Abstract
Background
Nkx2.5 is a homeodomain-containing nuclear transcription protein that has been associated with acute T-lymphoblastic leukemia. In addition, Nkx2.5 has an essential role in cardiomyogenesis. However, the expression of Nkx2.5 in the skin has not been investigated.
Objective
In an attempt to screen the differentially regulated genes involved in keratinocyte differentiation, using a cDNA microarray, we identified Nkx2.5 as one of the transcription factors controlling the expression of proteins associated with keratinocyte differentiation.
Methods
To investigate the expression of Nkx2.5 during keratinocyte differentiation, we used a calcium-induced keratinocyte differentiation model.
Results
RT-PCR and Western blot analysis revealed that the expression of Nkx2.5, in cultured human epidermal keratinocytes, increased with calcium treatment in a time-dependent manner. In normal skin tissue, the expression of Nkx2.5 was detected in the nuclei of the keratinocytes in all layers of the epidermis except the basal layer by immunohistochemistry. In addition, the expression of Nkx2.5 was significantly increased in psoriasis and squamous cell carcinoma, but was barely detected in atopic dermatitis and basal cell carcinoma.
Conclusion
These results suggest that Nkx2.5 may play a role in the change from proliferation to differentiation of keratinocytes and in the pathogenesis of skin disease with aberrant keratinocyte differentiation.
Keywords: Keratinocyte differentiation, Nkx2.5, Transcription factor
INTRODUCTION
Keratinocytes provide the rigid stratified structure of the skin through a highly complicated and tightly regulated process of differentiation1. Many differentiation-related genes, including those encoding transglutaminase 1 and 3 (TGase 1 and 3), involucrin, cornifin, loricrin, filaggrin, and small proline-rich proteins (SPRs), have been shown to be expressed in a temporally regulated manner during keratinocyte differentiation2-6. Despite intensive investigation to identify and determine the regulation of numerous candidate genes, the precise gene expression profile that governs the keratinocyte differentiation process remains to be determined. Recently, several molecular techniques such as cDNA microarray have enabled investigators to analyze global gene expression profiles. Using this technique, we attempted to identify the genes related to keratinocyte differentiation.
After we isolated hundreds of genes that may play a role during keratinocyte differentiation, we analyzed the promoter sequences of some of the differentially-regulated genes, and identified several candidate transcription factors for gene regulation during calcium-induced keratinocyte differentiation. Among them, we focused our attention on one interesting transcription factor, Nkx2.5, which has previously been designated as a cardiac specific homeobox protein (Csx) belonging to the gene family of Nkx2 of homeodomain-containing nuclear transcription factors7,8. Nkx-2.5 is a homolog of the Tinman protein expressed in Drosophila, and is essential for normal cardiomyogenesis; it is required for cardiac septation in which a single atrium and ventricle are separated into four chambers. Mutations that disrupt Nkx2.5 can result in atrial-septal defects, embryonic lethality and a variety of congenital heart abnormalities9-11. Nkx2.5 has also been associated with acute T-lymphoblastic leukemia12-14. However, the expression and putative role of Nkx2.5 has not been previously studied in skin keratinocytes.
In this study, we investigated the expression of Nkx2.5 during calcium-induced keratinocyte differentiation under in vitro culture conditions and in vivo with several skin diseases. The results suggest that Nkx2.5 may play a role in the change from proliferation to differentiation in keratinocytes and in the pathogenesis of skin diseases with aberrant keratinocyte differentiation.
MATERIALS AND METHODS
Patients and tissue specimens
Skin samples were obtained from normal donors and from patients with a variety of skin diseases including psoriasis, atopic dermatitis, actinic keratosis, basal cell carcinoma (BCC) and squamous cell carcinoma (SCC). All specimens were obtained after informed consent was obtained, in accordance with the ethics committee approval process of Chungnam National University, College of Medicine, Daejeon, Korea.
Cell culture
Normal human skin samples were briefly sterilized in 70% ethanol, minced, and then treated with dispase overnight at 4℃. The epidermis was separated and placed in a solution containing 0.05% trypsin and 0.025% EDTA at 37℃ for 15 min. After vigorous pipetting, the cells were pelleted and resuspended in serum-free keratinocyte growth medium (KGM) supplemented with bovine pituitary extract, recombinant human epidermal growth factor, insulin, and hydrocortisone (Clonetics, Walkersville, MD, USA). The cells were added to 100 mm collagen-coated dishes, incubated at 37℃ in 5% CO2. At 70~80% confluence, the cells were passaged.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated using the easy-BLUE™ Total RNA Isolation system kit (INTRON) following the manufacture's protocols. The quantity and quality of RNA were assessed by spectrometer and agarose gel electrophoresis. Two µg of total RNAs were reverse transcribed with M-MLV reverse transcriptase (Promega, Madison, WI, USA). Aliquots of RT mixture were subjected to PCR cycles with specific primer pairs derived from the corresponding GenBank sequences as follows: 94℃ for 30 s, 57℃ for 30 s, and 72℃ for 1 min for 30 cycles. Primers amplifying a control fragment from cyclophilin were included in each reaction.
