Abstract
The spindle checkpoint generates a “wait anaphase” signal at unattached kinetochores to prevent premature anaphase onset. Kinetochore-localized dynein is thought to silence the checkpoint by transporting checkpoint proteins from microtubule-attached kinetochores to spindle poles. Throughout metazoans, dynein recruitment to kinetochores requires the protein Spindly. Here, we identify a conserved motif in Spindly that is essential for kinetochore targeting of dynein. Spindly motif mutants, expressed following depletion of endogenous Spindly, target normally to kinetochores but prevent dynein recruitment. Spindly depletion and Spindly motif mutants, despite their similar effects on kinetochore dynein, have opposite consequences on chromosome alignment and checkpoint silencing. Spindly depletion delays chromosome alignment, but Spindly motif mutants ameliorate this defect, indicating that Spindly has a dynein recruitment-independent role in alignment. In Spindly depletions, the checkpoint is silenced following delayed alignment by a kinetochore dynein-independent mechanism. In contrast, Spindly motif mutants are retained on microtubule-attached kinetochores along with checkpoint proteins, resulting in persistent checkpoint signaling. Thus, dynein-mediated removal of Spindly from microtubule-attached kinetochores, rather than poleward transport per se, is the critical reaction in checkpoint silencing. In the absence of Spindly, a second mechanism silences the checkpoint; this mechanism is likely evolutionarily ancient, as fungi and higher plants lack kinetochore dynein.
Keywords: Centromere, aneuploidy, mitosis, kinetochore, microtubule, spindle, chromosome
Chromosome segregation requires the attachment of spindle microtubules to kinetochores, proteinaceous structures that assemble at the centromeric locus on each sister chromatid (Cheeseman and Desai 2008; Santaguida and Musacchio 2009). A surveillance mechanism known as the spindle checkpoint generates an inhibitory “wait anaphase” signal at unattached kinetochores, preventing premature anaphase onset (Musacchio and Salmon 2007). Microtubule attachments of the correct geometry are stabilized by tension experienced at sister kinetochores that have made bioriented connections to opposite poles (Nicklas 1997). Once all kinetochores are attached in a bioriented fashion to microtubule bundles, termed kinetochore fibers, the checkpoint signal is silenced and the cell proceeds to anaphase.
The spindle checkpoint regulates the E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C), which targets cyclin B and securin for destruction by the 26S proteasome. Specifically, the checkpoint components Mad2, BubR1, and Bub3 interact with and inhibit the essential APC/C cofactor Cdc20 by forming diffusible mitotic checkpoint complexes (Hwang et al. 1998; Sudakin et al. 2001; Nilsson et al. 2008). Additional components of the checkpoint pathway, including Mad1 and the kinases Bub1 and Mps1, are involved in the generation and amplification of the checkpoint signal (Hoyt et al. 1991; Li and Murray 1991; Abrieu et al. 2001).
The conserved KNL-1/Mis12 complex/Ndc80 complex (KMN) network constitutes the core attachment site for microtubules at the kinetochore and also recruits components that generate the checkpoint signal (Burke and Stukenberg 2008). Additional contacts to microtubules are made by the kinesin CENP-E (Weaver et al. 2003) and by the minus end-directed motor dynein and its cofactor, dynactin, which are recruited to the outer kinetochore via the Rod/Zwilch/Zw10 (RZZ) complex (Williams et al. 1992; Starr et al. 1998; Scaërou et al. 2001). In contrast to the KMN network and spindle checkpoint proteins, the RZZ complex, kinetochore-localized dynein/dynactin, and CENP-E are present in only the metazoan lineage.
A single unattached kinetochore can delay cell cycle progression (Rieder et al. 1995). Structural studies, reconstitution experiments, and checkpoint protein dynamics suggest that the checkpoint signal involves a catalytic step based on the conformational conversion of Mad2 (Howell et al. 2004; Luo et al. 2004; Shah et al. 2004; De Antoni et al. 2005). Current models envision a stably bound complex of Mad1 and “closed” Mad2 at unattached kinetochores that templates the conversion of “open” Mad2 in the soluble pool to the “closed” conformer competent to bind and inhibit Cdc20 (Mapelli and Musacchio 2007; Luo and Yu 2008).
Kinetochore-localized dynein/dynactin is thought to have a key role in checkpoint silencing by removing the catalytic Mad1/Mad2 scaffold and other checkpoint proteins from kinetochores upon microtubule attachment. Abrogation of dynein-mediated poleward transport by direct inhibitions of dynein/dynactin leads to the partial retention of Mad2 at aligned bioriented kinetochores with a normal microtubule complement and persistence of checkpoint signaling (Howell et al. 2001; Wojcik et al. 2001; Vergnolle and Taylor 2007; Mische et al. 2008; Sivaram et al. 2009). In addition to its proposed role in checkpoint silencing, kinetochore dynein/dynactin has been implicated in microtubule capture and transient poleward chromosome movement (Z Yang et al. 2007; Varma et al. 2008; Vorozhko et al. 2008).
The RZZ complex recruits dynein via a conserved kinetochore-specific dynein recruitment factor, called Spindly (Griffis et al. 2007; Gassmann et al. 2008; Yamamoto et al. 2008; Chan et al. 2009). Paradoxically, in contrast to direct dynein inhibitions, the spindle checkpoint appears to be silenced following microtubule attachment in Spindly-depleted human cells (Chan et al. 2009). This observation sheds doubt on the dominant model for checkpoint silencing in vertebrate somatic cells. There are three possible explanations of the difference between direct dynein inhibitions and Spindly depletion. First, checkpoint proteins may be removed from kinetochores by nonkinetochore dynein (Chan et al. 2009). Second, direct dynein inhibition may activate the checkpoint, as opposed to preventing its silencing. Third, kinetochore dynein may be essential for checkpoint silencing when Spindly is present, but may become dispensable in Spindly's absence. To distinguish between these possibilities, we generated single amino acid changes in a conserved motif in Spindly that do not affect its kinetochore targeting, but prevent it from recruiting dynein to kinetochores. Analysis of these mutants provided support for the third possibility, indicating that kinetochore dynein-mediated removal of Spindly is the critical reaction governing checkpoint silencing. In addition, we uncovered a dynein recruitment-independent function of Spindly at kinetochores that is central to the efficient alignment of chromosomes during prometaphase.
Results
Point mutations in the conserved Spindly motif uncouple kinetochore localization of Spindly from dynein/dynactin recruitment
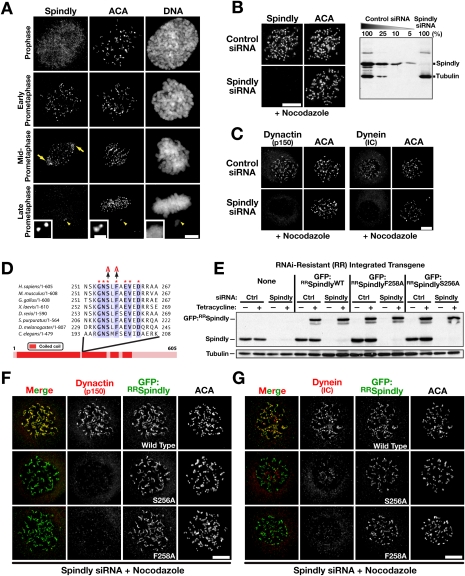
Immunofluorescence using an antibody generated against full-length human Spindly revealed that it is nuclear in interphase, concentrates at unattached kinetochores and spindle poles in prometaphase, and is no longer detectable at kinetochores or spindle poles by metaphase (Fig. 1A; Supplemental Fig. S1A). In the absence of microtubules, Spindly expanded to a crescent-like morphology, indicating that it is a component of the fibrous corona (Fig. 1B; Supplemental Fig. S1B), similar to dynein/dynactin and checkpoint proteins (Hoffman et al. 2001). Depletion of the RZZ subunit Zw10 confirmed that Spindly functions downstream from the RZZ complex (Supplemental Fig. S1C; Griffis et al. 2007; Gassmann et al. 2008; Chan et al. 2009). We tested four siRNAs to knock down Spindly in HeLa cells, and chose one siRNA that depleted the protein to undetectable levels on a single-cell basis by immunofluorescence and to >95% by immunoblot (Fig. 1B). In agreement with previous work in Caenorhabditis elegans and human cells (Gassmann et al. 2008; Chan et al. 2009), Spindly depletion prevented the kinetochore localization of both dynein and dynactin (Fig. 1C; Supplemental Fig. S1D). However, Spindly depletion did not affect dynein/dynactin localization to the spindle, spindle poles, and cell cortex, or to microtubule plus ends in interphase cells (Supplemental Fig. S1E–G; data not shown). Furthermore, the dynein/dynactin-binding partner NuMA was localized normally to spindle poles following Spindly depletion (Supplemental Fig. S1H). We conclude that Spindly depletion specifically perturbs the recruitment of dynein/dynactin to unattached kinetochores without affecting their localization to other structures in the cell.
