicity.
Results
Characterization of pediatric MSCs
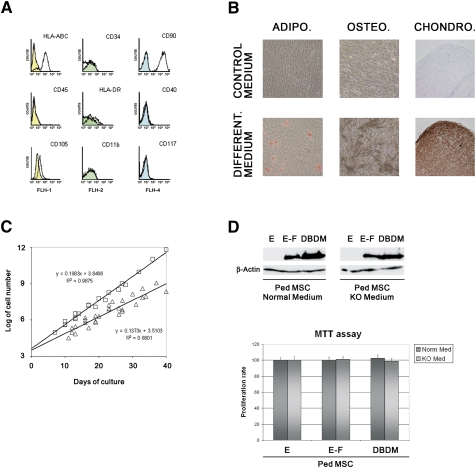
hpMSCs were isolated from surgically removed bone of healthy 6- to 14-yr-old patients undergoing corrective surgery. Isolation of hpMSCs was performed according to established protocols (Suva et al. 2004, 2008), and their cell surface phenotype—as defined by expression of HLA-ABC, CD90, and CD105, and absence of CD34, CD45, CD40, CD11b, and CD117—was verified (Fig. 1A). Cells were maintained in culture at subconfluent concentrations and were periodically assessed for their ability to differentiate into adipocytes, osteocytes, and chondrocytes (Fig. 1B). Cell surface marker expression and differentiation potential were comparable among pediatric and adult MSC populations. However, proliferation of pediatric MSCs was significantly higher than that of their adult counterparts (Fig. 1C). Expression of EWS-FLI-1 was stably maintained in these cells, just as it was in adult MSCs, and remained unaltered whether the cells were cultured in standard MSC medium (serum-supplemented) or in a defined KO medium (Fig. 1D, top panel). Similar to adult cells, hpMSCs cultured in both serum-supplemented and KO medium displayed marked rounding following expression of EWS-FLI-1. However, no difference in proliferation was observed between cells infected with retroviruses engineered to express wild-type EWS-FLI-1, EWS-FLI-1 containing a mutation within the DBD that precludes its binding to DNA (DBDM) (Welford et al. 2001), or empty vector (Fig. 1D, bottom panel; data not shown).
Figure 1.
Phenotype of hpMSCs and their permissiveness for EWS-FLI-1 expression. (A) FACS characterization of hpMSCs. Cells were labeled with phycoerythrin (PE)-conjugated antibodies against CD45, CD40, HLA-ABC, CD105, CD11b, CD34, CD90, HLA-DR, and CD117, or immunoglobulin isotype-matched control antibodies. (Colored area) Control antibody; (white area) specific antibody. (B) hpMSCs retain the ability to differentiate into adipogenic, osteogenic, and chondrogenic lineages upon stimulation with the appropriate cytokines (for details, see the Materials and Methods). (Bottom panels) Adipocytic differentiation, oil Red-O staining (control: medium only); osteoblastic differentiation, von Kossa staining (control: basal medium); chondrocytic differentiation, anti-collagen type II labeling counterstained with hematoxylin (control: unrelated, isotype-matched antibody). Magnification: adipocytes and osteocytes, 200×; chondrocytes, 100×. (C) Comparison of the proliferation rate between adult and pediatric hMSC during a 40-d culture period. (D, top panel) Western blot analysis of EWS-FLI-1 (E-F) and DBDM expression in hpMSCs cultured in FCS-containing or KO medium 12 d after the infection ([E] empty vector-infected cells). (Bottom panel) MTT assay analysis of the proliferation rate of hpMSCs expressing DBDM or EWS-FLI-1 compared with their empty vector-infected counterparts, cultured in KO or serum-supplemented conditions.
The response of pediatric MSCs to EWS-FLI-1 differs from that of adult MSCs
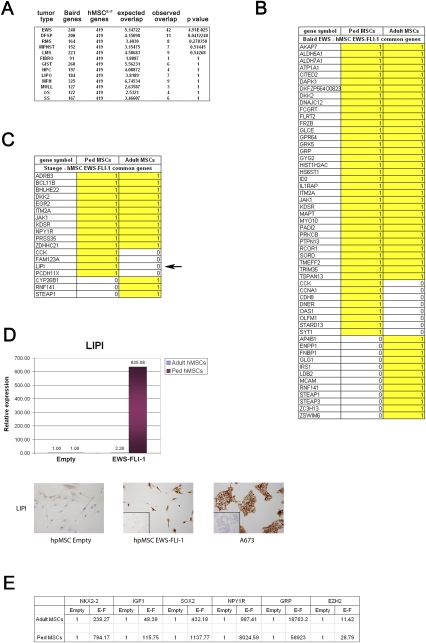
Gene expression analysis of hpMSCEWS-FLI-1s was performed on RNA from four independent hpMSC isolates grown in standard serum-supplemented medium 12 d following infection with retrovirus containing EWS-FLI-1. Comparison of the gene expression profile of these cells to a primary sarcoma gene expression database (Baird et al. 2005) revealed highly significant similarity with the transcriptome of Ewing sarcoma (Fig. 2A), comparable with that observed between the gene expression profiles of adult hMSCEWS-FLI-1s and primary ESFTs (Riggi et al. 2008). Qualitative comparison, however, revealed that, whereas the total number of up-regulated genes shared by hMSCEWS-FLI-1s and primary Ewing sarcoma did not change when adult MSCs were substituted by hpMSCs, the gene repertoire was not identical. Thus, several genes that were induced in hpMSCs were not induced in adult MSCs (Fig. 2B), and the same held true when comparison was made using other databases (Fig. 2C; Staege et al. 2004). Remarkably, the membrane-associated phospholipase A1 β LIPI that has been reported recently to be a specific ESFT marker (Foell et al. 2008) was induced in hpMSCs but not in adult MSCs (Fig. 2C). Quantitative real-time PCR analysis revealed that, whereas LIPI was up-regulated a little more than twofold in adult MSCs, it was induced >600-fold in hpMSCs (Fig. 2D, top panel). Accordingly, whereas LIPI protein expression was undetectable in empty vector-infected cells, almost all hpMSCEWS-FLI-1s displayed strong and homogeneous LIPI expression (Fig. 2D, bottom panel).
Figure 2.
EWS-FLI-1 expression in hpMSC induces a gene expression profile that is distinct from that induced in adult MSC. (A) Statistical analysis of genes common to hpMSCEWS-FLI-1 cultured in the presence of serum and a publicly available soft tissue tumor gene expression profile database (Baird et al. 2005). HpMSCEWS-FLI-1s display remarkable gene expression profile similarity to ESFTs but not to other sarcomas. (B) List of the 42 genes found to be shared by the hpMSCEWS-FLI-1 and ESFT gene expression profiles, and their comparison with the genes induced in adult hMSCEWS-FLI-1s, demonstrating the qualitative differences in the molecular response to EWS-FLI-1 expression between the two cell types. (C) List of the 15 common genes between hpMSCEWS-FLI-1s and the ESFT expression signature identified in another study (Staege et al. 2004), compared with that obtained using adult MSCs. (D, top panel) Real-time PCR analysis of LIPI expression levels in empty vector-infected and EWS-FLI-1-expressing pediatric and adult hMSCs, showing that the induction of LIPI is restricted to pediatric cells. (Bottom panel) hpMSCEWS-FLI-1s display strong homogeneous LIPI protein expression compared with their empty vector-infected counterparts and similar to the A673 cell line, as assessed by immunocytochemistry. Magnification, 100×; inset, 200×. (E) Real-time PCR assessment of selected EWS-FLI-1 target genes believed to play an important role in ESFT pathogenesis. Expression of all of these genes was more strongly induced by EWS-FLI-1 in hpMSCs than in adult MSCs. Real-time PCR experiments were normalized to cyclophyllin A and done in triplicate. Error bars represent the SD of three independent determinations. (EWS) Ewing's sarcoma; (DFSP) dermatofibrosarcoma; (Lipo) liposarcoma; (RMS) rhabdomyosarcoma; (MFH) malignant fibrous histiocytoma; (OS) osteosarcoma; (HPC) hemangiopericytoma; (GIST) gastrointestinal stromal tumor; (LMS) leiomyosarcoma; (SS) synovial sarcoma; (MULL) mixed mullerian; (MPNST) malignant peripheral nerve sheath tumor; (Fibro) fibrosarcoma.
A similar observation was made for the CCK gene that has been shown to play a role in the tumorigenic potential of ESFT cell lines (Supplemental Data 1A; Carrillo et al. 2007). Quantitative comparison, by real-time PCR, of the induction of a panel of EWS-FLI-1 target genes that are believed to participate in ESFT pathogenesis—including NKX2-2, IGF1, SOX2, NPY1R, GRP, and EZH2—revealed that all were more strongly induced in hpMSCs than in their adult counterparts (Fig. 2E). The stronger transcript induction could not be attributed to a difference in the EWS-FLI-1 protein expression level, which was comparable in adult and pediatric MSCs (data not shown). Moreover, among new candidate EWS-FLI-1 target genes induced in hpMSCEWS-FLI-1s, we identified NTRK1 and ALK that have been shown to play a critical role in the development of neuroblastoma (Janoueix-Lerosey et al. 2008; Mosse et al. 2008) and the MEIS1–PBX1 complex implicated in leukemogenesis (Supplemental Data 1B; Argiropoulos et al. 2007).