Western blot analysis
Semi-confluent cells growing in serum-free growth medium were suspended in trypsin/EDTA and centrifuged. The cell pellet was washed in ice-cold PBS, suspended in PRO-PREP™ protein extraction solution (iNtRON, Korea), and boiled briefly. The supernatant was collected and the protein concentration was determined with the BioRad assay (BioRad, Hercules, CA, USA). Typically, 20 or 30 µg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Pall corporation nitrocellulose membranes (Life sciences, FL, USA), and blocked by PBST with 5% non-fat milk. Protein was detected with peptide polyclonal anti-Nkx2.5 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Anti-species horseradish peroxidase-conjugated secondary antibodies were obtained from Santa Cruz (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and visual detection was performed using the enhanced chemoluminescence method (iNtRON, Korea).
Immunohistochemical staining
Paraffin sections of normal and affected human skin were de-waxed and re-hydrated, washed three times with phosphate-buffered saline, and treated with proteinase K (DAKO ready to use kit) for 5 min at 37℃, followed by the DAKO LSAV 2 System peroxidase kit. Briefly, sections were washed with phosphate-buffered saline 0.1% Tween 20 and treated with H2O2 for 10 min at room temperature (RT), blocked in phosphate-buffered saline with 0.1% Tween 20 and 1% bovine serum albumin for 20 min at RT, and washed three times with phosphate-buffered saline with 0.1% Tween 20, followed by reaction with anti-Nkx2.5 antibodies (1:200 dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4℃. After washing and color detection, the sections were counter stained with hematoxylin and mounted. Immunostaining was visualized and photographed under the microscope (Leica DMR). Sections without primary antibody were used as negative controls.
RESULTS
Reverse transcription-polymerase chain reaction (RT-PCR)
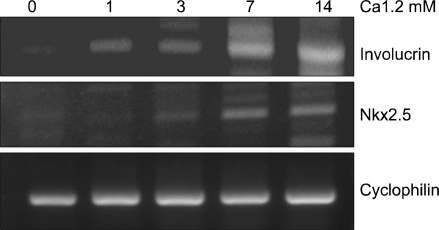
We used primary cultured human epidermal keratinocytes as a model system for investigating calcium-induced differentiation of keratinocytes. In the cultured keratinocytes, total RNA was isolated at four different times (1, 3, 7 and 14 days) after treatment with 1.2 mM calcium. We analyzed the mRNA level of Nkx2.5 during keratinocyte differentiation using RT-PCR. Involucrin was used as a terminal differentiation marker. Cyclophilin was used to compare the total amounts as an internal control (Fig. 1). Nkx2.5 mRNA was detected at 7 and 14 days after treatment with calcium of the NHK cultured in vitro. Therefore, Nkx2.5 appeared to increase in the upper layer of epidermis. However, it was expressed earlier than involucrin, a well known marker of early differentiation, suggesting that Nkx2.5 might be associated with early differentiation in the epidermis.
Fig. 1.
mRNA level of Nkx2.5 during calcium-induced keratinocyte differentiation. Two micrograms of total RNA were reverse transcribed with M-MLV reverse transcriptase and used for PCR amplification. Involucrin used as a control marker for differentiation was also increased with calcium treatment.
Western blot analysis
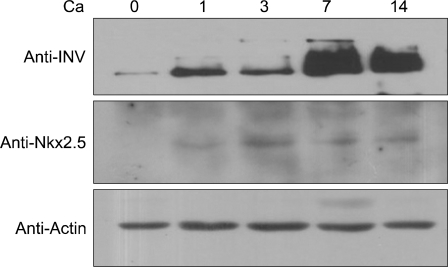
To investigate Nkx2.5 expression at the protein level, we performed immune blot analysis. Western blot analysis showed that the Nkx2.5 protein began to be expressed 1-day after calcium treatment and reached maximum levels at 3-days; then, its expression was slightly decreased but maintained until 14 days (Fig. 2). Involucrin used as a control marker for differentiation was also increased with calcium treatment. Actin was used as a loading control. These results indicated that this protein was involved in the early events of keratinocyte differentiation and in maintaining their differentiation before death to become the cornified layer.