Figure 1.
Single amino acid changes in Spindly uncouple its kinetochore localization from dynein/dynactin recruitment. (A) Mitotic HeLa cells fixed and immunostained for Spindly and centromere antigens (ACA). In addition to kinetochores, Spindly is also visible at spindle poles (arrows) during chromosome alignment. Spindly is absent from kinetochores at the metaphase plate, but is detectable at an unaligned kinetochore pair (arrowhead). (B) Immunoblot and immunofluorescence 48 h after transfection of HeLa cells with control and Spindly siRNA. α-Tubulin was used as a loading control for the immunoblot. Cells were treated with nocodazole for 4 h prior to fixation and immunostaining. (C) Localization of dynein intermediate chains and the dynactin subunit p150Glued at unattached kinetochores in control and Spindly siRNA-treated cells. Cells were incubated in nocodazole for 4 h to accumulate dynein/dynactin at kinetochores (see also Supplemental Fig. S1D). (D) Sequence alignment of the highly conserved motif in the Spindly protein family. The conserved serine and phenylalanine (S256 and F258 in human Spindly) that were individually mutated to alanine are indicated. (E) Immunoblot monitoring endogenous and RNAi-resistant (RR) transgenic Spindly expression. Cells were treated with control or Spindly siRNA for 22 h followed by induction with 0.2 μg/mL tetracycline for 8 h. α-Tubulin is used as a loading control. (F,G) Cell lines expressing GFP:RRSpindlyWT or the point mutants depicted in D immunostained for GFP, centromere antigens (ACA), and either the dynactin subunit p150Glued (F) or dynein intermediate chains using the monoclonal antibody 70.1 (G). Cells were treated with Spindly siRNA for 24 h, transgene expression was induced with tetracycline for 16 h, and nocodazole was added for 4 h prior to fixation. Bars, 5 μm; inset in A, 1 μm.
Human Spindly was reported in qualitative analysis to be dispensable for the removal of Mad2 from attached kinetochores (Chan et al. 2009). In contrast, direct inhibitions of dynein/dynactin result in the retention of Mad2 at aligned kinetochores (Howell et al. 2001; Vergnolle and Taylor 2007; Varma et al. 2008; Sivaram et al. 2009). To address this apparent contradiction and gain insight into the mechanism of checkpoint silencing in human cells, we investigated the relationship between Spindly and kinetochore-localized dynein/dynactin. Biochemical analysis of Spindly from mitotic HeLa cell extracts failed to reveal a clear association between soluble Spindly and dynein/dynactin components. We therefore focused on the only conserved region in Spindly—a short motif that was used to define this protein family (Fig. 1D). The absolute conservation of this motif against a near-complete divergence of the rest of the protein sequence (which is largely a predicted coiled-coil) indicates that it is part of a critical functional interaction. We mutated either of the two conserved residues S256 and F258 in human Spindly to alanine and assessed whether this change affected kinetochore localization of Spindly or its ability to recruit dynein/dynactin to kinetochores.
To analyze the Spindly motif point mutants, we integrated tetracycline-inducible RNAi-resistant (RR) GFP fusion constructs into a single genomic locus in HeLa cells by Flp-mediated DNA recombination. This approach made it possible to combine specific depletion of endogenous Spindly with expression of Spindly transgenes, either mutant or wild type, at identical, near-endogenous levels (Fig. 1E). GFP:RRSpindlyWT and the two mutant proteins GFP:RRSpindlyS256A and GFP:RRSpindlyF258A all localized robustly to kinetochores in cells depleted of endogenous Spindly (Fig. 1F,G). This indicates that the Spindly motif is not required for its kinetochore localization. GFP:RRSpindlyWT rescued dynein/dynactin recruitment to unattached kinetochores following depletion of endogenous Spindly, but expression of GFP:RRSpindlyF258A or GFP:RRSpindlyS256A did not (Fig. 1F,G). We conclude that mutations in the conserved Spindly motif uncouple its kinetochore localization from its ability to recruit dynein/dynactin to kinetochores.
Spindly-mediated targeting of dynein/dynactin to kinetochores is required for the poleward transport of checkpoint proteins
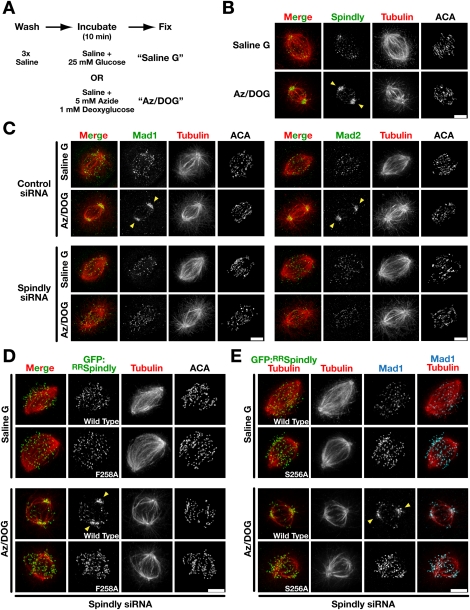
Both Spindly depletion and the Spindly motif mutants prevent recruitment of dynein/dynactin specifically to kinetochores without globally perturbing its function. If kinetochore-localized dynein, as opposed to nonkinetochore dynein, drives poleward transport of checkpoint proteins, this transport should be inhibited in both perturbations. To test this, we used a previously described assay (Howell et al. 2000) in which ATP levels are reduced by treatment with azide and deoxyglucose (Fig. 2A). Dynein transport still occurs under these conditions and moves checkpoint proteins to spindle poles, where they accumulate. Spindly itself also accumulates readily at spindle poles under these conditions (Fig. 2B; Chan et al. 2009). Mad1, Mad2, Zwilch, and CENP-E all accumulated at spindle poles after ATP reduction in control cells. In contrast, accumulation of these proteins at spindle poles was not observed in Spindly-depleted cells (Fig. 2C; Supplemental Fig. S2A). We also examined the localization in DLD-1 cells of a C-terminal CENP-E tail fragment (Chan et al. 1998), which is initially localized at kinetochores and subsequently prominently accumulates at spindle poles, even in the absence of azide/deoxyglucose treatment. In cells depleted of Spindly, the CENP-E tail fragment no longer accumulated at spindle poles, providing independent confirmation of the results obtained with the ATP reduction assay (Supplemental Fig. S2B; Supplemental Movie S1). Thus, Spindly is required for the minus end-directed transport of checkpoint proteins along kinetochore fibers.
Figure 2.
