Abstract
The loss of epithelial polarity is thought to play an important role during mammary tumor progression. Using a unique transgenic mouse model of ErbB2-induced mammary tumorigenesis, we demonstrated previously that amplification of ErbB2 is frequently accompanied by loss of the 14–3–3σ gene. Here, we demonstrate that ectopic expression of 14–3–3σ results in restoration of epithelial polarity in ErbB2-transformed mammary tumor cells. We further demonstrate that targeted deletion of 14–3–3σ within primary mammary epithelial cells increases their proliferative capacity and adversely affects their ability to form polarized structures. Finally, we show that 14–3–3σ can specifically form complexes with Par3, a protein that is essential for the maintenance of a polarized epithelial state. Taken together, these observations suggest that 14–3–3σ plays a critical role in retaining epithelial polarity.
Keywords: 14–3–3σ, ErbB2, conditional knockout, epithelial polarity
ErbB2 is a member of the epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases (RTKs). This family is comprised of four closely related type 1 RTKs: EGFR, ErbB2 (Neu, HER2), ErbB3 (HER3), and ErbB4 (HER4) (Hynes and Stern 1994). The importance of ErbB2 in primary human breast cancer is highlighted by the fact that 20%–30% of human breast cancers express elevated levels of ErbB2 due to genomic amplification of the erbB2 proto-oncogene (Slamon et al. 1987, 1989). Furthermore, its amplification and subsequent overexpression strongly correlate with a negative clinical prognosis in both lymph node-positive (Ravdin and Chamness 1995) and lymph node-negative (Andrulis et al. 1998) breast cancer patients. In an attempt to more closely mimic the events involved in ErbB2-induced mammary tumor progression, we derived and characterized transgenic mice that carry a Cre-inducible activated erbB2 under the transcriptional control of the endogenous erbB2 promoter (herein referred to as the ErbB2KI model) (Andrechek et al. 2000). We demonstrated that tumor progression in this strain was further associated with a dramatic elevation of both ErbB2 protein and transcript levels (Andrechek et al. 2000). Remarkably, the elevated expression of ErbB2 was further correlated with selective genomic amplification of the activated erbB2 allele (Montagna et al. 2002; Hodgson et al. 2005). Comparative genome hybridization (CGH) of the erbB2 amplicon suggests that the ErbB2KI model mimics ErbB2-initiated human breast cancer (Siegel et al. 1999).
In addition to erbB2 amplification, ErbB2KI tumors also demonstrated frequent deletions of chromosome 4 (Montagna et al. 2002), which encompasses 30 genes including the 14–3–3σ gene (Hodgson et al. 2005). Interestingly, loss of 14–3–3σ expression has been noted in a large proportion of primary human breast cancers (Ferguson et al. 2000; Vercoutter-Edouart et al. 2001). 14–3–3σ is a putative tumor suppressor that is transactivated by p53 in response to DNA damage (Hermeking et al. 1997). When up-regulated, 14–3–3σ induces S–G1 and G2–M cell cycle arrests (Chan et al. 1999; Laronga et al. 2000), and blocks Akt-mediated cell survival and proliferation (Yang et al. 2006). 14–3–3σ is also involved in the cytosolic sequestration of EGR2, a transcription factor that is critical for the up-regulation of erbB2 transcription (Dillon et al. 2007). Thus, ErbB2-induced tumors may ablate 14–3–3σ expression to prevent its inhibitory effects on erbB2 expression, although the role of 14–3–3σ in ErbB2-induced tumorigenesis remains unclear. Previous proteomic studies also indicate that 14–3–3σ is capable of associating with a number of polarity-regulating proteins (Benzinger et al. 2005), as well as molecules involved in mitotic translation machinery (Wilker et al. 2007).
Here, we further investigated the role of 14–3–3σ in mammary epithelial biology. We directly addressed the impact of restored 14–3–3σ expression on the growth of established ErbB2-transformed mammary tumor cell lines. Although re-expression of 14–3–3σ in these cells had little impact on tumor cell proliferation, it significantly decreased their invasive potential. This effect was further correlated with the restoration of both adherent and tight junction complexes. Conversely, inhibition of 14–3–3σ function by RNAi approaches in normal polarized epithelial cells resulted in the loss of epithelial polarity. To directly investigate the physiological relevance of these observations, we established transgenic mice that selectively delete the 14–3–3σ gene in the mammary epithelium. Loss of 14–3–3σ function was associated with an increase in the proliferative capacity of the mammary epithelium, and was correlated with a loss of epithelial polarity. We further demonstrate that 14–3–3σ is able to complex with the Par3 polarity protein. Taken together, these observations suggest that 14–3–3σ plays a critical role in the regulation of epithelial polarity through its interaction with Par3.
Results
Ectopic expression of 14–3–3σ in ErbB2-transformed mammary epithelial cells alters their metastatic and epithelial polarity properties
To examine the impact of 14–3–3σ restoration on ErbB2-induced tumor progression, we used an established breast cancer cell line, TM15, which is derived from mammary tumors of the ErbB2KI mouse model, carries amplified erbB2, and lacks 14–3–3σ expression (Dillon et al. 2007, 2009). TM15 cells were infected with retroviruses and ectopically expressing 14-3–3σ, 14–3–3σ mut, 14–3–3ζ, or the corresponding empty vector control (Supplemental Fig. S1A). 14–3–3σ mut is a truncated protein lacking the C-terminal 40 amino acids that constitute part of the ligand-binding domain of 14–3–3σ. A mouse model known as repeated epilation (Er) develops a skin differentiation defect due to germline expression of this mutant form of 14–3–3σ (Herron et al. 2005; Li et al. 2005). We first assessed whether expression of 14–3–3σ impacted the proliferative capacity of TM15 cells, both in vitro and in vivo. These studies revealed that re-expression of 14–3–3σ did not alter the growth of TM15 cells compared with empty vector controls (Supplemental Fig. S1B,C).