Fig. 2.
Nkx2.5 expression in calcium-induced keratinocyte differentiation. The protein from cultured keratinocyte extracts of 20 or 30 µg was loaded for SDS-PAGE, and was detected with peptide polyclonal anti-Nkx2.5 antibody, anti-involucrin. Secondary antibodies were obtained from Santa Cruz (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Involucrin used as a control marker for differentiation was also increased with calcium treatment. Actin was used as a loading control.
Immunohistochemical staining
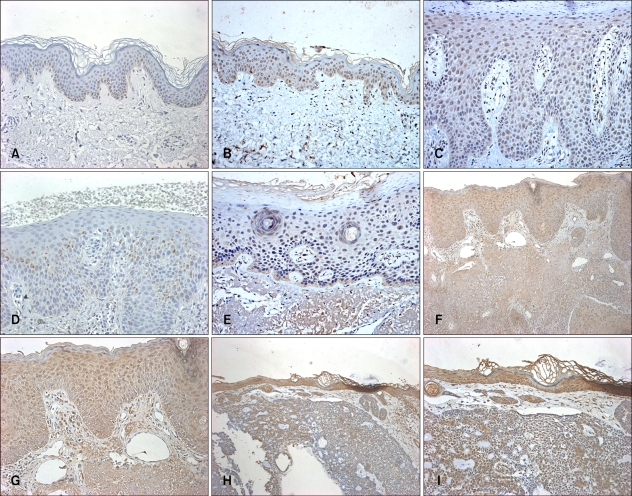
To verify expression of Nkx2.5 protein in the epidermis, we performed immunohistochemical staining in skin tissues including normal skin, inflammatory skin diseases and malignant tumors (Fig. 3). Nkx2.5 expression was generally observed in the tissues with various skin diseases as well as normal skin. In normal tissue, the nuclei were shown to be stained entirely in almost all layers of the epidermal keratinocytes except the basal layer. For the inflammatory diseases of the skin, two representative diseases including psoriasis and atopic dermatitis showed opposite patterns of Nkx2.5 expression in the epidermal keratinocytes. In psoriasis, not only the nuclei but also the cytoplasm was stained strongly in the epidermis, and these results were more significant in the upper layer. However, for atopic dermatitis, there was almost no staining in the epidermal keratinocytes except for a mild staining in the cytoplasm of keratinocytes in the focal spinous layer. For skin tumors, BCC, not only the nuclei but also the cytoplasm were mildly stained in the tumor components of the dermis compared to the staining of the normal epidermis. Although the cytoplasm stained mildly in the focal spinous layer of actinic keratosis, the nuclei and cytoplasm stained strongly in the tumor components of the dermis in SCC. The results of the immunohistochemical staining in normal skin were similar to the RT-PCR and Western blot results from cultured keratinocytes and provided more clues about the role of Nkx2.5 in keratinocyte differentiation.
Fig. 3.
Nkx2.5 expression in several skin diseases. Immunohistochemical staining was performed using Nkx2.5 antibody, at least 3 samples of each disease was studied. The Nkx2.5 expression generally observed in nuclei and/or cytoplasm in various skin tissues. Normal negative means that normal skin was stained in the same procedure only without Nkx2.5 antibody. (A) Normal negative (H&E, ×100), (B) Normal skin (H&E, ×100), (C) Psoriasis (H&E, ×200), (D) AD (H&E, ×200), (E) AK (H&E, ×200), (F) SCC (H&E, ×100), (G) SCC (H&E, ×200), (H) BCC (H&E, ×100), (I) BCC (H&E, ×200). AD: atopic dermatitis, AK: actinic keratosis, SCC: squamous cell carcinoma, BCC: basal cell carcinoma.
DISCUSSION
Several molecular techniques such as cDNA microarray analysis have allowed investigators to evaluate global gene expression profiles. Using this technique, we attempted to identify the genes associated with keratinocyte differentiation. For this purpose, we adopted a calcium-induced keratinocyte differentiation model. After we isolated hundreds of genes that may play a role in keratinocyte differentiation, we analyzed the promoter sequences of some of the differentially-regulated genes and identified several candidate transcription factors for gene regulation during calcium-induced keratinocyte differentiation. Among them, we focused our attention on one interesting transcription factor, Nkx2.5.