Spindly and its ability to target dynein/dynactin to kinetochores is required for poleward transport of checkpoint proteins. (A) Schematic of the ATP reduction assay used to analyze kinetochore dynein-mediated transport of checkpoint proteins (Howell et al. 2000). (B) Immunostaining of Spindly and centromere antigens (ACA) in the indicated states. (C) Immunostaining of Mad1 and Mad2 in control and Spindly siRNA-treated cells with normal or reduced ATP levels. Similar results were obtained for Zwilch and CENP-E (Supplemental Fig. S2A,B; Supplemental Movie S1). (D) The ATP reduction assay performed in cells expressing GFP:RRSpindlyWT or GFP:RRSpindlyF258A. Cells were treated with Spindly siRNA and tetracycline as described for Figure 1F and immunostained for GFP, α-tubulin, and centromere antigens (ACA). (E) Cells were treated as in D and immunostained for GFP, α-tubulin, and Mad1. For similar analysis of Mad2, see Supplemental Figure S2C. Arrowheads denote accumulation of the indicated proteins at spindle poles. Bars, 5 μm.
We next performed the ATP reduction assay with the Spindly motif mutants. As observed for endogenous Spindly, GFP:RRSpindlyWT accumulated at spindle poles upon treatment with azide/deoxyglucose (Fig. 2D). In contrast, the GFP:RRSpindlyF258A and GFP:RRSpindlyS256A mutants failed to accumulate at spindle poles (Fig. 2D,E). Furthermore, whereas GFP:RRSpindlyWT supported poleward transport, both motif mutants failed to facilitate transport of Mad1 and Mad2 to spindle poles (Fig. 2E; Supplemental Fig. S2C). We conclude that Spindly depletion and replacement of endogenous Spindly with Spindly motif mutants, both of which prevent dynein/dynactin recruitment to kinetochores, inhibit poleward transport of checkpoint proteins. As neither of these perturbations globally inhibits dynein/dynactin or perturbs its localization to other structures, these results support the model that it is specifically the kinetochore-localized pool of dynein/dynactin that transports checkpoint proteins from kinetochores to spindle poles.
A kinetochore dynein-independent mechanism is capable of removing checkpoint proteins from bioriented kinetochores in the absence of Spindly
Preventing poleward transport of checkpoint proteins by direct dynein/dynactin inhibitions causes their retention at bioriented kinetochores. As both Spindly and its ability to recruit dynein/dynactin are required for poleward transport, we next examined the effect of Spindly depletion and Spindly motif mutants on kinetochore levels of checkpoint proteins.
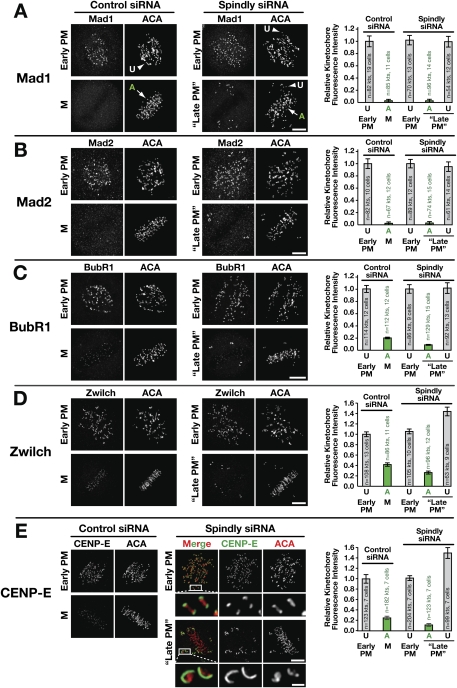
We began by quantifying the levels of Mad1, Mad2, BubR1, Zwilch, and CENP-E in immunofluoresence images of early prometaphase and metaphase kinetochores in control and Spindly-depleted cells. The majority of Spindly-depleted cells had a metaphase plate with several uncongressed chromosomes, and we quantified kinetochore levels of checkpoint proteins on both sets of chromosomes within a cell (Fig. 3A–E). In Spindly-depleted cells, checkpoint protein levels at early prometaphase kinetochores were equal to controls, suggesting that, as in Drosophila melanogaster and unlike in C. elegans, human Spindly is not required for checkpoint activation. Despite the inhibition of poleward transport, kinetochore levels of checkpoint proteins at congressed chromosomes were not increased in Spindly-depleted cells relative to control cells (Fig. 3A–E). Thus, in the absence of Spindly, a kinetochore dynein-independent mechanism is capable of removing checkpoint proteins from bioriented kinetochores. These conclusions are in agreement with a prior qualitative analysis of Mad2 and Zw10 localization in Spindly-depleted cells (Chan et al. 2009).
Figure 3.
Checkpoint proteins are not retained at bioriented kinetochores in Spindly-depleted cells. (A–E) Control or Spindly siRNA-treated cells immunostained for the checkpoint proteins Mad1 (A), Mad2 (B), BubR1 (C), Zwilch (D), and CENP-E (E). For each protein, staining is shown in an early prometaphase (PM) cell with no kinetochore–microtubule attachments, and in a cell where all (metaphase [M] in control siRNA) or most (“Late PM” in Spindly siRNA) chromosomes have congressed. Arrows and arrowheads point to examples of aligned (A) and unaligned (U) kinetochores, respectively, used for quantitation of checkpoint protein signals. Kinetochore intensity measurements for each protein were normalized relative to prometaphase of control siRNA-treated cells. Error bars represent the SEM with a 95% confidence interval. Bars, 5 μm; inset in E, 1 μm.
Spindly mutants defective in dynein/dynactin recruitment persist on kinetochores of aligned chromosomes
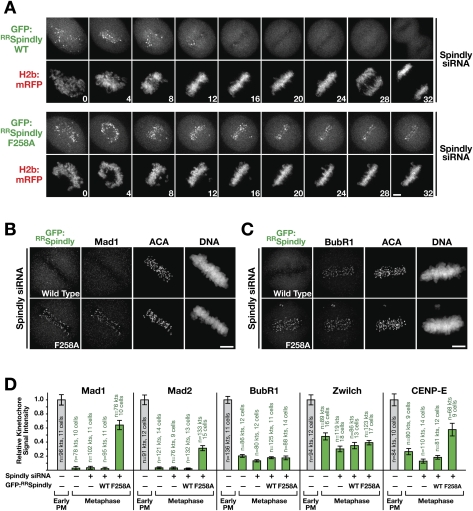
Checkpoint proteins were removed from attached bioriented kinetochores in Spindly-depleted cells, indicating the presence of a kinetochore dynein-independent mechanism for checkpoint protein loss. To test if the same mechanism can operate on Spindly itself in the absence of kinetochore dynein/dynactin, we monitored the localization of GFP:RRSpindlyWT and GFP:RRSpindlyF258A in a cell line stably coexpressing histone H2b:mRFP. In agreement with analysis of endogenous Spindly (Fig. 1A), GFP:RRSpindlyWT was removed rapidly from kinetochores during chromosome alignment and was undetectable by metaphase. In contrast, GFP:RRSpindlyF258A persisted at kinetochores even after all chromosomes had congressed to the metaphase plate (Fig. 4A). The same result was obtained in fixed analysis for GFP:RRSpindlyS256A (Supplemental Fig. S3A).
Figure 4.
Spindly motif mutants are retained together with Mad1 and Mad2 on kinetochores of aligned chromosomes. (A) Images from a time-lapse imaging sequence of cells expressing histone H2b:mRFP and either GFP:RRSpindlyWT or GFP:RRSpindlyF258A. Cells were treated with siRNAs for 32 h, and expression of the Spindly transgenes was induced for 16 h before filming. Bar, 5 μm. (B,C) Immunofluorescence images of cells with congressed chromosomes expressing GFP:RRSpindlyWT or GFP:RRSpindlyF258A stained for GFP and Mad1 (B) or BubR1 (C). Bars, 5 μm. (D) Quantitation of checkpoint protein levels at kinetochores of aligned chromosomes relative to unaligned kinetochores in early prometaphase control cells for the indicated cell lines, siRNA treatments, and transgenes.