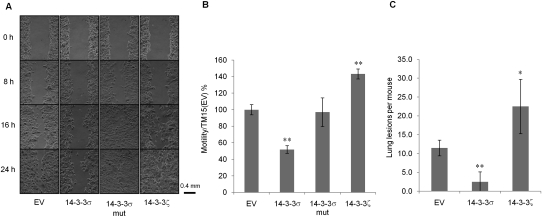
We next assessed whether the migratory behavior of tumor cells was affected as a consequence of ectopic expression of 14–3–3σ. We examined the motility of empty vector control, 14–3–3σ-, 14–3–3σ mut-, and 14–3–3ζ-expressing TM15 cells using Boyden chamber and wound healing assays. The results revealed that elevated expression of wild-type 14–3–3σ, but not the truncated 14–3–3σ, nor the closely related 14–3–3ζ isoform, resulted in a profound impairment of epithelial migration (Fig. 1A,B). To evaluate whether the observed migration defect was also associated with a reduced metastatic potential, TM15 cells were injected into mammary fat pads of nude mice, and the total tumor burden in the lungs was histologically evaluated. In contrast to empty vector control TM15 cells and 14–3–3ζ-expressing cells, the 14–3–3σ-expressing cells exhibited a dramatically impaired metastatic phenotype (Fig. 1C). Collectively, these data argue that restoration of 14–3–3σ function does not impact primary tumor growth, but significantly impairs ErbB2-induced breast cancer metastasis.
Figure 1.
Ectopic expression of 14–3–3σ in TM15 cells alters their migratory properties. (A) Wound healing assay of TM15 cell lines with empty vector (EV), 14–3–3σ-expressing, 14–3–3σ mut-expressing, and HA-14–3–3ζ-expressing. Pictures were taken at 0, 8, 16, and 24 h after scratching. (B) In vitro migration assay of the four aforementioned TM15 cell lines using transwell chambers. Results were normalized to TM15(EV) cells. Experiments were performed in triplicate. (C) Mammary metastatic lesions of nude mice after the injection of TM15 cell lines into their mammary fat pads. Each cell line was injected into four nude mice. Results significantly different from the control are indicated by asterisks: (*) P < 0.05; (**) P < 0.01.
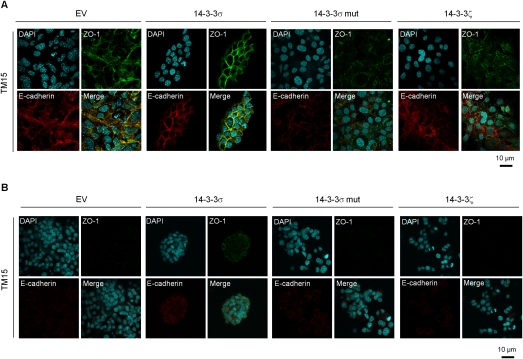
One possible explanation for these observations is that 14–3–3σ may affect the ability of tumor cells to form adherent and tight junction complexes. To study this, we cultured empty vector control, 14–3–3σ-, 14–3–3σ mut-, and 14–3–3ζ-expressing cells on plastic or in collagen and subjected them to immunostaining with E-cadherin and ZO-1 antisera as markers of adherens and tight junctions, respectively. Remarkably, re-expression of 14–3–3σ resulted in the restoration of adherent and tight junction complexes that were absent in parental cell lines on plastic (Fig. 2A). In contrast, elevated expression of 14–3–3σ mut or 14–3–3ζ had little impact on restoration of these cell junctional complexes. In three-dimensional (3D) collagen cultures, >80% of 14–3–3σ-expressing TM15 cells had partially restored cell junctions, which were completely lost in the other cell lines expressing the other 14–3–3 varients (Fig. 2B). Taken together, these data argue that 14–3–3σ plays a key role in assembly of these cell junctional complexes.
Figure 2.
Ectopic expression of 14–3–3σ in TM15 cells promotes cell junctions. (A) Representative immunofluorescent images of TM15 cell lines with empty vector (EV), 14–3–3σ-expressing, 14–3–3σ mut-expressing, and HA-14–3–3ζ-expressing in monolayer cultures. ZO-1 (green) was stained for tight junctions, and E-cadherin (red) was stained for adhesion junctions. Nuclei were counterstained with DAPI (blue). (B) Representative immunofluorescent images of all four aforementioned TM15 cell lines in 3D collagen cultures with the same staining. All experiments were performed in triplicate.
Loss of 14–3–3σ protein in MDCK and MCF10A cells results in loss of epithelial polarity
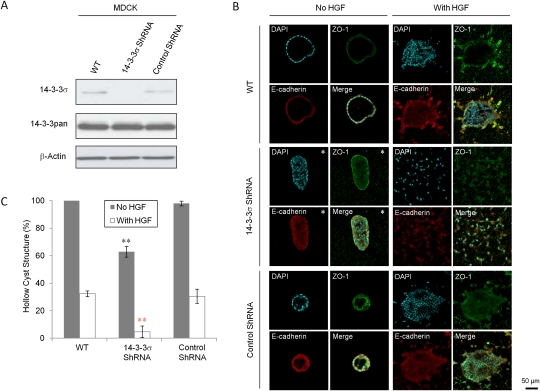
To further elucidate the role of 14–3–3σ in regulation of epithelial polarity, we generated stable MDCK cell lines that express siRNA against the endogenous canine 14–3–3σ and assessed their ability to form polarized epithelial cysts in collagen. Stable expression of a 14–3–3σ-specific siRNA resulted in a significant loss of 14–3–3σ protein expression (Fig. 3A). While wild-type or control siRNA-expressing MDCK cells formed hollow polarized epithelial cysts with ∼100% efficiency, only ∼60% of MDCK cells lacking endogenous 14–3–3σ were capable of forming hollow polarized cysts, while the other populations constituted depolarized structures (Fig. 3B,C). Following hepatocyte growth factor (HGF) stimulation, the 14–3–3σ knockdown epithelial cells also exhibited an eightfold reduction in formation of polarized epithelial structures, and the structures they formed were more depolarized when compared with the controls (Fig. 3B,C). On the other hand, under the treatment of HGF, MDCK cells that overexpress 14–3–3σ formed more hollow polarized cysts than those MDCK cells expressing 14–3–3σ mut, 14–3–3ζ, and empty vector control (Supplemental Fig. S2A,B). We next investigated whether loss of 14–3–3σ would impact the ability of MCF10A mammary epithelial cells to form cyst-like structures in Matrigel. Like the MDCK cells, MCF10A cells stably expressing a siRNA directed against 14–3–3σ exhibited efficient ablation of 14–3–3σ protein (Supplemental Fig. S3A). In contrast to parental MCF10A cells and those expressing a control siRNA, which formed hollow polarized cysts, loss of 14–3–3σ resulted in solid structures that lacked a discernable lumen (Supplemental Fig. S3B,C). Taken together, these observations suggest that down-regulation of 14–3–3σ results in the loss of epithelial polarity.