MSCs cultured in KO medium culture conditions display increased permissiveness for EWS-FLI-1 activity
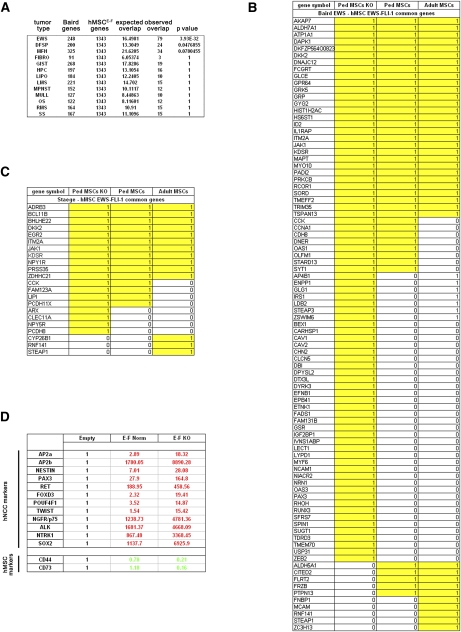
Recently, the possibility to genetically reprogram differentiated primary mouse and human cells toward iPSs by expression of a defined core set of transcription factors (OCT4–SOX2–MYC–KLF4 or OCT4–SOX2–NANOG–LIN28) followed by specific KO medium culture conditions of the infected cells has been reported (for review, see Jaenisch and Young 2008). Based on the ability of EWS-FLI-1 to induce at least two of the reprogramming factors in hMSC—namely, c-Myc (Dauphinot et al. 2001) and Sox2 (Riggi et al. 2008)—and the capacity of wild-type EWS to interact with Oct-4 in vivo and stimulate its transcriptional activity in ESC (Lee et al. 2005), we addressed the possibility that EWS-FLI-1 alone might be able to partially reprogram hpMSCs. Expression of EWS-FLI-1 in hpMSCs grown under KO culture medium conditions and subjected to the reported reprogramming protocol (Maherali and Hochedlinger 2008) induced a gene expression profile bearing the highest similarity to that of ESFTs observed in a primary cellular context thus far. Comparison of the transcriptome of hpMSCEWS-FLI-1s to that of the Baird et al. (2005) sarcoma database revealed the number of induced genes shared by ESFTs and hpMSCEWS-FLI-1s to almost double when hpMSCEWS-FLI-1s were cultured in KO rather than in serum-containing medium (79 vs. 42 genes) (Fig. 3A). The overlap with another database that identified a Ewing sarcoma gene expression signature (Staege et al. 2004) was also increased significantly (Fig. 3C). The observed quantitative and qualitative difference between KO medium-cultured hpMSCEWS-FLI-1 and adult MSCEWS-FLI-1 expression profiles was even more striking (Fig. 3B,C).
Figure 3.
hpMSCs cultured in KO stem cell culture conditions display increased permissiveness for EWS-FLI-1 activity and undergo genetic reprogramming to acquire a NCSC molecular phenotype. (A) Statistical analysis of the genes common to hpMSCEWS-FLI-1s and the soft tissue tumor gene expression profile database (Baird et al. 2005) presented in Figure 2, showing the marked increase in similarity between hpMSCEWS-FLI-1s and ESFTs when hpMSCs were cultured in KO conditions. (B) List of the 79 genes found to be shared by hpMSCEWS-FLI-1s grown in KO culture conditions and the ESFT gene expression profile, and their comparison with the genes induced in pediatric and adult hMSCEWS-FLI-1s cultured in presence of serum. (C) List of the 19 genes common to hpMSCEWS-FLI-1s cultured in KO conditions and the ESFT expression signature identified by Staege et al. (2004) compared with results obtained using pediatric and adult MSCs cultured in the presence of serum. (D) Expression levels of a selected panel of transcripts, currently used to define the NCSC phenotype, as assessed by real-time PCR, showing induction of all of the genes in hpMSCEWS-FLI-1s cultured in KO medium but not in serum-supplemented medium. In KO medium-cultured cells only, induction of hNCSC transcripts was accompanied by a concomitant repression of the two major MSC markers, CD73 and CD44. Real-time PCR experiments were normalized to cyclophyllin A and done in triplicate. Error bars represent the SD of three independent determinations. For abbreviations, see the legend for Figure 2.
HpMSCEWS-FLI-1s undergo genetic reprogramming toward a NCSC and primary ESFT phenotype
Primary ESFTs display both mesenchymal and neuroectodermal features that have fueled a long-standing debate as to their origin. In light of the recent demonstration that, under appropriate culture conditions, human primary NCSCs (hNCSCs) can give rise to MSCs (Takashima et al. 2007), induction by EWS-FLI-1 of the NCSC marker NGFR/p75 in hpMSCs prompted us to assess the expression of a panel of transcripts currently used to define the NCSC phenotype. The transcripts included AP2A, AP2B, NESTIN, PAX3, RET, FOXD3, POU4F1, TWIST, p75, ALK, and NTRK1. All of these genes were strongly induced in hpMSCEWS-FLI-1s cultured in KO medium, as assessed by real-time PCR, but were either more weakly induced or not induced at all when the same cells were cultured in the presence of serum. Their induction was dependent on direct binding of EWS-FLI-1 to DNA, as hpMSCs expressing the DBDM of the fusion gene displayed no similarity with either ESFT or hNCSC databases (data not shown). Some of the transcripts were induced 15-fold to 165-fold, whereas others, including NGFR/p75, ALK, NTRK1, and AP2B, were induced several-thousand-fold (Fig. 3D), which is virtually equivalent to de novo expression. Interestingly, in KO medium-cultured cells only, induction of hNCSC transcripts was accompanied by a concomitant repression of two prominent MSC markers, CD73 and CD44, consistent with the primary ESFT phenotype, where only a minority of tumor cells (5%–10%) express cell surface CD44 and/or CD73 (data not shown). Together, these observations suggest that the effect of EWS-FLI-1 expression in hpMSCs cultured in KO medium was not limited to a mere modulation of transcripts already present in these cells, but consisted in large part of promoting the generation of a new cellular phenotype characterized by the de novo appearance of hNCSC markers.
EWS-FLI-1 expression in hpMSCs induces generation of a cell subpopulation displaying an ESFT CSC phenotype
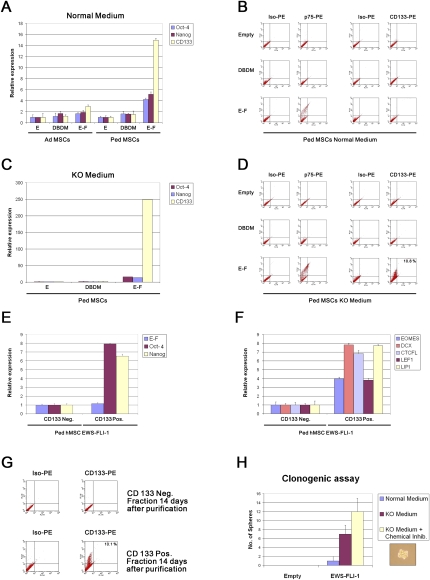
The core gene set required for reprogramming of differentiated cells into iPSs is composed of OCT4, SOX2, NANOG, C-MYC, KLF4, and LIN28. Because SOX2 and c-MYC are reported EWS-FLI-1 target genes (Dauphinot et al. 2001; Riggi et al. 2008), we assessed the expression level of the remaining genes required for reprogramming in three different adult and pediatric MSC populations upon infection with the wild-type or DBDM form of EWS-FLI-1 in the presence of serum or in KO medium. None of these transcripts was significantly induced by EWS-FLI-1 in adult MSCs, irrespective of culture conditions (Fig. 4A; data not shown). In contrast, a fourfold and fivefold induction of OCT4 and NANOG, respectively, was observed in all three pediatric populations tested in the presence of serum (Fig. 4A). Induction of these two genes was increased to 20-fold when the same cell populations were cultured in KO medium (Fig. 4B), confirming that the complete set of transcription factors required to orchestrate genetic reprogramming of primary human cells was induced in hpMSCEWS-FLI-1s, but not in their adult counterparts. Unlike its effect on OCT4, NANOG, MYC, and SOX2, EWS-FLI-1 did not induce KLF4 and LIN28 in hpMSCs (data not shown). No induction of any of these transcription factors was observed in cells expressing the DBDM form of EWS-FLI-1, irrespective of culture conditions.
Figure 4.
EWS-FLI-1 expression in hpMSCs grown in KO culture medium generates a cell subpopulation displaying the major in vitro features of ESFT CSCs. (A) Adult and pediatric hMSCDBDMs and hMSCEWS-FLI-1s were compared, by real-time PCR analysis, for expression of OCT4, NANOG, and CD133 that characterize the ESFT CSC molecular phenotype. No induction was observed in hMSCDBDMs. (B) FACS analysis of cell surface CD133 expression in hpMSCEWS-FLI-1s, showing that the 15-fold induction of the CD133 transcript was not accompanied by a concomitant appearance of its glycosylated, stem cell-related epitope, despite marked induction of the NGFR/p75 NCSC marker. (C,D) Experiments described in A and B were repeated using the same hpMSCEWS-FLI-1 population cultured in KO medium. In cells cultured in KO medium, EWS-FLI-1 strongly enhanced OCT4, NANOG, and CD133 transcription (C), and generated a hpMSCEWS-FLI-1 subpopulation expressing the glycosylated cell surface CD133 epitope (D). (E) Real-time PCR analysis of OCT4, NANOG, and EWS-FLI-1 gene expression levels, in CD133-positive and CD133-negative hpMSCEWS-FLI-1-derived RNA, showing that the CD133-positive fraction expresses a higher level of OCT4 and NANOG, but a nearly identical EWS-FLI-1 level. A single representative experiment is shown. All real-time PCR experiments were normalized to cyclophyllin A and done in triplicate. Error bars represent the SD of three independent determinations. (F) hpMSCEWS-FLI-1 CD133-positive fraction expresses a higher level of the ESFT marker LIPI, and of a panel of genes involved in stemness. (G) FACS analysis of cell surface CD133 expression of CD133-positive or CD133-negative hpMSCEWS-FLI-1-derived progeny, cultured in KO medium for 14 d following the initial sorting. (H) Clonogenic assays were performed in triplicate as described in the Materials and Methods. Error bars indicate the difference in the number of spheres formed in the three determinations. (Bottom right) Photomicrograph of a hpMSCEWS-FLI-1-derived sphere. Magnification, 400×.