One explanation for the persistence of the Spindly motif mutants at aligned kinetochores is that they exhibit a lower rate of exchange with the cytoplasmic pool, which would slow down their depletion following microtubule attachment. However, photobleaching analysis of GFP:RRSpindlyWT and the GFP:RRSpindlyF258A mutant in nocodazole-treated cells revealed similar turnover properties (Supplemental Fig. S3B–D). Thus, Spindly motif mutants, in contrast to wild-type Spindly, persist on kinetochores of aligned chromosomes, suggesting that Spindly requires kinetochore recruitment of dynein/dynactin to be removed following microtubule attachment.
Mad1 and Mad2 are retained on aligned kinetochores together with Spindly motif mutants
We next assessed the consequences of retaining Spindly motif mutants at aligned kinetochores by quantifying checkpoint protein levels at kinetochores. Spindly-depleted cells expressing the GFP:RRSpindlyF258A mutant retained significant levels of Mad1, Mad2, and CENP-E at aligned kinetochores (Figs. 4B,D, 5A). A similar result was obtained for the GFP:RRSpindlyS256A mutant (Supplemental Fig. S3A; data not shown). In contrast, increased retention was not observed for BubR1 or for Zwilch (Fig. 4C,D). Thus, single amino acid changes in Spindly that hamper its ability to recruit dynein/dynactin lead to the retention of a subset of checkpoint proteins, most notably the key checkpoint effectors Mad1 and Mad2, at kinetochores of congressed chromosomes. We conclude that the mechanism that facilitates checkpoint protein removal from aligned kinetochores in Spindly-depleted cells is suppressed when Spindly persists at kinetochores.
Figure 5.
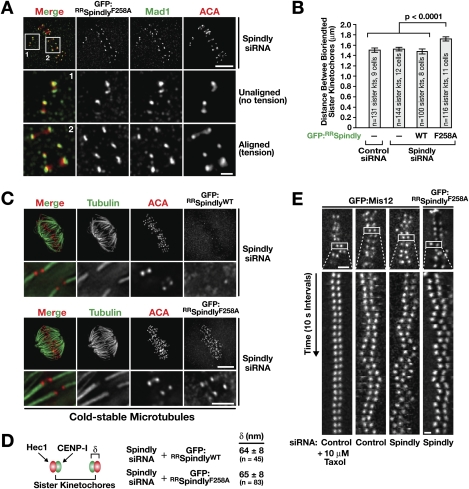
Aligned kinetochores retaining Spindly motif mutants have achieved stable, bioriented microtubule attachments. (A) Projection of three optical sections from an immunofluorescence Z-stack of a cell depleted of endogenous Spindly expressing GFP:RRSpindlyF258A stained for GFP and Mad1. Cells were treated with siRNAs for 32 h, and expression of the Spindly transgenes was induced for 16 h before fixation. Bar, 5 μm; inset, 1 μm. (B) Distance between the ACA signal of sister kinetochores at metaphase in the indicated states. Error bars represent the SEM with a 95% confidence interval. (C) Cold-stable kinetochore fibers visualized by immunofluorescence in cells depleted of endogenous Spindly expressing GFP:RRSpindlyWT or GFP:RRSpindlyF258A. Blowups of individual kinetochore fibers represent projections of selected sections of the image Z-stack. Bar, 5 μm; blowups, 2 μm. (D) Distance (δ) between the inner kinetochore component CENP-I and the outer kinetochore component Ndc80/Hec1, measured at bioriented metaphase kinetochores in the indicated states. The value for unperturbed metaphase kinetochores in HeLa cells was previously determined to be 62 ± 9 nm (Wan et al. 2009). Values are given as the mean ± standard deviation. (E) Kymographs of aligned sister kinetochore pairs marked by GFP:Mis12 (Kline et al. 2006) or GFP:RRSpindlyF258A after the indicated treatments. Bar, 2 μm; kymograph panels, 1 μm.
Aligned kinetochores harboring Spindly motif mutants have mature bioriented microtubule attachments
A straightforward explanation for the persistence of Mad1 and Mad2 at kinetochores of aligned chromosomes is that the presence of Spindly motif mutants causes defective kinetochore–microtubule attachments. To address this possibility, we probed the nature of microtubule attachment at aligned kinetochores harboring Spindly motif mutants using four independent criteria: sister kinetochore separation, kinetochore fiber formation, intrakinetochore stretch, and kinetochore motility on the spindle.
Aligned sister kinetochores enriched for Spindly motif mutants were clearly under tension, suggesting successful biorientation (Fig. 5A); in fact, sister kinetochore separation was slightly increased compared with bioriented kinetochores in cells expressing GFP:RRSpindlyWT, which experienced the same tension as sister kinetochores in control and Spindly-depleted cells (Fig. 5B). Consistent with the normal removal of BubR1 and Zwilch and the presence of tension, sister kinetochores harboring the GFP:RRSpindlyF258A mutant exhibited robust kinetochore fibers (Fig. 5C).
Recent work has suggested that checkpoint silencing correlates with increased physical separation of inner and outer kinetochore components following microtubule attachment (Maresca and Salmon 2009; Uchida et al. 2009). We therefore determined the distance between the inner kinetochore component CENP-I and the outer kinetochore component Hec1, which in unperturbed HeLa cells is 62 ± 9 nm at metaphase (Wan et al. 2009). We measured a similar distance between Hec1 and CENP-I at kinetochores of cells expressing GFP:RRSpindlyWT and the GFP:RRSpindlyF258A mutant (Fig. 5D). Thus, intrakinetochore stretch is not affected by the presence of the Spindly motif mutant, further indicating that the retention of Mad1 and Mad2 is not due to defective kinetochore–microtubule attachments.
Finally, we imaged aligned kinetochore pairs at high temporal resolution. Similar to controls, directional instability was observed for sister kinetochore pairs in GFP:RRSpindlyF258A-expressing cells, as well as in Spindly-depleted cells (Fig. 5E). In contrast, treatment with taxol, which perturbs kinetochore–microtubule interactions by stabilizing microtubules, abolished directional instability. Thus, kinetochores harboring a Spindly motif mutant exhibit mechanical behavior on the spindle, similar to kinetochores of control cells.
Cumulatively, the above experiments indicate that a defect in microtubule attachment is unlikely to be the cause for the persistence of Spindly motif mutants and spindle checkpoint proteins at kinetochores of aligned chromosomes. Instead, when Spindly cannot be removed by dynein/dynactin, it and a subset of checkpoint proteins are retained at kinetochores that have achieved mature bioriented microtubule attachments.
Spindly motif mutants, but not Spindly-depleted cells, exhibit a prolonged metaphase delay followed by aberrant mitotic progression
Having established that Spindly depletion and Spindly motif mutants have opposite effects on the removal of checkpoint proteins from bioriented kinetochores, we next examined the effect of these perturbations on mitotic progression.