Figure 3.
Loss of 14–3–3σ in MDCK cells results in loss of epithelial polarity. (A) Immunoblot analysis to detect 14–3–3σ and all 14–3–3 protein levels in wild-type (WT), 14–3–3σ–shRNA-expressing, and control shRNA-expressing MDCK cells. β-Actin was used as loading control. (B) Representative immunofluorescent images of aforementioned MDCK cells in 3D collagen cultures, with the exception of pictures having asterisks (*) showing the major abnormal structures. ZO-1 (green) was stained for tight junction, and E-cadherin (red) was stained for adhesion junction. Nuclei were counterstained with DAPI (blue). (C) Percentages of hollow cyst structures in the collagen 3D cultures of the aforementioned MDCK cells without (gray) and with (white) HGF treatment. All experiments were performed in triplicate. Results significantly different from wild-type cells are indicated by asterisks: (*) P < 0.05; (**) P < 0.01.
Generation of a conditional 14–3–3σ knockout mouse
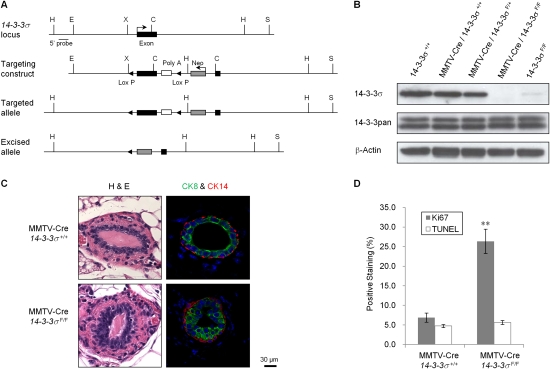
The above studies suggest that 14–3–3σ may be involved in the regulation of epithelial polarity in established epithelial cell lines. In order to verify this role for 14–3–3σ in vivo, we decided to conditionally target the mouse 14–3–3σ gene. Given that the entire coding region of 14–3–3σ is contained within a single coding exon, we created a targeting vector that carried the cDNA encoding 14–3–3σ flanked with LOXP recombination sites and inserted this into the single exon, followed by a PGK-neomycin cassette that was used for positive selection (Fig. 4A). Embryonic stem cells carrying the conditional 14–3–3σ allele were generated and were used to establish the mouse strain. After introgressing the conditional 14–3–3σ to a uniform FVB/N genetic background, we next interbred this strain to a separate strain carrying the MMTV-Cre transgene, which expresses Cre recombinase in mammary epithelium, to specifically delete 14–3–3σ in the luminal epithelial compartment (MMTV-Cre/14–3–3σF/F). We verified that the 14–3–3σ gene was excised (Supplemental Fig. S4A), and its expression was lost (Fig. 4B) in purified mammary epithelial cells from MMTV-Cre/14–3–3σF/F animals. Notably, mice with both conditional 14–3–3σ (14–3–3σF/F) alleles also showed significantly reduced 14–3–3σ levels, which may be due to the interference of LOXP sites and/or the selection cassette on 14–3–3σ transcription (Supplemental Fig. S4B). However, no apparent developmental or functional differences were observed in those mice.
Figure 4.
Targeted disruption of 14–3–3σ results in abnormal mammary epithelial development. (A) Schematic depicting the 14–3–3σ locus, the targeting construct, the targeted allele, and the excised allele. The Southern probe used for screening is indicated. (H) HindIII; (E) EcoRI; (X) XhoI; (C) ClaI; (S) SpeI. (B) Immunoblot analysis to detect 14–3–3σ and all 14–3–3 protein levels in the mammary gland protein extracts from mice of indicated genotypes. β-Actin was used as loading control. (C) Representative H&E and immunohistofluorescent stainings of mammary gland paraffin sections from 8-wk-old wild-type and conditional 14–3–3σ virgin mice, three for each strain. CK8 (green) was stained for luminal epithelium, and CK14 (red) was stained for myoepithelium. Nuclei were counterstained with DAPI (blue). (D) Percentages of positively Ki67-stained (gray) and TUNEL-stained (white) cells in the epithelium of aforementioned mouse mammary gland paraffin sections. All experiments were performed in triplicate. Results significantly different from wild-type cells are indicated by asterisks: (*) P < 0.05; (**) P < 0.01.
Disruption of 14–3–3σ in mammary epithelium results in increased epithelial proliferation and loss of epithelial polarity
Whole-mount analyses revealed that mammary ductal outgrowth was unaffected by 14–3–3σ ablation at different stages of mammary gland development (Supplemental Fig. S4D). However, histological examination of virgin mammary glands derived from wild-type and 14–3–3σ-deficient mice 8 wk old revealed that 14–3–3σ-proficient ducts retained a single layer of CK8+ luminal cells, while >60% of ducts from 14–3–3σ-deficient animals had multiple layers of CK8+ luminal cells contained within a single layer of CK14+ myoepithelial cells (Fig. 4C). Notably, the aforementioned 14–3–3σF/F mice, which have reduced mammary 14–3–3σ levels, also exerted a similar phenotype (Supplemental Fig. S4C). To ascertain whether this abnormal piling of luminal cells was due to increased proliferation or decreased apoptosis, we performed Ki-67 and TUNEL immunohistochemical staining. These analyses revealed that the rate of apoptotic cell death was unaffected in 14–3–3σ-deficient glands (Fig. 4D). Instead, the 14–3–3σ-deficient cells exhibited a threefold increase in their proliferative capacity (Fig. 4D). These data argue that multilayer epithelial structures observed in the 14–3–3σ-deficient glands result from increased proliferation of the luminal epithelial cell population.