Based on our recent demonstration that ESFTs contain CSCs that express the stem cell marker CD133 (Suva et al. 2009), and the notion that EWS-FLI-1 initiates ESFT development, we addressed the possibility that EWS-FLI-1 expression may generate a hpMSC subpopulation with ESFT CSC features. We first performed real-time PCR analysis of CD133 expression levels in adult and hpMSCEWS-FLI-1s cultured in the presence of serum or in KO medium. Slight induction of CD133 in adult cells and a 15-fold induction in pediatric populations were observed in the presence of serum (Fig. 4A). Strikingly, the same pediatric populations grown in KO medium displayed a 250-fold increase of the CD133 transcript, whereas no induction of CD133 was observed in cells expressing the DBDM form of the fusion protein. Based on these observations, hpMSCEWS-FLI-1s were tested for cell surface expression of CD133, and, more specifically, for its glycosylated epitope, AC133, which has been used for CSC identification (Suva et al. 2009). hpMSCEWS-FLI-1s grown in the presence of serum did not express the AC133 epitope, as assessed by flow cytometry, despite induction of the neural stem cell marker NGFR/p75 on their surface (Fig. 4B). In contrast, FACS analysis of the same cells cultured in KO medium revealed a stronger cell surface induction of NGFR/p75, and, more remarkably, the generation of a subpopulation of hpMSCEWS-FLI-1s expressing the glycosylated AC133 epitope of CD133 (Fig. 4D). The same result was obtained using three additional hpMSCEWS-FLI-1 populations, and the percentage of MSCEWS-FLI-1s observed to be positive for CD133 (6%–10%) was comparable with that of CD133+ CSCs found in primary ESFTs (Suva et al. 2009).
To verify that the appearance of CD133-positive cells was not a reflection of the expansion of a rare subpopulation of pre-existing CD133-positive cells that selectively respond to EWS-FLI-1, we performed CD133 expression analysis by flow cytometry in three different hpMSC populations cultured in KO medium before and after CD133 magnetic microbead separation. This technique is reported to allow identification of even very rare CD133-positive cells. Despite this approach, we could not identify a CD133-positive cell fraction (Supplemental Data 1D, left). In parallel, one of the hpMSC populations was used to perform a flow cytometric time-course analysis of CD133 protein detectability upon EWS-FLI-1 expression. Consistent with the reprogramming hypothesis, the CD133-positive subpopulation, comprising 6% of the bulk culture, was observed to appear abruptly at days 12–14 after EWS-FLI-1 expression, reaching a steady state rapidly thereafter. Thus, we did not observe a gradual emergence of CD133-positive cells that one might expect in the case of the expansion of a pre-existing CD133-positive population. In contrast, no CD133-positive cells were detectable in wild-type hpMSCs before or after CD133 magnetic microbead separation within this time frame (Supplemental Data 1D, right).
Identification of mechanisms that underlie the observed difference in susceptibility to reprogramming between adult and pediatric hMSCs would clearly be of interest, as our current understanding of the regulation of genetic reprogramming toward induced pluripotency is only partial. Among the reported pathways involved, the most relevant include those implicated in cell cycle and proliferation control. Several recent reports have suggested the cell cycle regulators Rb, p53, INK4a(p16), and ARF(p14) to provide major biological roadblocks for genetic reprogramming. We therefore compared the gene expression profile of three different batches of wild-type adult and pediatric MSCs (Supplemental Data 2), but recorded no difference in the expression level of Rb, p53, p16, or p14, as assessed by Affymetrix microarrays. However, to exclude possible subtle differences in their expression levels that may not have been detected by microarrays, we performed real-time PCR analysis of the same transcripts in adult and pediatric hMSCs. We observed a modest but significant reduction of p14/p16 (but not p53 or Rb) transcript levels when hpMSCs were cultured in KO compared with serum-containing medium, whereas the same transcripts displayed no difference in expression level among adult MSCs grown in the two culture conditions (Supplemental Data 3A).
In addition to displaying CD133 on their cell surface, primary ESFT CSCs were observed to express high levels of OCT4 and NANOG, generate both CD133-positive and CD133-negative progeny, retain MSC differentiation plasticity, and display sphere-forming ability in clonogenic assays (Suva et al. 2009). To determine whether CD133-positive hpMSCEWS-FLI-1s generated in KO medium display a CSC phenotype, we separated hpMSCEWS-FLI-1 CD133+ and CD133− fractions using magnetic beads, and compared OCT4 and NANOG expression in the two subpopulations. Consistent with the situation in primary ESFT CSCs, both genes were more highly expressed in CD133+ cells (Fig. 4E), despite an equal EWS-FLI-1 fusion gene expression level in the two cell populations (Suva et al. 2009). Microrarray gene expression comparison further revealed higher expression of numerous genes implicated in stem cell function and neuronal differentiation in the CD133+ population (Supplemental Data 2). Expression of several of these genes—including EOMES, DCX, CTCFL, LEF1, and LIPI—was validated by quantitative real-time PCR (Fig. 4F). To further assess the similarity between hpMSCEWS-FLI-1s and ESFT CSCs, hpMSCEWS-FLI-1 CD133+ and CD133− cells were separated, and both populations were plated in KO medium. After 2 wk of culture, cells derived from the two fractions were analyzed by flow cytometry for expression of CD133. Only the progeny of the CD133+ population contained CD133+ cells whose percentage was comparable with that in the initial unsorted population (Fig. 4G). These observations confirm that only the hpMSCEWS-FLI-1 CD133+ fraction possesses the ability, ascribed to CSCs, to generate both CD133-positive and CD133-negative progeny.
Although no currently available in vitro test is specific for CSCs, some growth properties, including spheroid formation in culture, constitute candidate CSC features (Singh et al. 2003; Todaro et al. 2007). We therefore tested the ability of empty vector-infected or EWS-FLI-1-expressing hpMSCs to form spheres in KO medium. hpMSCs were subjected to standard retroviral infection in the KO reprogramming protocol (Maherali and Hochedlinger 2008), then detached and seeded as single-cell suspensions in triplicate onto ultralow attachment 96-well plates. Because sphere formation is typically scored after a prolonged culture time (30 d in our experiments) following seeding, the KO culture medium was supplemented with three chemicals (PD0325901, CHIR99021, and A8301) that were reported recently to allow stable culture of reprogrammed hiPSs, precluding their senescence in vitro (Feng et al. 2009). Addition of the three chemicals to the culture medium did not modify the percentage of CD133+ cells generated by EWS-FLI-1 expression (data not shown). hpMSCs infected with empty vector could not form spheres, irrespective of the presence or absence of the chemicals. In contrast, hpMSCEWS-FLI-1s cultured in KO medium revealed spherogenic capacity similar to that displayed by ESFT CSCs, which was further enhanced by addition of the chemicals (Fig. 4H). Despite their spherogenicity, hpMSCs expressing EWS-FLI-1 failed to form tumors in vivo, suggesting either the requirement for a cooperating oncogenic event, or the possibility that the in vitro KO microenvironment, while allowing the emergence of a cellular subpopulation displaying CSC features, still lacks the capacity to fully reproduce the original cellular niche required for definitive transformation toward ESFTs.
It is noteworthy that the efficiency of EWS-FLI-1-mediated hpMSC reprogramming toward an ESFT CSC phenotype decreased with the successive hpMSC passages. The capacity of the hpMSCs to form CD133-positive cells upon EWS-FLI-1 expression was maximal between passages 1 and 5 in vitro. Thereafter, this ability progressively decreased between passages 5 and 10 and was definitively lost after passage 15. The reprogramming capacity was rapidly lost in cells submitted to freezing–thawing cycles, and was also dependent on the age of the pediatric donor, with younger patient-derived cells showing higher proclivity to form CD133-positive cells. Thus, compared with adult cells, the higher proliferation rate of hpMSCs associated with reduction in their p14/p16 transcript levels upon culture in KO conditions may at least partially explain their increased susceptibility to reprogramming toward ESFT CSC imposed by EWS-FLI-1 expression. These observations are consistent with the notion that cellular senescence impairs reprogramming, and that a high replicative potential is crucial to induce pluripotency in primary somatic cells (Banito et al. 2009).