As checkpoint proteins are released from attached kinetochores in Spindly-depleted cells, it is expected that the spindle checkpoint would be silenced once all chromosomes have aligned. This prediction is supported by progression into anaphase observed in Spindly-depleted cells in a prior study (Chan et al. 2009). To confirm this result under our experimental conditions, we filmed control and Spindly-depleted HeLa cells stably expressing histone H2b:YFP. Following a major delay in chromosome alignment, 94% of cells (n = 228) progressed to anaphase after spending ∼2.5-fold longer in a metaphase-like state relative to control cells (Supplemental Fig. S4A,C; Supplemental Movie 2). In a minority of cells (6%), chromosomes dispersed progressively from the metaphase plate and remained scattered for the duration of filming (see below). No significant defects were evident during chromosome segregation: Four percent of anaphases in Spindly-depleted cells exhibited lagging chromatids versus 2.7% in control cells, and the rate at which the chromosome masses separated was unaffected (Supplemental Fig. S4E). In agreement with eventual normal progression to anaphase, Spindly-depleted cells with mostly congressed chromosomes exhibited normal tension between sister kinetochores (Fig. 5B) and robust kinetochore fiber formation (Supplemental Fig S4F). These results indicate that, once Spindly-depleted cells have achieved complete chromosome alignment and established proper kinetochore–microtubule attachments, they silence the spindle checkpoint by a kinetochore dynein-independent mechanism and progress to a normal anaphase.
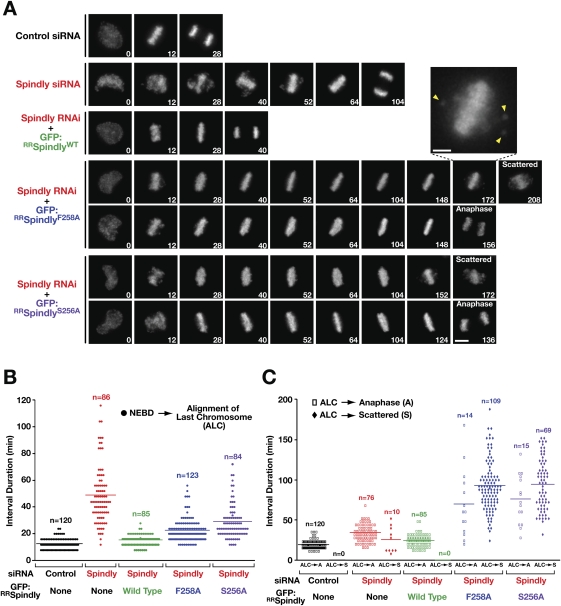
In contrast to what is observed in Spindly-depleted cells, the presence of Mad1 and Mad2 on bioriented kinetochores in cells expressing Spindly motif mutants predicts that checkpoint signaling persists despite congression of all chromosomes. We tested this by filming tetracycline-inducible HeLa Flp-In cell lines stably expressing histone H2b:mRFP. While the expression of Spindly motif mutants had no effect on cell viability in the presence of endogenous Spindly (Supplemental Table S1), the same mutants were toxic in Spindly-depleted cells (regardless of histone H2b:mRFP expression). Because a high proportion of viable interphase cells are needed at the start of time-lapse imaging experiments, we began filming 30 h after siRNA transfection, as opposed to the 48-h time point used in histone H2b:YFP imaging and all fixed cell analysis. Immunoblotting revealed a significant depletion of endogenous Spindly levels at the 30-h time point (Fig. 1E), and the Spindly depletion phenotype was generally similar to that observed at the 48-h time point (Supplemental Table S1).
Expression of GFP:RRSpindlyWT largely rescued endogenous Spindly depletion (Fig. 6A–C; Supplemental Table S1; Supplemental Movie S3). In contrast, Spindly-depleted cells expressing Spindly motif mutants spent an extended time in a metaphase-like state (Fig. 6A,C). The majority of cells (∼85%) eventually exhibited a chromosome “scattering” phenotype, characterized by partial loss of sister chromatid cohesion, high cyclin B1 levels, and terminal mitotic arrest (Supplemental Material; Supplemental Fig. S5; Supplemental Movies S4–S6). The cells that underwent scattering either stayed in this state for the duration of filming, or exhibited morphological changes characteristic of apoptosis, explaining the toxicity of the motif mutants following depletion of endogenous Spindly. The average time spent in metaphase with all chromosomes aligned before onset of scattering was 92 min and 94 min for GFP:RRSpindlyS256A and GFP:RRSpindlyF258A, respectively, compared with 16 min before anaphase onset in control cells and 34 min before anaphase onset in Spindly-depleted cells (Supplemental Table S1). Importantly, cells expressing nondegradable cyclin B1 from a single-copy integrated transgene, derived from the same parental line as the Spindly motif mutants, also scattered their chromosomes after an average of 87 min with all chromosomes aligned (Supplemental Table S1; Supplemental Movie S4). Therefore, the predominance of the scattering phenotype is likely due to the mitotic arrest induced by the Spindly motif mutants rather than a direct consequence of Spindly motif mutant expression (for detailed discussion of the scattering phenotype, see the Supplemental Material; Supplemental Fig. S5; Supplemental Movies S4–S6).
Figure 6.
Differential effects of Spindly motif mutants and Spindly depletions on chromosome alignment and spindle checkpoint silencing. (A) Selected images from a time-lapse series of cells expressing histone H2b:mRFP with or without Spindly transgenes (see also Supplemental Movie S3). The experimental protocol prior to the start of filming was identical to that for the immunoblot in Figure 1E. A blowup shows the onset of the scattering phenotype (see also the Supplemental Material; Supplemental Fig. S5; Supplemental Movies S4–S6). Bar, 5 μm; blowup, 2 μm. (B,C) Quantitative analysis of mitotic intervals for the experimental conditions shown in A. Interval averages are marked by horizontal bars (see also Supplemental Table S1). Two independent experiments were performed for each condition, and the number (n) of cells scored is indicated.
We conclude that, while Spindly depletion and Spindly motif mutants both affect kinetochore dynein/dynactin recruitment and poleward transport, only the Spindly motif mutants affect Mad1/Mad2 removal from aligned bioriented kinetochores and block progression to anaphase.
Spindly motif mutants significantly rescue the chromosome alignment defect observed in Spindly-depleted cells
The primary defect observed in Spindly depletions is in chromosome alignment: Quantitative analysis revealed that Spindly-depleted cells expressing histone H2b:YFP and imaged 48 h after siRNA transfection took, on average, six times longer than control cells to align all chromosomes at the metaphase plate (Supplemental Fig. S4A,C; Supplemental Movie S2); cells expressing histone H2b:mRFP imaged 30 h after siRNA transfection took four times longer (Fig. 6A,B; Supplemental Table S1).
The last few chromosomes to align in Spindly-depleted cells often displayed no directional movement for extended periods of time (Supplemental Fig. S4A,B; Supplemental Movie S2), and both the distribution of these chromosomes and the orientation of their sister kinetochores were random relative to the spindle axis (Fig. 3A–E; Supplemental Fig. S4D). Furthermore, both kinetochores of these unaligned chromosomes exhibited molecular signatures of being unattached: They had similar levels of checkpoint proteins as in early prometaphase (Fig. 3A–C) and, in the case of Zwilch and CENP-E, even showed significantly increased levels (Fig. 3D,E). Analysis of kinetochore composition in Spindly-depleted cells (Chan et al. 2009; this study), has not revealed a significant difference in kinetochore composition, aside from loss of dynein/dynactin; all 13 tested components (Hec1/Ndc80, CENP-E, CENP-F, Bub1, BubR1, Zwilch, Zw10, MCAK, Aurora B, Ska1, Nde1, Mad1, and Mad2) were localized normally. This observation is consistent with the result that all kinetochores in Spindly-depleted cells eventually make normal attachments and progress into anaphase without significant defects (Supplemental Fig. S4A,C). Thus, the delayed chromosome alignment in Spindly-depleted cells is due to a kinetic defect in microtubule association with kinetochores that results in a few chromosomes being trapped in an unattached state on the spindle.