Disruption of 14–3–3σ in mammary epithelium results in down-regulation of cell junctions and loss of epithelial polarity
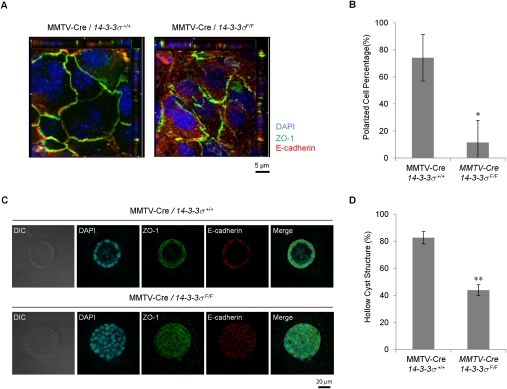
To investigate whether the multilayer epithelial phenotype observed in the 14–3–3σ-deficient gland is related to loss of epithelial polarity, we plated isolated mammary epithelial cells from control and 14–3–3σ-deficient mice on transwells and stained them with ZO-1 and E-Cadherin antibodies. In contrast to control cells, 14–3–3σ-deficient epithelial cell junctions were disrupted at both the basal and apical layers (Fig. 5A,B). Subsequently, we placed the cells in 3D Matrigel cultures. In contrast to the highly polarized acini derived from control cells, the 14–3–3σ-deficient epithelial cells formed structures that had filled lumens with depolarized epithelial growth (Fig. 5C). Careful quantification of the number of polarized structures revealed that the percentage of polarized cysts was reduced dramatically compared with wild-type controls (Fig. 5D).
Figure 5.
Primary mammary epithelial cells lacking 14–3–3σ have lost epithelial polarity. (A) Representative immunofluorescent images of control mammary epithelial cells (MMTV-Cre/14–3–3σ+/+) and 14–3–3σ-deleted cells (MMTV-Cre/14–3–3σF/F) in transwell monolayer cultures. ZO-1 (green) was stained for tight junctions, E-Cadherin (red) was stained for adhesion junctions. Nuclei were counterstained with DAPI (blue). (B) Quantification of polarized and nonpolarized epithelial cells. The ability of the cells to localize ZO-1 to the apical–lateral border was analyzed using XZ optical sections. A total of 189 out of 252 control cells (MMTV-Cre/14–3–3σ+/+) and 28 out of 259 14–3–3σ-deleted cells (MMTV-Cre/14–3–3σF/F) were classified as polarized. (C) Representative immunofluorescent images of those mammary cells in 3D Matrigel cultures with the same staining. (D) Percentages of hollow cyst structures of those cells in the 3D Matrigel cultures. Results significantly different from control cells are indicated by asterisks: (*) P < 0.05; (**) P < 0.01. All experiments were performed in triplicate.
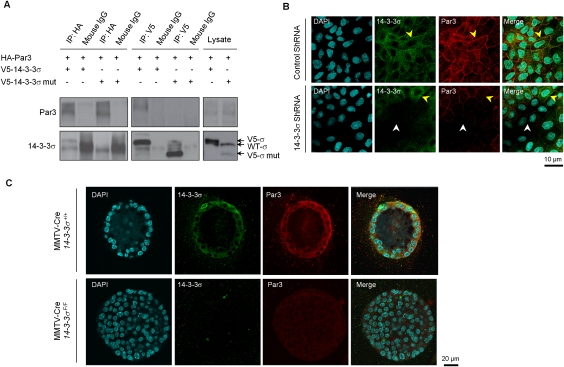
One possible explanation for the ability of 14–3–3σ to modulate epithelial polarity is through its interaction with components of the polarity (Par) complex. Indeed, the regulation of phosphorylation of key serine residues in Par3 that regulate binding of 14–3–3 proteins has been proposed recently as an important modulator of epithelial polarity (Hurd et al. 2003). To explore this possibility, we performed coimmunoprecipitation studies in MDCK cells to evaluate whether 14–3–3σ could interact with Par3. The results showed that Par3 could be detected readily in physical complexes with 14–3–3σ, but not with the truncated 14–3–3σ mutant (Fig. 6A). Immunofluoresent staining of MDCK cells showed colocalization of 14–3–3σ and Par3 at the membrane, while corresponding 14–3–3σ-deficient cells displayed lower Par3 levels at the membrane (Fig. 6B). The same phenomenon was also observed in the 3D Matrigel cultures of 14–3–3σ-deficient mammary epithelial cells, when compared with controls (Fig. 6C). Together, these observations suggest that 14–3–3σ can modulate epithelial polarity through its specific association with Par3.
Figure 6.
Association of 14–3–3σ and epithelial polarity protein Par3. (A) Coimmunoprecipitations of V5-14–3–3σ and HA-Par3 proteins in MDCK cells, with secondary antibody precipitations and straight cell lysates as controls. (B) Representative immunofluorescent images of MDCK cells with control shRNA and 14–3–3σ shRNA in monolayer cultures. 14–3-–3σ was stained green. Par3 was stained red. Nuclei were counterstained with DAPI (blue). Both yellow and white arrows indicate cell membranes. (C) Representative immunofluorescent images of mammary epithelial cells (MMTV-Cre/14–3–3σ+/+) and 14–3–3σ-deleted cells (MMTV-Cre/14–3–3σF/F) in 3D Matrigel cultures with the same staining. All experiments were performed in triplicate.
Discussion
The pathogenesis of breast cancer is thought to involve multiple genetic events. Karyotypic and epidemiological analyses of mammary tumors at various stages suggest that breast carcinomas become increasingly aggressive through the stepwise accumulation of genetic changes. The majority of genetic changes found in human breast cancer fall into two categories: gain-of-function mutations in proto-oncogenes involved in supporting cell growth, division, and survival; and loss-of-function mutations in so-called tumor suppressor genes involved in preventing unrestrained cellular growth. Genetic and biochemical analyses of ErbB2-induced mammary tumors have revealed that amplification of ErbB2 involves coamplification and coexpression of several closely linked genes, including Grb7 (Siegel et al. 1999; Kauraniemi et al. 2001; Montagna et al. 2002; Andrechek et al. 2003). In addition to the elevated expression of these components of the erbB2 amplicon, the loss of a region of mouse chromosome 4 spanning the 14–3–3σ tumor suppressor is frequently observed in mouse models of ErbB2 tumor progression (Dillon et al. 2007). Consistent with these observations, 90% of mammary tumors that arise in the ErbB2KI transgenic mouse model fail to express the 14–3–3σ protein (Montagna et al. 2002). In human breast cancers, loss of 14–3–3σ expression is also frequently observed (Andrechek et al. 2003), and involves the deletion of the distal end of chromosome 1p, which is syntenic with mouse chromosome 4 (Genuardi et al. 1989; Tsukamoto et al. 1998). Whereas these genetic studies suggest that the loss of 14–3–3σ is a common event in ErbB2-induced mammary tumor progression, its role in this process is poorly understood. Here, we demonstrate that the loss of 14–3–3σ plays a critical role in positively regulating epithelial polarity.