EWS-FLI-1 induces higher SOX2, OCT-4, and NANOG transcript levels in KO conditions through repression of miRNA145
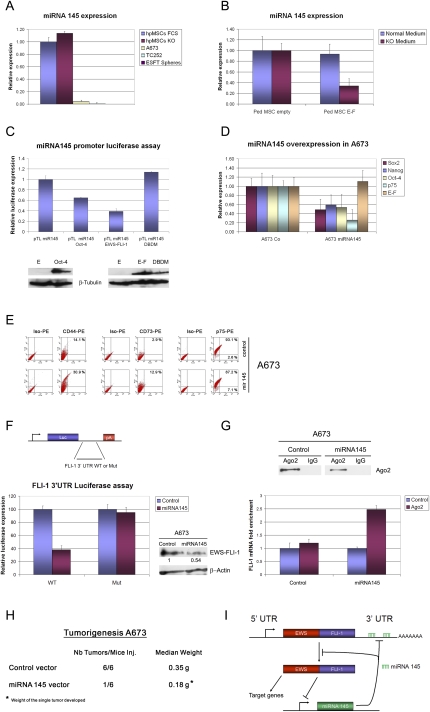
Enhanced induction of OCT4, NANOG, and SOX2 in hpMSCEWS-FLI-1s cultured in KO medium could not be explained by a mere difference in EWS-FLI-1 protein expression levels, since they were found to be equal in both culture conditions (Fig. 1D). We therefore addressed the possible existence of a regulatory mechanism that coordinately modulates expression of these genes in hpMSCEWS-FLI-1s grown in the absence of serum. A recently reported regulator of OCT4 and SOX2 expression in human ESCs is miRNA145 (Chivukula and Mendell 2009; Xu et al. 2009). miRNA145 expression is low to absent in undifferentiated human ESCs and progressively increases during their differentiation (Chivukula and Mendell 2009; Xu et al. 2009). miRNA145 itself is reported to act directly on the 3′ untranslated regions (UTRs) of OCT4 and SOX2, resulting in repression of both genes, thereby favoring the transition from ESCs to embryonic bodies (Xu et al. 2009). To assess the putative involvement of miRNA145 in the stronger OCT4, SOX2, and NANOG induction observed in hpMSCEWS-FLI-1s grown in the absence of serum, we compared miRNA145 expression in wild-type hpMSCs, cultured in the presence or absence of serum, ESFT A673 and TC252 cell lines, and primary ESFT CSC sphere cultures. Expression of miRNA145 was found to be 95%–99% lower in the ESFT cell lines A673 and TC252, respectively, than in hpMSCs, whereas virtually no expression was detected in primary ESFT CSC sphere cultures (Fig. 5A). Given that hMSCs are candidate ESFT cells of origin (Riggi et al. 2008), we addressed the possibility that the marked difference in miRNA145 expression observed between wild-type hpMSCs and ESFT cells could be due to the presence of the EWS-FLI-1 fusion protein in the latter. hpMSCs were grown in the presence or absence of serum, and the expression level of miRNA145 was compared between EWS-FLI-1-expressing and empty vector-infected cells. Remarkably, introduction of EWS-FLI-1 resulted in significant repression of miRNA145 transcript levels in hpMSC cultured in KO but not serum-supplemented medium (Fig. 5B), suggesting that the stronger induction of OCT4, SOX2, and NANOG observed in hpMSCEWS-FLI-1s grown in the absence of serum may result from repressive activity of EWS-FLI-1 on the miRNA145 promoter.
Figure 5.
miRNA145 and EWS-FLI-1 exert a reciprocal control mechanism on their mutual expression, as well as on their common target genes, SOX2 and OCT-4, which regulate ESFT cell differentiation and tumorigenicity. (A) Real-time PCR comparison of miRNA145 expression in hpMSC, A673, and TC252 ESFT cell lines and primary ESFT spheres. (B) EWS-FLI-1 expression in hpMSCs cultured in KO but not serum-containing conditions causes a significant decrease in miRNA145 expression, as assessed by real-time PCR analysis. (C, top panel) miRNA145 promoter luciferase assay reveals that wild-type EWS-FLI-1 but not its DBDM represses miRNA145 promoter activity. Oct-4 expression was used as a positive control. (Bottom panel) Western blot analysis in the corresponding cellular lysates of EWS-FLI-1, DBDM, and Oct-4 protein expression. (D,E) Retroviral-mediated overexpression of miRNA145 in A673 cells resulted in a significant decrease in SOX2, NANOG, and OCT4 expression levels, compared with their empty vector-infected counterparts (D), and a concomitant increase in the mesenchymal markers CD44 and CD73 on the surface of these cells (E), as assessed by real-time PCR and FACS analysis, respectively. (F, left panels) miRNA145 specifically represses its target gene, FLI-1, in a luciferase assay performed in HEK-293 cells engineered to express a luciferase reporter vector containing the human wild-type FLI-1 3′UTR. No repression in luciferase expression was observed when a mutant form (Mut) of the FLI-1 3′UTR harboring a deletion of miRNA145-binding sites was used. Error bars represent the SD from three independent experiments. (Right panel) miRNA145 expression in A673 cells results in a significant reduction of EWS-FLI-1 protein levels at 72 h post-infection. (G) Endogenous Ago2 protein (top panel) and EWS-FLI-1 mRNA (bottom panel) show significant enrichment in Ago2 immunoprecipitated material derived from cellular extracts of miRNA145-expressing but not empty vector-infected A673 cells, as assessed by Western blot and real-time PCR analysis, respectively. (H) miRNA145 overexpression in A673 cells causes a striking reduction in tumor growth, compared with cells infected with the corresponding empty vector. (I) Schematic representation of the reciprocal negative regulatory loop between EWS-FLI-1 and miRNA145. Real-time PCR experiments were normalized to either cyclophyllin A or Rnu6b, and done in triplicate. Error bars represent the SD of three independent determinations.
miRNA145 expression in human ESCs is tightly regulated in vivo through direct binding and repression of its promoter by OCT-4 (Xu et al. 2009). This negative feedback loop allows maintenance of the high OCT4 and, probably, SOX2 expression levels required to maintain ESC in an undifferentiated state, a mechanism designated recently as “repression of the repressor” (Xu et al. 2009). To determine whether a similar mechanism might allow EWS-FLI-1 to directly repress miRNA145 expression in hpMSCs, we subjected the putative miRNA145 promoter sequence to bioinformatics analysis using the TFSEARCH tool (http://www.cbrc.jp/research/db/TFSEARCH.html). Four putative ETS-binding motifs in the selected genomic region were found. A luciferase reporter assay was then performed using 293T cells engineered to express luciferase driven by the miRNA145 promoter as described previously (Xu et al. 2009). Introduction of EWS-FLI-1 into these cells resulted in 60% reduction of luciferase expression, consistent with the ability of EWS-FLI-1 to repress miRNA145 promoter activity, whereas introduction of Oct-4 reduced expression by <40%. No effect was observed when the EWS-FLI-1 DBDM was introduced into the same cells (Fig. 5C).
To determine its role in ESFTs, miRNA145 was overexpressed in A673 and TC252 cells using a retroviral system (Supplemental Data 3B), and the effect on their gene expression profile, differentiation status, and tumorigenic potential was assessed. In A673 cells, miRNA145 overexpression resulted in significant reduction of SOX2, OCT4, and NANOG transcript levels, with concomitant induction of the mesenchymal markers CD44 and CD73, as assessed by real-time PCR and flow cytometry (Fig. 5D,E). In contrast, down-regulation of these stem cell genes was less significant in TC252 cells and was not associated with any changes in their mesenchymal marker profile (Supplemental Data 3C; data not shown), conceivably due to their lower baseline miRNA145 expression.
Several recent studies have reported the ability of miRNAs to act through a negative regulatory feedback loop by binding to the 3′UTR of the genes that modulate their own expression (Ordonez et al. 2009). We therefore assessed the presence of putative miRNA145-binding sites within the wild-type FLI-1 3′UTR sequence, using the TargetScan miRNA target prediction software. Three highly and one poorly conserved putative miRNA145-binding sites were identified (UTR nucleotide positions 88–94, 473–479, 513–519, and 265–271, respectively). (For further details, see the Materials and Methods section and TargetScan analysis at http://www.targetscan.org/cgi-bin/targetscan/vert_50/view_gene.cgi?taxid=9606&gs=FLI1.)
To determine whether FLI-1 could be directly targeted by miRNA145, we engineered the PGL3 luciferase reporter vector to contain either the human wild-type full-length human FLI-1 3′UTR or its mutant form harboring a 6-base deletion in the four putative miRNA145 target sites (Fig. 5F, top panel). In HEK293 cells, cotransfection of the luciferase reporters with a retroviral vector expressing the miRNA145 resulted in a 60% reduction in luciferase expression from the wild-type FLI-1 3′UTR reporter. In contrast, no significant reduction in luciferase activity was observed from the mutated reporter compared with control cells transfected with an empty vector (Fig. 5F, bottom panel). No difference in EWS-FLI-1 transcript levels was detected by real-time PCR in A673 cells after miRNA145 overexpression (Fig. 5D), suggesting that the effect of miRNA145 on EWS-FLI-1 expression may occur at the translational level. EWS-FLI-1 protein levels were therefore assessed by Western blot analysis in A673 cells 72 h post-infection with an empty vector or a miRNA145-expressing retroviral vector. Significant reduction of the EWS-FLI-1 protein level was observed upon expression of miRNA145 (Fig. 5F), consistent with the ability of miRNAs to repress their target gene expression through either mRNA degradation or translation inhibition mechanisms.
To rigorously probe for miRNA145 pairing to EWS-FLI-1 mRNA, Argonaute2 (Ago2), a component of the RISC complex, was immunoprecipitated, along with isotype-matched control antibody, from extracts of A673 cells infected with either empty vector or miRNA145-expressing retroviral vector. The immunoprecipitated material was analyzed by Western blot and real-time PCR to detect enrichment in Ago2 protein and FLI-1 mRNA, respectively, as described previously (Ordonez et al. 2009). Endogenous FLI-1 mRNA was significantly enriched in the Ago2 immunoprecipitated fraction derived from the A673 cells expressing miRNA145, compared with the control empty vector-infected cells (Fig. 5G), confirming direct interaction between miRNA145 and the EWS-FLI-1 3′UTR in vivo.
Finally, we assessed the tumorigenicity of A673 cells overexpressing miRNA145 and found a striking reduction in their tumor-forming capacity following subcutaneous injection into NOD-SCID mice (Fig. 5H), a result that could be explained, at least in part, by miRNA145 repression of EWS-FLI-1 protein expression.