We next compared the kinetics of chromosome alignment between Spindly depletions and the Spindly motif mutants, both of which prevented recruitment of dynein/dynactin to kinetochores. Surprisingly, the Spindly motif mutants significantly ameliorated the chromosome alignment defect of Spindly-depleted cells (Fig. 6A,B; Supplemental Table S1; Supplemental Movie S3). Thus, the inefficient capture of microtubules and delayed chromosome alignment observed in Spindly-depleted cells cannot be explained solely by a lack of kinetochore-localized dynein/dynactin. In addition, the significant rescue of the alignment defect in Spindly depletions by the Spindly motif mutants argues against the possibility that the motif mutants simply exacerbate the depletion and cause a more penetrant Spindly loss-of-function phenotype. We conclude that a dynein/dynactin recruitment-independent function of Spindly at kinetochores contributes to the efficient alignment of chromosomes in prometaphase.
Discussion
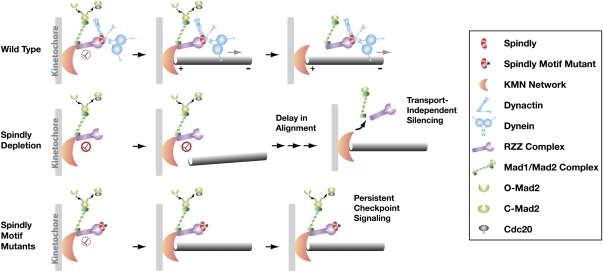
The motor dynein was the first microtubule-associated protein localized to the kinetochore region of mitotic chromosomes (Pfarr et al. 1990; Steuer et al. 1990). The significance of dynein localization at this site has remained a topic of intensive study. Direct perturbations of dynein and its cofactor, dynactin, have revealed a role in silencing of the mitotic checkpoint through removal of checkpoint signaling complexes (Howell et al. 2001). Dynein is also implicated in initial capture of microtubules, but its function in chromosome alignment and segregation is debated (Howell et al. 2001; Z Yang et al. 2007). Here, we confirm in human cells that dynein/dynactin recruitment and function at kinetochores requires the conserved Spindly protein. Our results revealed striking differences between removal of Spindly and single amino acid changes in the highly conserved Spindly motif—two perturbations that prevent recruitment of dynein/dynactin to kinetochores (Fig. 7). The observed differences indicate that the key step in checkpoint silencing in human cells is dynein-dependent removal of Spindly from microtubule-attached kinetochores. In addition, our results highlight the existence of a conserved dynein/dynactin-independent mechanism involving Spindly that is important for chromosome alignment during prometaphase.
Figure 7.
Model explaining the different consequences of Spindly depletion and Spindly motif mutants. Spindly depletion and replacement of endogenous Spindly with Spindly motif mutants abrogate recruitment of dynein/dynactin to kinetochores, but have differential effects on chromosome alignment and checkpoint silencing. See the text for details.
Single amino acid substitutions in the Spindly motif: a precise means of preventing dynein/dynactin recruitment to kinetochores
Spindly targets dynein/dynactin specifically to kinetochores in C. elegans embryos and human cells (Gassmann et al. 2008; Chan et al. 2009); in D. melanogaster, dynactin recruitment to kinetochores has been suggested to be Spindly-independent (Griffis et al. 2007). In all organisms where it has been analyzed, Spindly is recruited to kinetochores by the heterotrimeric RZZ complex. Weak association between Spindly and RZZ subunits has been reported in both C. elegans and human cells, but the interacting regions remain to be defined. Spindly family proteins are primarily predicted coiled-coil, with the only conserved sequence feature being a short motif located near a break in the coiled-coil. Our results establish that the conserved Spindly motif is central to the kinetochore recruitment of dynein/dynactin. Motif mutants are normally kinetochore-localized and exhibit turnover properties similar to those of wild-type Spindly, but fail to recruit dynein/dynactin. Whether the Spindly motif is sufficient for dynein/dynactin recruitment is not clear, and the target of this motif on dynein/dynactin remains to be defined. The multisubunit structure and large size of both dynein and dynactin has challenged efforts to understand cargo interactions of this widely used motor complex. The requirement for Spindly to target dynein/dynactin specifically to kinetochores, and the subtle change in the Spindly motif that dramatically affects this localization, will provide a precise tool to elucidate the mechanism targeting dynein/dynactin to kinetochores in future work.
Checkpoint silencing in the absence of Spindly and kinetochore-localized dynein
Mad1/Mad2 kinetochore localization is correlated with the generation of a checkpoint signal, and dynein-mediated stripping of Mad1/Mad2 is the only mechanism proposed to directly link attachment status with checkpoint silencing. Two other mechanisms, involving the Mad2 mimic p31comet (Habu et al. 2002; M Yang et al. 2007) and protein phosphatase 1 (PP1) (Pinsky et al. 2009; Vanoosthuyse and Hardwick 2009a), have also been linked to checkpoint silencing. p31comet does not appear to be conserved in all species with Mad2-like proteins (Habu et al. 2002), and the involvement of PP1 in checkpoint silencing has only recently been demonstrated in fungi (Pinsky et al. 2009; Vanoosthuyse and Hardwick 2009a). Importantly, whether the p31comet and PP1-dependent mechanisms are sensitive to kinetochore–microtubule attachment status is currently unclear, and it is possible that these mechanisms operate to limit or inactivate the checkpoint signal in the cytoplasm (Vanoosthuyse and Hardwick 2009b). For PP1, one study has suggested a potential role in dynein-mediated removal of checkpoint proteins (Whyte et al. 2008). However, in this study, PP1 was globally inhibited using a dominant-negative mutant, and whether checkpoint silencing was prevented was not addressed.
In Spindly-depleted cells, where kinetochore dynein is absent, Mad1 and Mad2 dissociate from kinetochores following microtubule attachments, and the spindle checkpoint is silenced without poleward transport (Fig. 7). Thus, there exists a kinetochore dynein-independent mechanism capable of promoting attachment status-dependent removal of checkpoint proteins from kinetochores. Kinetochore dynein/dynactin is absent in fungi with closed mitoses, and dynein/dynactin has been lost altogether in higher plants (Yeh et al. 1995; Wickstead and Gull 2007). Database searches failed to reveal Spindly or RZZ orthologs in these species, with the exception of Zw10 (Starr et al. 1997), which performs double duty as a subunit of a distinct complex involved in membrane trafficking (Hirose et al. 2004). We speculate that, when Spindly is depleted from cells, as well as in organisms that naturally lack the Spindly–RZZ–dynein/dynactin kinetochore module, the spindle checkpoint is silenced via the KMN network, which provides the core microtubule-binding activity of the kinetochore and acts as the platform for spindle checkpoint activation (Kiyomitsu et al. 2007; Burke and Stukenberg 2008; Essex et al. 2009). We suggest that microtubule engagement by the KMN network has the capacity to feed back on the checkpoint activation reaction(s), and thereby couple silencing to attachment. In support of this idea, in budding yeast a specific mutant allele of the Ndc80 subunit of the KMN network constitutively activates the checkpoint without affecting kinetochore–microtubule interactions (Kemmler et al. 2009).
Dynein-mediated removal of Spindly from kinetochores: the key step in silencing the spindle checkpoint in metazoans
The failure of checkpoint silencing in Spindly motif mutants despite the presence of mature bioriented attachments suggests that, when Spindly is present at kinetochores, the dynein-independent mechanism for silencing is ineffective, and dynein-mediated removal of Mad1 and Mad2 becomes essential for checkpoint silencing (Fig. 7). Timely removal of Spindly from attached kinetochores depends on poleward transport by kinetochore-localized dynein/dynactin, in agreement with previous work showing that direct inhibitions of dynein/dynactin result in Mad2 retention at bioriented kinetochores (Howell et al. 2001; Wojcik et al. 2001; Vergnolle and Taylor 2007; Mische et al. 2008; Varma et al. 2008; Chan et al. 2009; Sivaram et al. 2009).