ErbB2 tumor cells engineered to re-express 14–3–3σ indicate that this protein can suppress several key features of ErbB2-induced tumor progression. Specifically, we demonstrated that, while restoration of functional 14–3–3σ had little impact on in vitro and in vivo growth properties of these tumor cells (Supplemental Fig. S1B,C), these tumor cells had impaired migratory behavior that was further correlated with dramatic reduction in their metastatic capacity (Fig. 1C). The impairment of the metastatic phenotype by re-expression of 14–3–3σ was further correlated with restoration of both tight junctional and adherens complexes (Fig. 2A,B). Previous studies have suggested that elevated expression of 14–3–3σ may impact cell cycle progression in certain cancer cell lines (Hermeking et al. 1997; Laronga et al. 2000). The observed lack of an effect of re-expression of 14–3–3σ on cell proliferation in TM15 tumor cell lines may reflect the loss of checkpoint controls in these cells. However, the dramatic impact on re-expression of 14–3–3σ on impairing metastatic progression (Fig. 1C) indicates that 14–3–3σ may target other steps in tumor progression. The observation that loss of 14–3–3σ is a frequent event in a number of ErbB2 mammary tumor models and primary human breast cancers (Ferguson et al. 2000; Vercoutter-Edouart et al. 2001) is consistent with the notion that loss of 14–3–3σ is a rate-limiting step in ErbB2-induced tumor induction.
Consistent with the importance of 14–3–3σ in ErbB2-driven tumor induction, loss of 14–3–3σ in MDCK and MCF10A epithelial cells resulted in a dramatic disruption of epithelial polarity (Fig. 3C; Supplemental Fig. S3C). Remarkably, the disorganized structures that are formed through down-regulation of 14–3–3σ closely resemble the structures that are formed by these cells upon induction of ErbB2 expression (Muthuswamy et al. 2001; Khoury et al. 2005). More recently, this ErbB2-dependent disruption of epithelial polarity has been linked to the interaction of the Par6/aPKC complex with ErbB2 (Aranda et al. 2006). The capacity of 14–3–3σ to restore epithelial polarity in ErbB2-expressing tumor cells (Fig. 2A,B) suggests that it can override ErbB2-dependent signals. The observed capacity of 14–3–3σ to complex with Par3 (Fig. 6A) may be partly responsible for this effect. Indeed, the modulation of epithelial polarity by interaction of 14–3–3 proteins has been noted previously in several different epithelial cell types (Benton and Johnston 2003; Hurd et al. 2003).
The concept that 14–3–3σ plays an instrumental role in the regulation of epithelial polarity in vivo is supported by the mammary epithelial disruption of this key tumor suppressor. Although mammary epithelial-specific disruption of 14–3–3σ had little impact on the initial stages of ductal outgrowth (Supplemental Fig. S4D), histological analysis of 14–3–3σ-deficient epithelium revealed an increase in the number of luminal epithelial cells that correlated with an increase in the proliferative capacity of these cells (Fig. 4D). Furthermore, in contrast to well-organized single-layer epithelial organoids derived from wild-type mammary epithelium, the 14–3–3σ-deficient organoids were filled structures that bore a remarkable similarity to mammary organoids that ectopically express ErbB2. Consistent with the importance of 14–3–3σ in controlling polarity of luminal mammary epithelium, the mammary glands derived from the hypomorphic conditional 14–3–3σ strain (Fig. 4B; Supplemental Fig. S4B) exhibited an identical luminal hyperproliferation phenotype (Supplemental Fig. S4C), suggesting that a critical threshold of 14–3–3σ is required to restrict luminal epithelial proliferation.
While the precise mechanisms that modulate epithelial polarity are unclear, proteomic analyses have revealed that 14–3–3σ can interact with several proteins that play important roles in regulating cell contacts (Benzinger et al. 2005). Indeed, we showed that 14–3–3σ interacts with the Par3 component of the polarity complex, although the association with Par3 might not be direct. Loss of 14–3–3σ leads to dislocation of Par3 from cell membranes, while it has little impact on Par3 protein levels in TM15, mouse primary mammary epithelium, or MDCK cells (Supplemental Figs. S1A, S5A,B). Previous studies in Drosophila have indicated that 14–3–3 protein plays a crucial role in establishing epithelial polarity through its action on Par3 function (Benton and Johnston 2003). There is also recent evidence to suggest that Par3 plays a crucial function in regulating mammary epithelial biology. Disruption of Par3 in the mammary epithelium results in mammary epithelial hyperplasia with luminal filling (McCaffrey and Macara 2009). Further evidence supporting the role of 14–3–3σ in epithelial polarity derives from studies of a mouse mutant bearing a truncated 14–3–3σ protein. Mice heterozygous for this truncated 14–3–3σ gene have disrupted epithelial stratification in the skin, and homozygous fetuses die shortly after birth with severe skin abnormality (Herron et al. 2005; Li et al. 2005). In addition, primary corneal epithelial cells expressing this dominant-negative protein failed to differentiate and form tight junctions (Xin et al. 2010). Taken together with our observations, these data suggest that 14–3–3σ plays a critical role in regulating epithelial polarity. In contrast to the positive effects of 14–3–3σ on epithelial polarity, ectopic expression of 14–3–3ζ results in disruption of epithelial polarity of MDCK cells through binding Par3 (Hurd et al. 2003). Moreover, elevated expression of 14–3–3ζ can enhance the invasiveness of mammary tumor cell lines (Fig. 1C; Lu et al. 2009). It is conceivable that the opposing effects on epithelial polarity of these two closely related 14–3–3 proteins is due to their ability to localize Par3 to distinct apical and basal compartments within the cell.