Taken together, these results demonstrate the existence of a direct regulatory loop between miRNA145 and EWS-FLI-1 that may allow the two partners to exert a reciprocal control mechanism on their mutual expression, as well as on the fusion protein target gene repertoire during the initial steps of ESFT development (Fig. 5I).
SOX2 plays a critical role in ESFT differentiation and pathogenesis
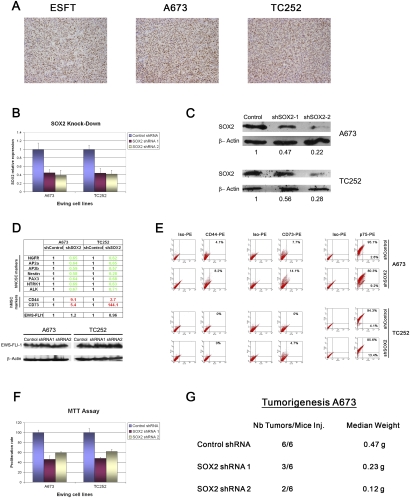
Although induction of a mesenchymal gene expression profile in A673 cells expressing miRNA145, along with reduction of their tumorigenic potential, could be explained by the decrease in EWS-FLI-1 protein levels, it is conceivable that one or several miRNA145 target genes may also contribute to the observed phenotypic changes. Among genes found to be modulated by miRNA145 in ESFT cells, SOX2, a core reprogramming factor and EWS-FLI-1 target gene, was induced more than a thousand-fold during the emergence of a NCSC phenotype in hpMSCEWS-FLI-1s. SOX2 plays a key role in CNS development, neuronal differentiation, and neural stem cell maintenance (Graham et al. 2003; Ellis et al. 2004; Fong et al. 2008), in addition to genetic reprogramming of terminally differentiated cells into hiPSs. Immunohistochemical analysis revealed elevated Sox2 expression in a subpopulation of primary ESFT cells, and a similar Sox2 expression pattern was observed in tumors derived from subcutaneous injection into immunocompromised mice of the A673 and TC252 cell lines (Fig. 6A). Therefore, based on the notions that Sox2 fulfils a role in both neural stem cell maintenance and genetic induction of pluripotency, that ESFTs display features of NCSCs and MSCs, and that primary hNCSCs have the ability to give rise to MSCs, we addressed the possibility that Sox2 might be functionally implicated in the reduced tumorigenic potential as well as the differentiation changes along the neuroectodermal–mesenchymal axis observed in A673 cells upon miRNA145 expression. To this end, we selected two ESFT cell lines that are representative of the two extremities of the ESFT differentiation spectrum: A673, which displays a partial mesenchymal phenotype with 5%–30% CD44+, 4%–20% CD73+, and 0%–4% CD133+ cells; and TC252, which displays a more pronounced neural phenotype with 0% CD44+ or CD73+ cells and almost 100% CD133+ cells (data not shown). Consistent with their tendency toward neuroectodermal differentiation, TC252 cells were found to express a fivefold higher level of SOX2 than A673 cells (Supplemental Data 1C). To address the potential involvement of Sox2 in ESFT cell fate determination, SOX2 expression was decreased in both cell lines using two distinct SOX2-specific shRNAs, and corresponding changes in NCSC and MSC differentiation marker expression were assessed. In response to the 50%–70% reduction of SOX2 transcript and protein levels that was obtained (Fig. 6B,C), expression of all of the NCSC markers tested—including NGFR/p75, AP2A, AP2B, NESTIN, PAX3, NTRK1, and ALK—underwent a significant decrease, as assessed by real-time PCR (Fig. 6D). In contrast, expression of the MSC markers CD44 and CD73 was strongly enhanced (Fig. 6D). Importantly, no difference in EWS-FLI-1 mRNA and protein expression levels was noted between control and SOX2-depleted cells, confirming that, in this context, the observed induction of mesenchymal markers could not result from a concomitant decrease in EWS-FLI-1 protein expression, as observed upon miRNA145 expression or shRNA-mediated EWS-FLI-1 depletion (Fig. 6D; Tirode et al. 2007). The increase in CD44 and CD73 and the decrease in NGFR/p75 transcript levels in A673 cells were further confirmed by flow cytometry (Fig. 6E), with CD44+ and CD73+ cells rising from 4.1% to 8.2% and 7.7% to 14.1%, respectively, and NGFR/p75+ cells decreasing from 95.1% to 80.3%. The same trend was observed in TC252 cells, where SOX2 depletion resulted in NGFR/p75 expression decrease and the concomitant appearance of a CD73+ cell subpopulation (Fig. 6E). The absence of CD44 expression in TC252 may be linked to the significantly higher baseline SOX2 expression level than in A673 cells, and is consistent with the possibility that the partial SOX2 depletion by shRNA may have been insufficient for its induction. Induction of SOX2 by EWS-FLI-1 expression in hpMSCs may therefore play a critical role in the generation of the observed NCSC profile. Finally, the relationship between the observed differentiation changes and the proliferative and tumorigenic properties of the two cell lines was assessed. Depletion of SOX2 resulted in a comparable decrease in proliferation of both cell lines (Fig. 6F). More importantly, tumorigenicity of both cell lines was reduced (Fig. 6G; Supplemental Data 3A), although not as strongly as by miRNA145 overexpression. Taken together, these data suggest that SOX2 may be a EWS-FLI-1 target gene acting downstream from the fusion protein to regulate ESFT cell differentiation and tumorigenicity.
Figure 6.
SOX2 is directly involved in ESFT differentiation and pathogenesis. (A) Immunohistochemical assessment of SOX2 expression in primary ESFTs and cell line-derived tumors grown in immunocompromised mice. Magnification, 100×. (B,C) Real-time PCR (B) and Western blot analysis (C) of SOX2 expression following stable SOX2-specific shRNA expression in A673 and TC252 ESFT cell lines. The experiments were performed using two different SOX2-specific (SOX2 shRNA 1 and 2) sequences and an unrelated shRNA control. (D,E) Depletion of SOX2 in A673 and TC252 cells significantly decreased expression of numerous NCSC markers, but not of EWS-FLI-1 protein levels, and was accompanied by an increased expression of the hMSC markers CD44 and CD73, as assessed by real-time PCR (D, top panel), Western blot (D, bottom panel), and FACS analysis (E). (F) A673 and TC252 cell proliferation was significantly reduced after the stable knockdown of SOX2 shown in C. Means of triplicate values ± SD of three separate determinations are shown. (G) Depletion of SOX2 in A673 cells causes a striking reduction in tumor growth compared with cells expressing unrelated RNAi sequences. Real-time PCR experiments were normalized to cyclophyllin A and done in triplicate. Error bars represent the SD of three independent determinations.
Discussion
Consistent with the notion that ESFTs are primarily a pediatric and adolescent malignancy, pediatric hMSCs infected with EWS-FLI-1-containing retroviral vectors displayed a quantitatively and qualitatively different response than adult MSCs to comparable expression levels of EWS-FLI-1. In both pediatric and adult MSCs, EWS-FLI-1 induced a gene expression repertoire highly reminiscent of that of primary Ewing sarcoma. However, many of the genes induced in adult MSCs and believed to be implicated in ESFT pathogenesis were more strongly induced in hpMSCs. More importantly, numerous genes that are among the most prominent ESFT markers were induced in hpMSCEWS-FLI-1s but not in their adult counterparts.
Identification of novel candidate EWS-FLI-1 target genes in pediatric MSCs
EWS-FLI1-induced transcriptome changes were far more pronounced when the cells were cultured in KO medium according to the protocol used to reprogram human primary cells into iPSs. Not only was the level of induction of ESFT-relevant genes higher, but several genes that were not significantly induced in cells cultured in standard medium were strongly up-regulated in cells cultured in KO medium. Several of these genes—including ALK, NTRK1, PBX1, and MEIS1 (Argiropoulos et al. 2007; Janoueix-Lerosey et al. 2008)—are implicated in transformation and tumorigenesis, and may bear relevance to ESFT pathogenesis.
The anaplastic lymphoma kinase (ALK) is reported to be expressed in ESFTs (Dirks et al. 2002), and has been shown recently to be implicated in genetic predisposition to neuroblastoma development (Mosse et al. 2008). Similar to ALK, the nerve growth factor receptors p75/NGFR and NTRK1 are expressed in ESFTs in addition to a variety of human cancers, including prostate, pancreas, and breast carcinoma, as well as gliomas (Reichardt 2006). In breast cancer, NTRK1 is reported to promote growth, migration, invasion, and metastasis (Lagadec et al. 2009), and its overexpression alone is observed to result in constitutive induction of its tyrosine kinase activity and engagement of the downstream PI3K–Akt and MAP kinase signaling pathways (Lagadec et al. 2009). It is noteworthy that neither NTRK1 nor ALK expression was detectable in wild-type hpMSCs. Their induction by EWS-FLI-1 and expression in primary ESFT may, along with IGF1R engagement by IGF-1, be responsible for the observed activation of PI3K–Akt and MAP kinase pathways in ESFT. Therapeutic targeting of IGF-1R with monoclonal antibodies has shown promise for the control of ESFT growth in some patients, but resistance to IGF-1R blockade is observed, possibly because of maintenance of PI3K–Akt and MAP kinase pathway activity by other receptor tyrosine kinases (RTKs). ALK, NGFR/p75, and NTRK1 provide plausible candidate RTKs whose signaling could help bypass the requirement for IGF1 by ESFT cells. Their inhibition may therefore have potential therapeutic relevance as well as feasibility, in view of the recent emergence of selective kinase inhibitors as an important class of anti-cancer drugs (for review, see Janne et al. 2009; Montagut and Settleman 2009).