Why has the dynein-independent checkpoint silencing mechanism been supplanted in metazoans by Spindly and dynein-dependent poleward transport? We note that, despite dissociation of checkpoint proteins from bioriented kinetochores, Spindly-depleted cells remain in metaphase with all chromosomes aligned ∼2.5 times longer than control cells. While we cannot exclude the possibility that some of the aligned kinetochores have aberrant microtubule attachments that produce a residual checkpoint signal, an attractive alternative explanation for the extended metaphase state is that dynein-mediated poleward transport promotes a switch-like transition into anaphase not only by removing checkpoint signaling complexes from kinetochores, but also by transporting them to a site that efficiently deactivates them. Consistent with this view, prior work has shown that reactions governing anaphase entry are spatially localized on the spindle (Clute and Pines 1999; Huang and Raff 1999; Raff et al. 2002).
The observation that Spindly, the RZZ complex, and Mad1/Mad2 all move to the poles suggests that these proteins interact directly with each other to form a motor–cargo complex for poleward transport. The RZZ complex is required to recruit Mad1/Mad2 to kinetochores throughout metazoans (Buffin et al. 2005; Kops et al. 2005; Gassmann et al. 2008; Yamamoto et al. 2008), although no evidence for a direct interaction between the RZZ complex and Mad1/Mad2 has been reported. In C. elegans, SpindlySPDL-1 is also required to recruit Mad1/Mad2 to kinetochores (Gassmann et al. 2008; Yamamoto et al. 2008), and Mad1/Mad2 can be immunoprecipitated with SpindlySPDL-1 (Yamamoto et al. 2008). Although human Spindly is dispensable for initial Mad1/Mad2 recruitment, it controls Mad1/Mad2 release after microtubule attachment. Thus, the contrasting effect of Spindly depletion on Mad1/Mad2 recruitment in C. elegans versus other organisms is likely to reflect variations on a similar underlying mechanism that involves direct physical connections between Spindly, the RZZ complex, and Mad1/Mad2. The definition of these physical interactions is a central goal of future work.
Cross-talk between the RZZ–Spindly–dynein/dynactin module and the KMN network during chromosome alignment
The most striking phenotype of Spindly-depleted cells is a prolonged delay in chromosome alignment. Spindly-depleted kinetochores are slow to make productive attachments to microtubules. This reflects a kinetic delay rather than a permanent impairment, as is observed after inhibition of KMN network components. Dynein is proposed to accelerate the establishment of kinetochore–microtubule attachments (Rieder and Alexander 1990; Z Yang et al. 2007; Gassmann et al. 2008; Vorozhko et al. 2008), so it is tempting to conclude that the delayed chromosome alignment in Spindly depletions is due to the lack of kinetochore dynein. However, Spindly motif mutants defective in dynein and dynactin recruitment significantly ameliorate the alignment defect observed in Spindly-depleted cells (Fig. 7). The motif mutants lead to increased sister kinetochore separation, an effect observed previously following injection of a monoclonal antibody targeting the microtubule-binding Hec1(Ndc80) subunit of the KMN network (DeLuca et al. 2006). These findings are reminiscent of prior work in the C. elegans embryo, where different phenotypic outcomes of Spindly and RZZ complex inhibitions provided evidence for a kinetochore dynein recruitment-independent role of the Spindly–RZZ complex in regulating the activity of the KMN network (Gassmann et al. 2008). These results, together with the contrasting effects on checkpoint silencing observed in Spindly depletions and Spindly motif mutants, raise caution against interpreting the phenotypic consequences of perturbing Spindly/RZZ as solely reflecting dynein/dynactin function at kinetochores. Cumulatively, the work in both C. elegans embryos and human cells suggests that there is cross-talk at the kinetochore between the Spindly–RZZ–dynein/dynactin module and the KMN network that is important for establishing timely kinetochore–microtubule attachments and promoting rapid chromosome alignment and biorientation during prometaphase. Thus, by linking checkpoint activation, efficient alignment, and checkpoint silencing following microtubule attachment, the Spindly–RZZ–dynein/dynactin module ensures both the rapid kinetics and the high fidelity of chromosome segregation in metazoans.
Materials and methods
Cells lines and antibodies
Stable isogenic cell lines expressing Spindly constructs, the CENP-ETail fragment, and nondegradable cyclin B1 (lacking the N-terminal 86 amino acids) were generated by FRT/Flp-mediated recombination as described previously (Tighe et al. 2004). Full-length Spindly cDNA and cDNA corresponding to amino acids 1569–2603 of CENP-E were cloned into a pcDNA5/FRT/TO-based vector (Invitrogen) modified to contain an N-terminal Myc-LAP epitope tag. The LAP tag consists of GFP-TEV-S-peptide (Cheeseman et al. 2004). The Δ86 cyclin B1 construct was cloned into pcDNA5/FRT/TO with an N-terminal Myc tag. For Spindly constructs, site-directed mutagenesis (QuickChange, Stratagene) was used to introduce four silent mutations conferring RNAi resistance (gaaGggAtcCcaGactgaa; changes in capital letters), and to generate the S256A and F258A mutants by changing the appropriate codons to GCT. Vectors were cotransfected into HeLa or DLD-1 Flp-In T-Rex cells (a kind gift from Steven S. Taylor, University of Manchester, UK) with pOG44 encoding the Flp recombinase. After selection in hygromycin, colonies were pooled and transgene expression was induced with 0.2 μg/mL tetracycline. HeLa and DLD-1 Flp-In T-Rex cell lines stably expressing histone H2b:mRFP and HeLa cells stably expressing YFP:α-tubulin were generated by retroviral delivery as described previously (Shah et al. 2004).
Affinity-purified antibodies against full-length Spindly and Zwilch were generated as described previously (Desai et al. 2003).
Cell culture and RNAi
Cells were maintained at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% tetracycline-free fetal bovine serum (Clontech), 100 U/mL penicillin, 100 U/mL streptomycin, and 2 mM L-glutamine. For immunofluorescence, cells were seeded on 12-mm poly-L-lysine-coated coverslips in 12-well plates 24 h prior to transfection with siRNAs. For live-cell imaging experiments, cells were seeded in a 35-mm glass-bottom dish coated with poly-D-lysine (MatTek). Cells were transfected using Oligofectamine and reduced-serum Opti-MEM (Invitrogen) according to the manufacturer's instructions. A predesigned (Thermo Scientific) siRNA for Spindly (GAAAGGGUCUCAAACUGAA) or a nontargeting control siRNA (UGGUUUACAUGUCGACUAA) was used at a final concentration of 100 nM. After incubation for 5–6 h, 1 vol of medium and fetal bovine serum (10% final) was added. After 24 h, the transfection mixture was replaced with fresh medium. For immunofluorescence of HeLa Flp-In T-Rex cells, transgene expression was induced with tetracycline 24 h post-transfection and cells were fixed 20–24 h later. For live-cell imaging of HeLa Flp-In T-Rex cells (and for the immunoblot shown in Fig. 1E), transgene expression was induced 22 h after transfection, and the filming session was initiated 8 h later.
Live-cell imaging
For live-cell imaging, medium was replaced with CO2-independent medium (Gibco) supplemented as described above. Tetracycline (0.2 μg/mL) was added to Flp-In T-Rex cells to maintain transgene expression, and the medium was covered with mineral oil immediately before filming. Detailed information about imaging conditions for individual cell lines is provided in Supplemental Table S3.