The observation that ErbB2 tumors are associated with loss of 14–3–3σ has important implications in understanding the genetic events involved in ErbB2-induced tumor progression. The fact that 14–3–3σ is a major positive regulator of epithelial polarity suggests that loss of polarity may be an important step in tumorigenesis and metastasis. Consistent with this concept, it has been demonstrated recently that loss of the Scribble polarity regulator plays an important role in c-Myc-induced mammary tumors (Zhan et al. 2008). Whether 14–3–3σ plays a comparable role in ErbB2 mammary tumor progression remains to be addressed.
Materials and methods
Plasmid constructs
14–3–3σ cDNA, 14-3-3σ mut cDNA, V5-tagged 14–3–3σ cDNA, and V5-tagged 14–3–3σ mut cDNA were cloned into EcoRI/XhoI sites of pcDNA3 (Invitrogen), and 14–3–3σ cDNA and 14-3-3σ mut cDNA were subsequently cloned into BglII/HpaI sites of pMSCVhygro (Clontech). The primer sequences used were SfnF, 5′-CGGAATTCATGGAGAGAGCCAGTCTGATC-3′; V5SfnF, 5′-CCCAAGCTTATGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGGGATCCATGGAGAGAGCCAGTCTGATC-3′; SfnR, 5′-CCGCTCGAGTCAGCTCTGGGGCTCCTCCG-3′; SfnMR, 5′-CCGCTCGAGTCACTGAAGGGTGTGCAGGTCGG-3′. shRNA oligo pairs specific to canine and human 14–3–3σ and control shRNA pairs were inserted into the HindIII/BglII sites of pSUPER.retro.puro (OligoEngine). The primer sequences used were SfnDF, 5′-GATCCCCTTGAGGAGTGTCCCACCCTTTCAAGAGAAGGGTGGGACACTCCTCAATTTTTGGAAA-3′; SfnDR, 5′-AGCTTTTCCAAAAAGTGACCCTGTTTCCTCTCATCTCTTGAATGAGAGGAAACAGGGTCACGGG-3′; SfnHF, 5′-GATCCCCGTGACCATGTTTCCTCTCATTCAAGAGATGAGAGGAAACATGGTCACTTTTTGGAAA-3′; SfnHR, 5′-AGCTTTTCCAAAAAGTGACCATGTTTCCTCTCATCTCTTGAATGAGAGGAAACATGGTCACGGG-3′; SfnCF, 5′-GATCCCCGAGTAGGAGAGATATAGAGTTCAAGAGACTCTGTGTCTCTCCTGCTCTTTTTGGAAA-3′; SfnCR, 5′-AGCTTTTCCAAAAAGAGTAGGAGAGATATAGAGTCTCTTGAACTCTGTGTCTCTCCTGCTCGGG-3′.
Conditional knockout of 14–3–3σ
The 14–3–3σ locus was isolated by screening the Lambda FIX II Library (Stratagene) by PCR. A 6-kb EcoRI–SpeI fragment containing the only 14–3–3σ exon was cloned into the pGem-9Zf vector (Promega), and was used to build the targeting construct. A cDNA encoding mouse 14–3–3σ was obtained by RT–PCR. The PCR product with a 5′ loxP site was followed by SV40 PolyA signal with a downstream loxP site, and followed by a PGK-neomycine selection cassette (Invitrogen). The whole 2.9-kb XhoI–ClaI fragment was used to replace the corresponding part of the exon, which included the first 434 coding base pairs. The primer sequences used for targeting construct were LoxSfnF, 5′-CCGCTCGAGATAACTTCGTATAGCATACATTATACGAGGTTATACAGCCAGTTCGCCCGTC-3′; SfnR, 5′-CCCAAGCTTGCTGCTTCAGCTCTGGGGC-3′; PolyAF, 5′-CCCAAGCTTGGGTTCGAAATGACCGACC-3′; LoxPolyAR, 5′-GAAGATCTATAACTTCGTATAATGTATGCTATACGAAGTTATAAGCTCTAGCTAGAGGTCG-3′; NeoF, 5′-CCATCGATGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCGAATTCTACC-3′; NeoR, 5′-GAAGATCTGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCAAGCTCTAGCTAGAGGTCG-3′. The vector was linearized with NotI, and gene targeting was performed following standard techniques using the embryonic stem cell line R1. Embryonic stem cell clones containing the correct integration event were identified by Southern blotting with a 5′ external probe. The primer sequences used were ProbeF, 5′-TTCATCTTGCCATTGCCTGG-3′; ProbeR, 5′-TCAACAGTCTGACCAGAGGG-3′. Two embryonic stem cell clones were then used to generate chimeras and subsequent lines of mice heterozygous for 14–3–3σ (14–3–3σF/+) by the McGill Transgenic Facility. The mice were introgressed in a FVB/N background. 14–3–3σF/+ animals were interbred with the MMTV-Cre transgenic to generate MMTV-Cre/14–3–3σF/F mice, which have a mammary-specific 14–3–3σ knockout.
Cell lines
MDCK cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Wisent). TM15 cells were maintained in DMEM with 10% fetal bovine serum with MEGS supplements (Cascade Biologics). Cell lines were generated by retroviral infection, followed by 2 μg/mL and 4 μg/mL puromycine selection for MDCK and TM15, respectively. Primary mammary epithelial cells were obtained from virgin mouse mammary glands. Mammary glands were harvested and processed with the McILWAN Tissue Chopper, and were disassociated in DMEM with 2.4 mg/mL Collagenase B (Roche) and 2.4 mg/mL Dispase II (Roche) for 3 h at 37°C. Disassociated cells were washed with PBS/EDTA (1 mM), plated on plastic, and maintained in DMEM with 2% fetal bovine serum with MEGS supplements.