The results of this study showed that Nkx2.5 was up-regulated during normal keratinocyte differentiation using RT-PCR and Western blot analysis. In addition, Nkx2.5 was usually observed in the nuclei of the keratinocytes throughout the epidermis of normal skin on immunohistochemical staining. Of note, was that Nkx2.5 expression differed depending on the skin condition. The expression was significantly increased in psoriasis and squamous cell carcinoma (SCC), but decreased in atopic dermatitis and basal cell carcinoma (BCC). These findings suggest that Nkx2.5 plays different roles in the pathogenesis of skin diseases with aberrant keratinocyte differentiation.
SCC and BCC are the non-melanoma skin cancers that represent the most common malignancies in humans. SCC is derived from suprabasal epidermal keratinocytes; these tumor cells have the characteristics of squamous cells that proliferate faster than the cells in normal tissue. In SCC, Nkx2.5 was shown to be stained strongly both in the nuclei and cytoplasm of the entire tumor using immunohistochemical staining. This is the first report of Nkx2.5 associated with the development of SCC. Although it is possible that Nkx2.5 might play a role in the development of SCC similar to T-cell acute lymphoblastic leukemia, there is no data to support that Nkx2.5 plays a significant role in carcinogenesis from normal keratinocytes; the observation that Nkx2.5 was barely detected in BCC, another common skin tumor, does not support such a role.
Psoriasis is a chronic relapsing skin disease with keratinocyte hyperproliferation and altered epidermal differentiation. Previous studies have shown that psoriatic keratinocytes proliferate about 27 times faster than normal keratinocytes. Although it is not a cancerous lesion, it has a faster rate of differentiation than normal tissue, showing some characteristics of a tumor. For psoriasis, Nkx2.5 was stained strongly both in the nuclei and cytoplasm throughout the epidermis; this tendency was more remarkable in the upper layer using immunohistochemical staining. To date, it has not been demonstrated that Nkx2.5 is related to the pathophysiology of psoriasis. However, considering the hyperproliferation of keratinocytes and altered epidermal differentiation, that appears to be similar to cancer, one could speculate that increased Nkx2.5 plays a role in the pathogenesis of psoriasis. Therefore, Nkx2.5 may play a role in the change from proliferation to differentiation in keratinocytes.
Nkx2.5 is an essential transcription factor involved in normal cardiovascular development and an important factor in myocardiogenesis. Many studies have shown that mutation of Nkx2.5 increases the risk of congenital heart disease. Psoriasis has been demonstrated to increase the risk of cardiovascular disease. That is, patients with psoriasis have an increased risk of atherosclerosis and myocardial infarction15-18. Although several studies previously assumed that psoriasis was an independent risk factor for cardiovascular disease, the precise relationship between cardiovascular diseases and psoriasis has not been elucidated15-17. We speculate that increased expression of Nkx2.5 in psoriasis may cause derangement of the cardiovascular system making patients with psoriasis more susceptible to atherosclerosis and/or myocardial infarction; however, evidence is needed to confirm these possibilities. Many prior studies investigated the relationship between the aberration of Nkx2.5 expression and heart diseases as well as T-cell acute lymphoblastic leukemia. However, there has been little research regarding the relationship between Nkx2.5 and psoriasis and non-melanoma skin cancer.
However, different from psoriasis, another inflammatory skin disease (atopic dermatitis) showed opposite patterns of Nkx2.5 expression in the immunohistochemical staining. For atopic dermatitis, there was almost no staining in the epidermal keratinocytes; there was mild staining in the cytoplasm of keratinocytes in the focal spinous layer. As mutations or polymorphisms of the genes involved in the physical barrier function of the skin have recently been identified as risk factors for atopic dermatitis19, it is interesting that Nkx2.5, a transcription factor, was initially reported to be expressed in the keratinocytes, and was shown to be down-expressed in the epidermis of atopic dermatitis. Although it is not understood why Nkx2.5 expression was decreased in atopic dermatitis and increased in psoriasis, it is possible that this difference could be very important to understanding the pathogenesis of both diseases similar to antimicrobial peptides20.
Using RT-PCR, Western blot analysis, and immunohistochemical staining, we showed that Nkx2.5 was expressed and had a tendency to increase with keratinocyte differentiation. In addition, promoter analysis using a cDNA microarray predicted that Nkx2.5 regulates decreased gene expression. Therefore, it is likely that Nkx2.5 exerts its role as a transcriptional repressor of early differentiation-related genes such as K5 and K14 during the terminal differentiation process. The precise functional role of Nkx2.5 in keratinocyte differentiation, however, remains to be further investigated.
Footnotes
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (KRF-2008-314-E00152). This study was also supported by a research fund from the Amore-Pacific Corporation, Yongin, Korea.