Genetic reprogramming of hpMSCs by EWS-FLI-1 toward ESFT CSCs and miRNA145 modulation
Culture of hpMSCs expressing EWS-FLI-1 in the appropriate KO medium allowed not only identification of hitherto unrecognized candidate EWS-FLI-1 target genes, but resulted in the generation of a cellular subpopulation recapitulating all of the major in vitro features of ESFT CSCs. These include cell surface CD133 expression, acquisition of asymmetric division capacity, and expression of a panel of genes involved in stem cell maintenance, cellular reprogramming, and transformation. Interestingly, the emergence of this subpopulation was preceded by the robust induction of the core reprogramming genes OCT-4, NANOG, and SOX2, driven, in part, by the EWS-FLI-1-mediated miRNA145 repression observed when hpMSCEWS-FLI-1s are grown in the appropriate microenvironment.
miRNAs represent a class of small (20- to 25-nucleotide [nt]) noncoding RNAs that are key regulators of numerous cellular events, including the balance between proliferation and differentiation during tumorigenesis and development. They generally inhibit target mRNAs by repressing translation or reducing mRNA stability. Among miRNAs that play a major role in supporting stem cell properties, miRNA145 is of particular interest because its expression has been shown recently to inhibit human ESC pluripotency and self-renewal and to favor lineage-restricted differentiation through repression of SOX2 and OCT4. In hpMSCs, SOX2 may constitute both a direct and an indirect EWS-FLI-1 target gene whose induction, along with that of the other core reprogramming factors OCT4 and NANOG in hpMSCEWS-FLI-1s grown in KO medium, appears to be fine-tuned by the suppressive activity of EWS-FLI-1 on the miRNA145 promoter. Thus, modulation of miRNA expression provides an attractive link between stem cell maintenance, induced pluripotency, and CSC biology. It is tempting to speculate that the appearance of a subpopulation of hpMSCEWS-FLI-1s bearing an ESFT CSC phenotype observed in the present study may be a consequence of the synergy between the direct action of EWS-FLI-1 on stem cell genes and on miRNA-145. Only within the appropriate microenvironment, represented by KO culture medium here, can the expression level of the target reprogramming genes attain the putative threshold required to initiate genetic reprogramming of hpMSCs into ESFT CSCs.
Identification of several miRNA145 target sites in the 3′UTR of the human FLI-1 transcript suggests that miRNA145, in addition to modulating OCT4, NANOG, and SOX2 expression levels after their initial induction by EWS-FLI-1, may be further involved in a feedback regulatory loop controlling EWS-FLI-1 expression itself. Our experiments confirmed the existence of such a reciprocal control mechanism, which seems to provide tight regulation of expression of both partners as well as that of their common target genes. The observation that EWS-FLI-1 constitutes both a miRNA145 transcriptional regulator and one of its direct targets offers new insight into ESFT initiation mechanisms. The requirement for an appropriate culture microenvironment that allows EWS-FLI-1 to exert its repressing activity on the miRNA145 promoter indicates a link between EWS-FLI-1, miRNA145, and the permissiveness of cells from which ESFTs originate. Thus, it is conceivable that initial expression of the fusion protein during the very early steps of ESFT development requires an environment that allows miRNA145 repression, resulting in stabilization of the EWS-FLI-1 protein and in enhancement of OCT4, NANOG, and SOX2 expression. These events could endow the ESFT cell of origin with CSC properties and ultimately result in the conversion of cellular permissiveness for oncogene expression to tumor-initiating ability.
SOX2 involvement in ESFT differentiation and tumorigenesis
The changes in tumorigenic potential and differentiation observed upon miRNA145 expression in ESFT cells may be partially explained by the concomitant reduction in EWS-FLI-1 protein levels. However, changes in expression of one or several EWS-FLI-1/miRNA145 target genes could also be involved in the appearance of the observed phenotype.
Based on its physiological role in neural stem cell maintenance and differentiation (Graham et al. 2003; Ellis et al. 2004; Fong et al. 2008), its participation in genetic reprogramming (Jaenisch and Young 2008), and its induction by EWS-FLI-1, Sox-2 provides an attractive candidate to explain how expression of EWS-FLI-1 might induce a partial neuroectodermal phenotype in a tumor arising from transformed hpMSCs. Consistent with this notion, even suboptimal depletion of SOX2 in two distinct ESFT cell lines that represent the two extremities of the Ewing sarcoma differentiation spectrum attenuated their neuroectodermal phenotype with a concomitant enhancement of mesenchymal markers. Interestingly, SOX2 was also observed to participate in ESFT cell proliferation and tumorigenesis, suggesting that, among candidate EWS-FLI-1 target genes, it may play a central role in ESFT pathogenesis.
Sox-2 has been identified recently as a lineage preservation oncogene that is able to promote squamous identity in lung and esophageal squamous cell carcinomas (Bass et al. 2009), but the mechanism whereby it may promote tumorigenicity in ESFT cells is unclear. It should be noted that Sox-2 depletion did not have as strong an effect on abrogation of ESFT cell tumorigenicity as did overexpression of miRNA-145, consistent with the notion that other genes subject to miRNA-145 control may be implicated as well. However, the ability of Sox-2 to control differentiation may help generate a window of opportunity for the activity of some of the survival and growth-promoting genes induced by EWS-FLI-1. Numerous candidate EWS-FLI-1 target genes have been proposed to participate in primary cell transformation and maintenance of the resulting tumor cells, but which of these genes are essential for ESFT pathogenesis remains unclear. A plausible scenario is that EWS-FLI-1-mediated transformation requires cooperation between several EWS-FLI-1 target genes that can occur only in a permissive cellular context. hpMSCs can provide the microenvironment required for EWS-FLI-1-mediated repression of miRNA-145 and induction of SOX2, OCT4, and other reprogramming genes whose combined activity may ensure maintenance of stem cell properties that facilitate subsequent generation of CSCs. Concomitant induction of the polycomb group gene product EZH2 may abolish the response to oncogenic stress triggered by overexpression of c-Myc, Alk, or Ntrk1, whereas IGF-1 and possibly other growth factors may secure cell survival. The combined effect of these target gene products may provide a reasonable basis for MSC transformation toward ESFT CSCs.
Taken together, our observations indicate that, in the appropriate MSC microenvironment, a single aberrant transcription factor generated by a reciprocal chromosomal translocation is able to induce genetic reprogramming toward a CSC phenotype by combined action on its target genes and by directly modulating miRNA-145 expression. To the best of our knowledge, this is the first report of a mechanism whereby expression of a single oncogene in a primary cell population generates the features of a CSC.
Materials and methods
Cell culture
hpMSCs were obtained from femoral head bone marrow of four pediatric patients undergoing limb axis correction surgery as described previously (Suva et al. 2004, 2008; Riggi et al. 2008). MSCs were cultured at low confluence in IMDM, 10% FCS, and 10 ng/mL PDGF-BB (PeProtechEC), and were tested for multilineage differentiation into adipocytes, chondrocytes, and osteoblasts (Riggi et al. 2008). TC252 (kindly provided by Dr. T. Triche) and A673 cell lines (American Type Culture Collection) were cultured in RPMI 1640 containing 10% FBS.
Cloning and RT–PCR
Retroviral vectors containing the V5-tagged cDNA clone encoding the human EWS-FLI1 type 2 fusion and the EWS-FLI-1 R340N DBDM were described previously (Riggi et al. 2008). Oct-4 cDNA was amplified from hpMSCs using SuperScript one-step RT–PCR with the platinum Taq kit (Invitrogen). The amplified fragment was digested with BglII and XhoI and inserted into a pMSCV Puro retroviral vector (BD Biosciences Clontech). Primer sequences for Oct-4 were forward BglII, GAAGATCTGCCACCATGGCGGGACACCTGGCTT, and reverse XhoI, CCGCTCGAGCGGTCAGTTTGAATGCATGGGAGAGCCCA.
Retroviral infection
Expression of EWS-FLI-1V5 and DBDM in hMSCs was achieved using Retroviral Gene Transfer and Expression (BD Biosciences Clontech), according to the manufacturer's recommendations. Expression of the fusion genes and corresponding proteins were tested at each time point in all of the batches of cells by RT–PCR and Western blot analysis, respectively, using the mouse anti-V5 antibody. Infected cells were selected with 0.75 μg/mL puromycin for 5 d, and the bulk of the resistant cells was used in subsequent experiments. miRNA overexpression was performed using HEK-293T cells. Cells were cotransfected with p-hTR or miR145 (kindly provided by Dr. R. Agami), along with VSV-G and the packaging vector, using FuGene6 (Roche) according to the manufacturer's recommendations. Supernatants of transfected HEK-293T cells were collected after 72 h, filtered, and used to infect target cells in the presence of polybrene (10 μg/mL). Infected cells were selected with 1 μg/mL blasticidin for 5 d and used for further experiments.
Western blot
Cell lysis, SDS-PAGE, blotting, and immunostaining were performed by standard procedures, and protein bands were detected using a chemoluminescent substrate kit (Pierce) according to the manufacturer's recommendations. Primary mouse anti-V5 epitope (Invitrogen), mouse anti-FLI-1 (Calbiochem; Merck), mouse anti-βactin (Sigma), mouse anti-Oct-4 (Santa Cruz Biotechnologies), and mouse anti-α-tubulin (Calbiochem) monoclonal antibodies and rabbit anti-SOX2 polyclonal antibody (Millipore; Chemicon) were used. Secondary antibodies were HRP-conjugated goat anti-mouse (Bio-Rad) and goat anti-rabbit immunoglobulin (Dako).