3D cell culture
For 3D collagen cultures, 24-well plates (NUNC) were used. For each well, 350 μL of 80% collagen I (Inamed) in PBS (pH 7.4) was plated and solidified for 45 min at 37°C without CO2. Subsequently, 3000 monodispersed cells mixed with 500 μL of collagen were seeded on top and incubated for 30 min at 37°C without CO2. One milliliter of Leibovitz's L-15 medium (Invitrogen) was applied into each well and changed every 4 d. For HGF stimulation, after 12 d of culture, HGF (gift from Dr. Morag Park) was added in the medium to a final concentration of 10 U/mL, and cells were cultured with HGF for an additional 2 d before subsequent staining. For 3D Matrigel cultures, eight-well Chamber slides (NUNC) were used. For each well, 100 μL of Matrigel (BD Biosciences) was plated and solidified for 30 min at 37°C. Later, 1500 monodispersed cells mixed in 300 μL of DMEM with 2% fetal bovine serum (BSA), 2% Matrigel, and MEGS supplements were seeded on top. Medium was changed every 4 d. Cells were stained after 10 d of culture.
Immunoblotting
For immunoblotting, membranes were blocked in TBS with 3% skim milk, and incubated overnight with primary antibodies in 3% skim milk: 14–3–3σ (1:1000; Santa Cruz Biotechnologies); 14–3–3pan (1: 1000; Millipore); β-actin (1:5000; Sigma); HA-11 (1:5000; Covance); and Par3 (1:1000; Millipore). Later, membranes were treated by 1 h of incubation at room temperature with HRP-conjugated mouse, rabbit, and goat secondary antibodies (1:5000 in TBS/3% milk; Jackson Laboratories). Blots were developed with enhanced chemiluminescence (Amersham).
Immunofluorescence (IF)
Cells were fixed for 15 min at room temperature with 2% paraformaldehyde in PBS, permeabilized for 10 min in 0.5% Triton X-100 in PBS, followed by washes of 100 mM glycine in PBS. Blocking was performed with IF buffer (PBS, 0.1% BSA, 0.2% Triton X-100, 0.05% Tween-20) plus 2% BSA for 1 h followed by 1 h of incubation at room temperature with the following primary antibodies diluted in IF buffer: Par3 (1:100; Millipore); 14–3–3σ (1:100; Millipore); E-Cadherin (1:100; BD Transduction Laboratories); and ZO-1 (1:100; Zymed). Cells were washed in IF buffer and incubated in a humidified chamber with the appropriate Alexa-fluor-conjugated secondary antibodies (1:1000; Molecular Probes) diluted in IF buffer for 45 min at room temperature. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Jackson Laboratories) for 5 min.
Immunohistofluorescence (IHF)/immunohistochemistry (IHC)
Paraffin-embedded sections were deparaffinized in three changes of xylenes. Sections were heated in 10 mM sodium citrate (pH 6), followed by incubation in 3% H2O2 for 20 min. Samples were incubated in primary antibodies of CK8, CK14 (1:100; Covance), and Ki67 (1:100; Cedarlane) diluted in PBS/2% BSA for 1 h at room temperature. For IHF, samples were incubated in a humidified chamber with the appropriate Alexa-fluor-conjugated secondary antibodies (1:1000; Molecular Probes) diluted in IF buffer for 45 min at room temperature, followed by nuclei counterstaining with DAPI for 5 min. For IHC, samples were subject to incubation at room temperature with HRP-conjugated secondary antibody (1:1000 in PBS/2% BSA; Jackson Laboratories). Immunoreactivity was visualized using the DAB+ substrate chromagen system (DAKO), and the tissues were counterstained with hematoxylin. For TUNEL IHC staining, the ApopTag Fluorescein in Situ Apoptosis Detection Kit (Serological) was used.
Coimmunoprecipitation
Forty-eight hours after transient transfection of MDCK cells with pcDNA3-V5-14–3–3σ, pcDNA3-V5-14–3–3σmut, and pKH3-Par3 using Fugene6 (Roche), cells were lysed with modified TNE buffer supplemented with protease inhibitors and Na3VO4. Immunoprecipitations were performed overnight at 4°C with antibodies to V5 tag (1:150; Genway) and HA tag (1:150; Covance). Immunoprecipitations were then incubated for 2 h with protein G-agarose (Amersham Biosciences). The reaction products were washed with lysis buffer, and the immune complexes were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were electrotransferred to polyvinylidene difluoride membranes, and the membranes were immunoblotted with antibodies to Par3 (1:1000; Millipore), and 14–3–3σ (1:1000; Santa Cruz Biotechnologies), followed by secondary antibody incubation and subsequent developed with enhanced chemiluminescence (Amersham).
Confocal imaging
Confocal imaging was performed using an Axiovert 200M microscope (Carl Zeiss MicroImaging, Inc.) with 40×, 63×, and 100×/1.4 plan-APOCHROMAT objectives equipped with a confocal microscope system (LSM 510 Meta confocal microscope; Carl Zeiss MicroImaging, Inc.). Image analysis was carried out using the LSM 5 image browser (Empix Imaging).
Statistics
Statistically significant differences were determined using the Student's t-test. Differences were considered significant if P < 0.05, and dramatically significant if P < 0.01.
Acknowledgments
We are grateful to Dr. Morag Park, Dr. Dihua Yu, Dr. Luke McCaffrey, Dr. Graeme Hodgson, Dr. Michael Tremblay, Dr. Xiangjiao Yang, and Dr. Giuseppina Ursini-Siegel for providing reagents and advice. C.L. receives scholarship from the DoD Breast Cancer Research Program (W81XWH-06-1-0700). These studies were supported by funding from the CIHR (CIHR MOP-10594).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1896810.
Supplemental material is available at http://www.genesdev.org.