References
- 1.Kalinin AE, Kajava AV, Steinert PM. Epithelial barrier function: assembly and structural features of the cornified cell envelope. Bioessays. 2002;24:789–800. doi: 10.1002/bies.10144. [DOI] [PubMed] [Google Scholar]
- 2.Rice RH, Green H. The cornified envelope of terminally differentiated human epidermal keratinocytes consists of cross-linked protein. Cell. 1977;11:417–422. doi: 10.1016/0092-8674(77)90059-9. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs E. Epidermal differentiation and keratin gene expression. J Cell Sci Suppl. 1993;17:197–208. doi: 10.1242/jcs.1993.supplement_17.28. [DOI] [PubMed] [Google Scholar]
- 4.Steinert PM, Marekov LN. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J Biol Chem. 1995;270:17702–17711. doi: 10.1074/jbc.270.30.17702. [DOI] [PubMed] [Google Scholar]
- 5.Steinert PM, Marekov LN. Direct evidence that involucrin is a major early isopeptide cross-linked component of the keratinocyte cornified cell envelope. J Biol Chem. 1997;272:2021–2030. doi: 10.1074/jbc.272.3.2021. [DOI] [PubMed] [Google Scholar]
- 6.Nemes Z, Steinert PM. Bricks and mortar of the epidermal barrier. Exp Mol Med. 1999;31:5–19. doi: 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- 7.Harvey RP. NK-2 Homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 8.Ma CM. Spatial and temporal expression patterns of Xenopus Nkx-2.3 gene in skin epidermis during metamorphosis. Gene Expr Patterns. 2004;5:129–134. doi: 10.1016/j.modgep.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol Cell Biol. 2003;23:9222–9232. doi: 10.1128/MCB.23.24.9222-9232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003;42:1650–1655. doi: 10.1016/j.jacc.2003.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacol Ther. 2005;107:252–268. doi: 10.1016/j.pharmthera.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Nagel S, Scherr M, Kel A, Hornischer K, Crawford GE, Kaufmann M, et al. Activation of TLX3 and NKX2-5 in t(5;14)(q35;q32) T-cell acute lymphoblastic leukemia by remote 3'-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 2007;67:1461–1471. doi: 10.1158/0008-5472.CAN-06-2615. [DOI] [PubMed] [Google Scholar]
- 13.Przybylski GK, Dik WA, Grabarczyk P, Wanzeck J, Chudobska P, Jankowski K, et al. The effect of a novel recombination between the homeobox gene NKX2-5 and the TRD locus in T-cell acute lymphoblastic leukemia on activation of the NKX2-5 gene. Haematologica. 2006;91:317–321. [PubMed] [Google Scholar]
- 14.Nagel S, Kaufmann M, Drexler HG, MacLeod RA. The cardiac homeobox gene NKX2-5 is deregulated by juxtaposition with BCL11B in pediatric T-ALL cell lines via a novel t(5;14)(q35.1;q32.2) Cancer Res. 2003;63:5329–5334. [PubMed] [Google Scholar]
- 15.Kremers HM, McEvoy MT, Dann FJ, Gabriel SE. Heart disease in psoriasis. J Am Acad Dermatol. 2007;57:347–354. doi: 10.1016/j.jaad.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig RJ, Herzog C, Rostock A, Ochsendorf FR, Zollner TM, Thaci D, et al. Psoriasis: a possible risk factor for development of coronary artery calcification. Br J Dermatol. 2007;156:271–276. doi: 10.1111/j.1365-2133.2006.07562.x. [DOI] [PubMed] [Google Scholar]
- 17.Markuszeski L, Bissinger A, Janusz I, Narbutt J, Jedrzejowska AS, Zalewska A. Heart rate and arrhythmia in patients with psoriasis vulgaris. Arch Med Res. 2007;38:64–69. doi: 10.1016/j.arcmed.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Han C, Robinson DW, Jr, Hackett MV, Paramore LC, Fraeman KH, Bala MV. Cardiovascular disease and risk factors in patients with rheumatoid arthritis, psoriatic arthritis, and ankylosing spondylitis. J Rheumatol. 2006;33:2167–2172. [PubMed] [Google Scholar]
- 19.Zeeuwen PL, de Jongh GJ, Rodijk-Olthuis D, Kamsteeg M, Verhoosel RM, van Rossum MM, et al. Genetically programmed differences in epidermal host defense between psoriasis and atopic dermatitis patients. PLoS One. 2008;3:e2301. doi: 10.1371/journal.pone.0002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho SH, Seo SJ, Hong CK. Expression of antimicrobial peptides according to changes of transepidermal water loss levels in patients with atopic dermatitis. Ann Dermatol. 2007;19:47–54. [Google Scholar]