Reprogramming protocol
hpMSCs were cultured in standard FCS-containing medium and infected following the standard retroviral protocol. Forty-eight hours after the retroviral infection, hpMSCs were washed and incubated in KO medium with or without addition of the chemicals PD0325901, CHIR99021, and A8301 (Stemgent). KO medium was composed of IMDM, 20% KO Serum Replacement (Invitrogen, GIBCO), 10 ng/mL PDGF-BB, 1% penicillin–streptomycin, and 10 ng/mL human recombinant leukemia inhibitory factor (LIF) (Millipore; Chemicon). For clonogenic assays, PD0325901 (0.5 μM), CHIR99021 (3 μM), and A8301 (0.25 μM) were added to the KO medium.
Magnetic cell sorting and FACS analysis
Single MSC suspensions were incubated with CD133 microbeads (Miltenyi) and separated using an autoMACS device (Miltenyi) according to the manufacturer's instructions. Prior to and following separation, samples were analyzed by flow cytometry using CD133/2-PE (293C3, 1/10) antibody (Miltenyi) and isotype control mouse IgG2b-PE (1/10) (Miltenyi) in a FACScalibur apparatus (Becton Dickinson). Further flow cytometry was performed using rat anti-human CD44-PE (eBioscience), mouse anti-human CD73-PE (BD Pharmingen), mouse anti-human NGFR/p75 (Miltenyi), and isotype control rat IgG2b-PE (BD Pharmingen) and mouse IgG2b-PE antibodies (BD Pharmingen).
Affymetrix GeneChip arrays analysis and statistical analysis of the expression data
Total RNA was extracted 12 d after infection and selection, from each of the EWS-FLI-1 and vector-containing hMSC batches, using the RNeasy Mini Kit (Qiagen) according to the manufacturer's recommendations. RNA quality was verified by an Agilent RNA 600 nanoassay and by measuring the 260/280 absorbance ratio. Quality-tested total RNA was used by the Lausanne DNA Array Facility (DAFL) to perform gene expression profile analysis on Affymetrix HG-U133 Plus 2.0 Arrays following the manufacturer's recommendations (http://www.unil.ch/dafl). For each of the four freshly isolated batches of hMSCs, Affymetrix analysis was performed on EWS-FLI-1V5-expressing cells and on empty vector control cells. Gene expression data were obtained with the RMA method (Irizarry et al. 2003) using the Affymetrix Power Tools suite. By comparing each EWS-FLI-1V5-expressing batch with the same batch transfected with the empty vector, we obtained, for each probe set in the chip, four measurements of the expression ratio. Differentially expressed genes were then identified with the rank product method (Breitling et al. 2004) as implemented in the “RankProd” Bioconductor package (Hong et al. 2006). Probe sets identified as differentially expressed with a false discovery rate (FDR) of 1% were retained for further analysis. For cells cultured in KO medium, probe sets with an expression fold change >2 or <0.5 were considered as differentially expressed. To compare our differentially expressed genes with the ones identified in other studies, we first used the annotation files provided by Affymetrix (version 29) to translate the lists of probe sets into lists of Entrez gene IDs. Such lists were used to compare our results with the ones in the published databases (Staege et al. 2004; Baird et al. 2005), translated when necessary into Entrez gene IDs using the GenBank accession numbers reported by Staege et al. (2004) and Baird et al. (2005). Statistical significance of the overlap between lists of genes was evaluated with a two-sided Fisher test and Bonferroni correction for multiple testing.
Real-time quantitative RT–PCR
cDNA was obtained using M-MLV reverse transcriptase and RNase H minus (Promega). Typically, 500 ng of template total RNA and 250 ng of random hexamers were used per reaction. Real-time PCR amplification was performed using a TaqMan Universal PCR mastermix and Assays-On-Demand gene expression products or Power SYBR mix and specific PCR primers in an ABI Prism 7900 instrument (Applied Biosystems). Relative quantification of each target, normalized to an endogenous control (Cyclophyllin A), was performed using a comparative Ct method (Applied Biosystems). Probes used included human IGF1, NKX2-2, GRP, NPY1R, EZH2, SOX2, NTRK1, ALK, NGFR/p75, OCT-4, NANOG, CD133, CCK, MEIS1, PBX1, CD73, and CD44 (Assays-On-Demand gene expression). SYBR Green gene expression quantification for EWS-FLI-1, AP2a, AP2b, NESTIN, PAX3, RET, FOXD3, POUF4F1, TWIST, EOMES, DCX, CTCFL, LEF1, and LIPI was achieved using published primer sequences (Peter et al. 2001; Thomas et al. 2008). SYBR Green primer sequences for the quantification of human p53, Rb, p16, and p14 transcripts were as follows: p53: fwd, 5′-CCCAGCCAAAGAAGAAACCA-3′, and rev, 5′-TTCCAAGGCCTCATTCAGCT-3′; Rb: fwd, 5′-GAGCTTGGTTAACTTGGGAGAA-3′, and rev, 5′-CAGATTCCCCACAGTTCCTTT-3′; p14: fwd, 5′-GGCCCTCGTGCTGATGCTAC-3′, and rev, 5′-TGGAGCAGCAGCAGCTCCGC-3′; p16: fwd, 5′-CACCGAATAGTTACGGTCGG-3′, and rev, 5′-GCACGGGTCGGGTGAGAGTG-3′. For miRNA quantification, total RNA containing miRNAs was extracted using a miRNAeasy kit (Qiagen) according to the manufacturer's instructions. miRNA was amplified using a miRNA reverse transcription kit (Applied Biosystems). RT–PCR amplification was performed as described and was normalized using RNU6B as an endogenous control. Probes included RNU6B and hsa-miR-145 (Applied Biosystems).
Clonogenic assay
Empty vector-infected or EWS-FLI-1-expressing hpMSCs were plated as single cells in four 96-well plates and cultivated for 30 d in standard FCS-containing or stem cell KO medium. Sphere formation was scored 30 d later.
SOX2 knockdown and in vitro MTT assay
The sequence of the 19-nt shRNA 1 targeting the human SOX2 sequence was described previously as sequence A (Fong et al. 2008); the sequence of shRNA 2 was 5′-AGACTAGGACTGAGAGAAA-3′. Sense and antisense oligonucleotides were annealed to form duplexes and were inserted into the pSIREN-Retro Q retroviral vector (BD Biosciences Clontech) according to the manufacturer's recommendations. Briefly, plasmids containing SOX2 or control shRNA were transfected into GP2 packaging cells to produce the virus used to infect the TC252 and A673 target cell lines. Infected TC252 and A673 cells were selected for 5 d with 1 μg/mL and 2 μg/mL puromycin, respectively, and the level of SOX2 knockdown was assessed by real-time quantitative PCR and Western blot analysis. Densitometric analysis was performed using the public domain image processing and analysis program NIH Image J (http://rsb.info.nih.gov/ij). For the MTT assay, TC252 and A673 cells infected with pSIREN containing SOX2 or control shRNA were incubated in 24-well plates, and their proliferation was tested with the CellTiter Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer's recommendations.
Luciferase reporter assay
A luciferase reporter assay was performed as described previously with some modifications (Cironi et al. 2008). Briefly, for the miRNA145 promoter assay, HEK-293T cells were cotransfected with empty pTransluc (kindly provided by Dr. K.S. Kosik) or pTransluc miR-145 promoter (purchased from Addgene) along with empty pMSCV puro, pMSCV EWS-FLI1, pMSCV EWS-FLI1 R340N DBDM, or pMSCV OCT-4 using FuGene 6 (Roche). For the assessment of miRNA145 binding to the FLI-1 3′UTR, a 600-base-pair (bp) PCR fragment of the human FLI-1 3′UTR was amplified and cloned into the XbaI site of the pGL3 control vector (Promega). A mutant form of this construct, harboring the deletion of the four miRNA145-binding sites, was obtained by site-directed mutagenesis (QuickChange kit; Stratagene), and HEK-293T cells were then cotransfected with either control or miR145 expression vectors. Forty-eight hours after transfection, luciferase activity was measured using the Luciferase Assay System (Promega) according to the manufacturer's recommendations. Normalization was performed as described previously (Cironi et al. 2008). All experiments were performed in triplicate.
Tumorigenicity assays
Six NOD/SCID IL2 receptor common γ chain knockout mice were anaesthetized and 2 × 106 empty vector-infected or EWS-FLI-1-expressing hpMSCs were injected under the renal capsule. All mice were sacrificed 3 mo later, and the kidneys were removed at autopsy for histological analysis. For the assessment of the tumorigenic potential of SOX2-depleted and miRNA145-overexpressing ESFT cell lines, six NOD/SCID mice were injected subcutaneously with 2 × 106 or 1 × 106 of control, shRNA-infected, or miRNA145-overexpressing TC252 or A673 cells, respectively, and the animals were sacrificed 3–4 wk after injection. All tumors were resected at autopsy and sectioned for histological analysis. Experimental protocols involving mice were approved by the Etat de Vaud, Service Vétérinaire, authorization number VD1477.2.
Immunohistochemistry
Paraffin-embedded sections of primary Ewing's sarcoma, TC252-, and A673-derived tumors were stained with rabbit anti-human SOX2 polyclonal antibody (1:1000 dilution; Millipore, Chemicon). For the detection of LIPI expression in hpMSCs, empty vector-infected or EWS-FLI-1-infected cells were selected with puromycin, seeded onto histological slides, fixed with PFA, and stained with mouse polyclonal anti-LIPI antibody (Abnova Corporation). Horseradish peroxidase staining was performed using biotin-conjugated goat anti-rabbit or rabbit anti-mouse Ig (Vector Laboratories), and was revealed with a DAKO DAB Kit (Dako).