References
- Andrechek ER, Hardy WR, Siegel PM, Rudnicki MA, Cardiff RD, Muller WJ 2000. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc Natl Acad Sci 97: 3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrechek ER, Laing MA, Girgis-Gabardo AA, Siegel PM, Cardiff RD, Muller WJ 2003. Gene expression profiling of neu-induced mammary tumors from transgenic mice reveals genetic and morphological similarities to ErbB2-expressing human breast cancers. Cancer Res 63: 4920–4926 [PubMed] [Google Scholar]
- Andrulis I, Bull S, Blackstein M, Sutherland D, Mak C, Sidlofsky S, Pritzker K, Hartwick R, Hanna W, Lickley L, et al. 1998. neu/erbB-2 amplification identifies a poor-prognosis group of women with node-negative breast cancer. Toronto Breast Cancer Study Group. J Clin Oncol 16: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T, Muthuswamy SK 2006. Par6–aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol 8: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Benton R, Johnston DS 2003. Drosophila PAR-1 and 14–3–3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115: 691–704 [DOI] [PubMed] [Google Scholar]
- Benzinger A, Muster N, Koch HB, Yates JR III, Hermeking H 2005. Targeted proteomic analysis of 14–3–3ς, a p53 effector commonly silenced in cancer. Mol Cell Proteomics 4: 785–795 [DOI] [PubMed] [Google Scholar]
- Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B 1999. 14–3–3σ is required to prevent mitotic catastrophe after DNA damage. Nature 401: 616–620 [DOI] [PubMed] [Google Scholar]
- Dillon RL, Brown ST, Ling C, Shioda T, Muller WJ 2007. An EGR2/CITED1 transcription factor complex and the 14–3–3σ tumor suppressor are involved in regulating ErbB2 expression in a transgenic-mouse model of human breast cancer. Mol Cell Biol 27: 8648–8657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, Muller WJ 2009. Akt1 and Akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res 69: 5057–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA, Stampfer MR, et al. 2000. High frequency of hypermethylation at the 14–3–3σ locus leads to gene silencing in breast cancer. Proc Natl Acad Sci 97: 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuardi M, Tsihira H, Anderson DE, Saunders GF 1989. Distal deletion of chromosome Ip in ductal carcinoma of the breast. Am J Hum Genet 45: 73–82 [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B 1997. 14–3–3σ is a p53-regulated inhibitor of G2/M progression. Mol Cell 1: 3–11 [DOI] [PubMed] [Google Scholar]
- Herron BJ, Liddell RA, Parker A, Grant S, Kinne J, Fisher JK, Siracusa LD 2005. A mutation in stratifin is responsible for the repeated epilation (Er) phenotype in mice. Nat Genet 37: 1210–1212 [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Malek T, Bornstein S, Hariono S, Ginzinger DG, Muller WJ, Gray JW 2005. Copy number aberrations in mouse breast tumors reveal loci and genes important in tumorigenic receptor tyrosine kinase signaling. Cancer Res 65: 9695–9704 [DOI] [PubMed] [Google Scholar]
- Hurd TW, Fan S, Liu C-J, Kweon HK, Hakansson K, Margolis B 2003. Phosphorylation-dependent binding of 14–3–3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol 13: 2082–2090 [DOI] [PubMed] [Google Scholar]
- Hynes NE, Stern DF 1994. The biology of erbB-2/nue/HER-2 and its role in cancer. Biochim Biophys Acta 1198: 165–184 [DOI] [PubMed] [Google Scholar]
- Kauraniemi P, Barlund M, Monni O, Kallioniemi A 2001. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res 61: 8235–8240 [PubMed] [Google Scholar]
- Khoury H, Naujokas MA, Zuo D, Sangwan V, Frigault MM, Petkiewicz S, Dankort DL, Muller WJ, Park M 2005. HGF converts ErbB2/Neu epithelial morphogenesis to cell invasion. Mol Biol Cell 16: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laronga C, Yang H-Y, Neal C, Lee M-H 2000. Association of the cyclin-dependent kinases and 14–3–3σ negatively regulates cell cycle progression. J Biol Chem 275: 23106–23112 [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Estepa G, Verma IM 2005. Identification of 14–3–3σ mutation causing cutaneous abnormality in repeated-epilation mutant mouse. Proc Natl Acad Sci 102: 15977–15982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Guo H, Treekitkarnmongkol W, Li P, Zhang J, Shi B, Ling C, Zhou X, Chen T, Chiao PJ, et al. 2009. 14–3–3ζ cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer Cell 16: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG 2009. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes & Dev 23: 1450–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna C, Andrechek ER, Padilla-Nash H, Muller WJ, Ried T 2002. Centrosome abnormalities, recurring deletions of chromosome 4, and genomic amplification of HER2/neu define mouse mammary gland adenocarcinomas induced by mutant HER2/neu. Oncogene 21: 890–898 [DOI] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS 2001. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol 3: 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravdin PM, Chamness GC 1995. The c-erbB-2 proto-oncogene as a prognostic and predictive marker in breast cancer: A paradigm for the development of other macromolecular markers—A review. Gene 159: 19–27 [DOI] [PubMed] [Google Scholar]
- Siegel PM, Ryan ED, Cardiff RD, Muller WJ 1999. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: Implications for human breast cancer. EMBO J 18: 2149–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon D, Clark G, Wong S, Levin W, Ullrich A, McGuire W 1987. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235: 177–182 [DOI] [PubMed] [Google Scholar]
- Slamon D, Godolphin W, Jones L, Holt J, Wong S, Keith D, Levin W, Stuart S, Udove J, Ullrich A, et al. 1989. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244: 707–712 [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Ito N, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G, Nakamura Y, Emi M 1998. Allelic loss on chromosome 1p is associated with progression and lymph node metastasis of primary breast carcinoma. Cancer 82: 317–322 [PubMed] [Google Scholar]
- Vercoutter-Edouart A-S, Lemoine J, Le Bourhis X, Louis H, Boilly B, Nurcombe V, Revillion F, Peyrat J-P, Hondermarck H 2001. Proteomic analysis reveals that 14–3–3σ is down-regulated in human breast cancer cells. Cancer Res 61: 76–80 [PubMed] [Google Scholar]
- Wilker EW, van Vugt MATM, Artim SC, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, et al. 2007. 14–3–3σ controls mitotic translation to facilitate cytokinesis. Nature 446: 329–332 [DOI] [PubMed] [Google Scholar]
- Xin Y, Lu Q, Li Q 2010. 14–3–3σ controls corneal epithelial cell proliferation and differentiation through the Notch signaling pathway. Biochem Biophys Res Commun 392: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wen Y-Y, Zhao R, Lin Y-L, Fournier K, Yang H-Y, Qiu Y, Diaz J, Laronga C, Lee M-H 2006. DNA damage-induced protein 14–3–3 σ inhibits protein kinase B/Akt activation and suppresses Akt-activated cancer. Cancer Res 66: 3096–3105 [DOI] [PubMed] [Google Scholar]
- Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK 2008. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 135: 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]