Indirect immunofluorescence and fixed-cell assays
The ATP reduction assay was performed as described previously for PtK2 cells (Howell et al. 2000), with the exception that the incubation time in azide/deoxyglucose was reduced from 30 min to 10 min. To visualize kinetochore fibers, cells were treated as described in Lampson and Kapoor (2005). For immunofluorescence, cells were fixed immediately after aspiration of the medium with 4% formaldehyde in Phem buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgCl2 at pH 6.9) for 5 min at room temperature, then permeabilized for 2 min with 0.1% Triton X-100 in Phem buffer and rinsed three times in Phem buffer. Alternatively, cells were fixed at −20°C in methanol for 45 min, then rehydrated twice for 5 min in phosphate-buffered saline. Cells were processed further as described previously (Kline et al. 2006). Primary antibody information is listed in Supplemental Table S2. Images were recorded on a DeltaVision microscope at 1 × 1 binning with a 100× NA 1.3 U-planApo objective (Olympus). Z-stacks (0.2-μm sections) were deconvolved using softWoRx (Applied Precision), and maximum intensity projections were imported into Adobe Photoshop CS4 (Adobe) for further processing. For quantitation of kinetochore signals, 0.5 μm Z-stacks were acquired at 1 × 1 binning using the 100× NA 1.3 U-planApo objective. Maximum intensity projections of five to 10 Z-sections of the primary 12-bit image were analyzed with MetaMorph software as described in detail by Hoffman et al. (2001). Cells were costained with ACA for definition of kinetochore regions, which were then transferred to the other channels for intensity measurements. Interkinetochore stretch was determined for sister ACA spots whose maximum intensities were in the same Z-plane using the “Measure Distance” tool in softWoRx. Intrakinetochore stretch was measured between CENP-I (Rhodamine Red-X) and Hec1 (Cy5) using previously published methods (Wan et al. 2009).
Acknowledgments
We thank Andrea Musacchio, Kevin T. Vaughan, Song-Tao Liu, and Stephen S. Taylor for antibodies; Stephen S. Taylor for the parental DLD-1 and HeLa Flp-In T-Rex cell lines; Jennifer Meerloo of the University of California at San Diego Neuroscience Microscopy Shared Facility (NINDS P30 NS047101) for help with live-cell imaging; and Jagesh Shah as well as members of the Desai, Cleveland, and Salmon laboratories for helpful discussions. This work was supported by a National Science Foundation of Switzerland fellowship (R.G.) and a European Molecular Biology Organization (EMBO) Long-Term Fellowship (A.J.H.); the Nando Peretti Foundation (F.Ç.); grants from the NIH to A.D. (GM074215), D.W.C. (GM29513), and E.D.S. (GM024364); and funding from the Ludwig Institute for Cancer Research to A.D., K.O., and D.W.C.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1886810.
Supplemental material is available at http://www.genesdev.org.
References
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe JC 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 106: 83–93 [DOI] [PubMed] [Google Scholar]
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess R 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15: 856–861 [DOI] [PubMed] [Google Scholar]
- Burke D, Stukenberg PT 2008. Linking kinetochore-microtubule binding to the spindle checkpoint. Dev Cell 14: 474–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GK, Schaar BT, Yen TJ 1998. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol 143: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y, Fava LL, Uldschmid A, Schmitz MH, Gerlich DW, Nigg E, Santamaria A 2009. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J Cell Biol 185: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A 2008. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates J, Oegema K, Desai A 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes & Dev 18: 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P, Pines J 1999. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol 1: 82–87 [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol 15: 214–225 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127: 969–982 [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Müller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes & Dev 17: 2421–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex A, Dammermann A, Lewellyn L, Oegema K, Desai A 2009. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol Biol Cell 20: 1252–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Essex A, Hu JS, Maddox P, Motegi F, Sugimoto A, O'Rourke S, Bowerman B, McLeod I, Yates J, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes & Dev 22: 2385–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu T, Kim SH, Weinstein J, Matsumoto T 2002. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J 21: 6419–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose H, Arasaki K, Dohmae N, Takio K, Hatsuzawa K, Nagahama M, Tani K, Yamamoto A, Tohyama M, Tagaya M 2004. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J 23: 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon E 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12: 1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon E 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol 150: 1233–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon E 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155: 1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon E 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol 14: 953–964 [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517 [DOI] [PubMed] [Google Scholar]
- Huang J, Raff JW 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J 18: 2184–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW 1998. Budding yeast Cdc20: A target of the spindle checkpoint. Science 279: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J 2009. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J 28: 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell 13: 663–676 [DOI] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 173: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Kim Y, Weaver BA, Mao Y, Mcleod I, Yates J, Tagaya M, Cleveland D 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol 169: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M, Kapoor T 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol 7: 93–98 [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW 1991. Feedback control of mitosis in budding yeast. Cell 66: 519–531 [DOI] [PubMed] [Google Scholar]
- Luo X, Yu H 2008. Protein metamorphosis: The two-state behavior of Mad2. Structure 16: 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H 2004. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol 11: 338–345 [DOI] [PubMed] [Google Scholar]
- Mapelli M, Musacchio A 2007. MAD contortions: Conformational dimerization boosts spindle checkpoint signaling. Curr Opin Struct Biol 17: 716–725 [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED 2009. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol 184: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische S, He Y, Ma L, Li M, Serr M, Hays TS 2008. Dynein light intermediate chain: An essential subunit that contributes to spindle checkpoint inactivation. Mol Biol Cell 19: 4918–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED 2007. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nicklas RB 1997. How cells get the right chromosomes. Science 275: 632–637 [DOI] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J 2008. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol 10: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Nelson CR, Biggins S 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol 19: 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR 1990. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 345: 263–265 [DOI] [PubMed] [Google Scholar]
- Raff JW, Jeffers K, Huang JY 2002. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J Cell Biol 157: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol 110: 81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol 130: 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A 2009. The life and miracles of kinetochores. EMBO J 28: 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaërou F, Starr DA, Piano F, Papoulas O, Karess RE, Goldberg ML 2001. The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J Cell Sci 114: 3103–3114 [DOI] [PubMed] [Google Scholar]
- Shah J, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland D 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol 14: 942–952 [DOI] [PubMed] [Google Scholar]
- Sivaram MV, Wadzinski TL, Redick SD, Manna T, Doxsey SJ 2009. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J 28: 902–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Li Z, Etemad-Moghadam B, Dawe RK, Goldberg ML 1997. Conservation of the centromere/kinetochore protein ZW10. J Cell Biol 138: 1289–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Hays TS, Goldberg ML 1998. ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol 142: 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer ER, Wordeman L, Schroer TA, Sheetz MP 1990. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature 345: 266–268 [DOI] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 154: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Taylor SS 2004. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci 117: 6339–6353 [DOI] [PubMed] [Google Scholar]
- Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T 2009. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol 184: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Hardwick KG 2009a. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol 19: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Hardwick KG 2009b. Overcoming inhibition in the spindle checkpoint. Genes & Dev 23: 2799–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Monzo P, Stehman SA, Vallee RB 2008. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J Cell Biol 182: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle MA, Taylor SS 2007. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr Biol 17: 1173–1179 [DOI] [PubMed] [Google Scholar]
- Vorozhko V, Emanuele M, Kallio M, Stukenberg PT, Gorbsky G 2008. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma 117: 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O'Quinn R, Pierce H, Joglekar AP, Gall W, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell 137: 672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland D 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol 162: 551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J, Bader J, Tauhata SB, Raycroft M, Hornick J, Pfister KK, Lane WS, Chan GK, Hinchcliffe E, Vaughan PS, et al. 2008. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J Cell Biol 183: 819–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K 2007. Dyneins across eukaryotes: A comparative genomic analysis. Traffic 8: 1708–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Karr TL, Montgomery JM, Goldberg ML 1992. The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J Cell Biol 118: 759–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E, Basto R, Serr M, Scaërou F, Karess R, Hays T 2001. Kinetochore dynein: Its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol 3: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Watanabe S, Essex A, Kitagawa R 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J Cell Biol 183: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X 2007. p31comet blocks Mad2 activation through structural mimicry. Cell 131: 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder C 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol 17: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol 130: 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]