Ago2 immunoprecipitation
Ago2 immunoprecipitation was performed as described previously with some modifications (EZH2 mol cell). Briefly, A673 were infected with either empty control hTR- or miR145-expressing vectors. After 72 h, cells were harvested and lysed, and the cell extracts were incubated with 2 μg of Ago2 (Wako Chemicals) or mouse IgG (Sigma) antibodies for 2 h. The immunoprecipitated complex was subsequently incubated with precleared protein-G sepharose beads for 1 h, after which beads were washed and total RNA was extracted using Trizol reagent. Real-time PCR was performed using the following primers: hFLI-1 Fwd, 5′-GGGCACAAACGATCAGTAAGA-3′, and hFLI1 Rev, 5′-GCTAGGCGACTGCTGGTC-3′, and normalized to RNA 18S (Assays-On-Demand gene expression; Applied Biosystems).
Acknowledgments
We thank Dr. Timothy Triche for kindly providing the TC252 cell line, Dr Reuven Agami for providing the miRNA145-expressing vector, Dr Kenneth S. Kosik for providing the vector expressing luciferase under the miRNA145 promoter, Professor André Kaelin for providing surgical samples, Professor Pierre Hoffmeyer for fruitful collaboration, and Carlo Fusco for insightful discussions. We are grateful to the Lausanne DNA Array Facility (DAFL) for their continued support and expertise. This work was supported by FNRS grant 3100A0-105833, Oncosuisse grant 02158, and a grant from NCCR Molecular Oncology to I.S.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1899710.
Supplemental material is available at http://www.genesdev.org.
References
- Argiropoulos B, Yung E, Humphries RK 2007. Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes & Dev 21: 2845–2849 [DOI] [PubMed] [Google Scholar]
- Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS 2005. Gene expression profiling of human sarcomas: Insights into sarcoma biology. Cancer Res 65: 9226–9235 [DOI] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes & Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. 2009. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 41: 1238–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P 2004. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Carrillo J, Garcia-Aragoncillo E, Azorin D, Agra N, Sastre A, Gonzalez-Mediero I, Garcia-Miguel P, Pestana A, Gallego S, Segura D, et al. 2007. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin Cancer Res 13: 2429–2440 [DOI] [PubMed] [Google Scholar]
- Chivukula RR, Mendell JT 2009. Abate and switch: miR-145 in stem cell differentiation. Cell 137: 606–608 [DOI] [PubMed] [Google Scholar]
- Cironi L, Riggi N, Provero P, Wolf N, Suva ML, Suva D, Kindler V, Stamenkovic I 2008. IGF1 is a common target gene of Ewing's sarcoma fusion proteins in mesenchymal progenitor cells. PLoS One 3: e2634. doi: 10.1371/journal.pone.0002634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM 2006. Cancer stem cells—Perspectives on current status and future directions: AACR Workshop on Cancer Stem Cells. Cancer Res 66: 9339–9344 [DOI] [PubMed] [Google Scholar]
- Dauphinot L, De Oliveira C, Melot T, Sevenet N, Thomas V, Weissman BE, Delattre O 2001. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2 and c-Myc expression. Oncogene 20: 3258–3265 [DOI] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. 1992. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359: 162–165 [DOI] [PubMed] [Google Scholar]
- Deneen B, Denny CT 2001. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene 20: 6731–6741 [DOI] [PubMed] [Google Scholar]
- Dirks WG, Fahnrich S, Lis Y, Becker E, MacLeod RA, Drexler HG 2002. Expression and functional analysis of the anaplastic lymphoma kinase (ALK) gene in tumor cell lines. Int J Cancer 100: 49–56 [DOI] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L 2004. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci 26: 148–165 [DOI] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC, Ng HH 2009. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4: 301–312 [DOI] [PubMed] [Google Scholar]
- Foell JL, Hesse M, Volkmer I, Schmiedel BJ, Neumann I, Staege MS 2008. Membrane-associated phospholipase A1 β (LIPI) is an Ewing tumour-associated cancer/testis antigen. Pediatr Blood Cancer 51: 228–234 [DOI] [PubMed] [Google Scholar]
- Fong H, Hohenstein KA, Donovan PJ 2008. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 26: 1931–1938 [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L 2003. SOX2 functions to maintain neural progenitor identity. Neuron 39: 749–765 [DOI] [PubMed] [Google Scholar]
- Helman LJ, Meltzer P 2003. Mechanisms of sarcoma development. Nat Rev Cancer 3: 685–694 [DOI] [PubMed] [Google Scholar]
- Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J 2006. RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22: 2825–2827 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janne PA, Gray N, Settleman J 2009. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov 8: 709–723 [DOI] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, et al. 2008. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455: 967–970 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadec C, Meignan S, Adriaenssens E, Foveau B, Vanhecke E, Romon R, Toillon RA, Oxombre B, Hondermarck H, Le Bourhis X 2009. TrkA overexpression enhances growth and metastasis of breast cancer cells. Oncogene 28: 1960–1970 [DOI] [PubMed] [Google Scholar]
- Lee J, Rhee BK, Bae GY, Han YM, Kim J 2005. Stimulation of Oct-4 activity by Ewing's sarcoma protein. Stem Cells 23: 738–751 [DOI] [PubMed] [Google Scholar]
- Lessnick SL, Dacwag CS, Golub TR 2002. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 1: 393–401 [DOI] [PubMed] [Google Scholar]
- Liu Y, Clem B, Zuba-Surma EK, El-Naggar S, Telang S, Jenson AB, Wang Y, Shao H, Ratajczak MZ, Chesney J, et al. 2009. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell 4: 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K 2008. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell 3: 595–605 [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagut C, Settleman J 2009. Targeting the RAF–MEK–ERK pathway in cancer therapy. Cancer Lett 283: 125–134 [DOI] [PubMed] [Google Scholar]
- Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, et al. 2008. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez JL, Osuna D, Herrero D, de Alava E, Madoz-Gurpide J 2009. Advances in Ewing's sarcoma research: Where are we now and what lies ahead? Cancer Res 69: 7140–7150 [DOI] [PubMed] [Google Scholar]
- Peter M, Gilbert E, Delattre O 2001. A multiplex real-time PCR assay for the detection of gene fusions observed in solid tumors. Lab Invest 81: 905–912 [DOI] [PubMed] [Google Scholar]
- Reichardt LF 2006. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci 361: 1545–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N, Stamenkovic I 2007. The biology of Ewing sarcoma. Cancer Lett 254: 1–10 [DOI] [PubMed] [Google Scholar]
- Riggi N, Cironi L, Provero P, Suva ML, Kaloulis K, Garcia-Echeverria C, Hoffmann F, Trumpp A, Stamenkovic I 2005. Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res 65: 11459–11468 [DOI] [PubMed] [Google Scholar]
- Riggi N, Cironi L, Suva ML, Stamenkovic I 2007. Sarcomas: Genetics, signalling, and cellular origins. Part 1: The fellowship of TET. J Pathol 213: 4–20 [DOI] [PubMed] [Google Scholar]
- Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, Joseph JM, Stehle JC, Baumer K, Kindler V, et al. 2008. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 68: 2176–2185 [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB 2003. Identification of a cancer stem cell in human brain tumors. Cancer Res 63: 5821–5828 [PubMed] [Google Scholar]
- Staege MS, Hutter C, Neumann I, Foja S, Hattenhorst UE, Hansen G, Afar D, Burdach SE 2004. DNA microarrays reveal relationship of Ewing family tumors to both endothelial and fetal neural crest-derived cells and define novel targets. Cancer Res 64: 8213–8221 [DOI] [PubMed] [Google Scholar]
- Suva D, Garavaglia G, Menetrey J, Chapuis B, Hoffmeyer P, Bernheim L, Kindler V 2004. Non-hematopoietic human bone marrow contains long-lasting, pluripotential mesenchymal stem cells. J Cell Physiol 198: 110–118 [DOI] [PubMed] [Google Scholar]
- Suva D, Passweg J, Arnaudeau S, Hoffmeyer P, Kindler V 2008. In vitro activated human T lymphocytes very efficiently attach to allogenic multipotent mesenchymal stromal cells and transmigrate under them. J Cell Physiol 214: 588–594 [DOI] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Stehle JC, Baumer K, Tercier S, Joseph JM, Suva D, Clement V, Provero P, Cironi L, et al. 2009. Identification of cancer stem cells in Ewing's sarcoma. Cancer Res 69: 1776–1781 [DOI] [PubMed] [Google Scholar]
- Takashima Y, Era T, Nakao K, Kondo S, Kasuga M, Smith AG, Nishikawa S 2007. Neuroepithelial cells supply an initial transient wave of MSC differentiation. Cell 129: 1377–1388 [DOI] [PubMed] [Google Scholar]
- Thomas S, Thomas M, Wincker P, Babarit C, Xu P, Speer MC, Munnich A, Lyonnet S, Vekemans M, Etchevers HC 2008. Human neural crest cells display molecular and phenotypic hallmarks of stem cells. Hum Mol Genet 17: 3411–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O 2007. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 11: 421–429 [DOI] [PubMed] [Google Scholar]
- Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, et al. 2007. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 1: 389–402 [DOI] [PubMed] [Google Scholar]
- Welford SM, Hebert SP, Deneen B, Arvand A, Denny CT 2001. DNA binding domain-independent pathways are involved in EWS/FLI1-mediated oncogenesis. J Biol Chem 276: 41977–41984 [DOI] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS 2009. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137: 647–658 [DOI] [PubMed] [Google Scholar]