Abstract
Reports of centromere pairing in early meiotic cells have appeared sporadically over the past thirty years. Recent experiments demonstrate that early centromere pairing occurs between non-homologous centromeres. As meiosis proceeds, centromeres change partners, becoming arranged in homologous pairs. Investigations of these later centromere pairs indicate that paired homologous centromeres are actively associated rather than positioned passively, side-by-side. Meiotic centromere pairing has been observed in organisms as diverse as mice, wheat and yeast, indicating that non-homologous centromere pairing in early meiosis and active homologous centromere pairing in later meiosis might be themes in meiotic chromosome behavior. Moreover, such pairing could have previously unrecognized roles in mediating chromosome organization or architecture that impact meiotic segregation fidelity.
We were waltzin’ together to a dreamy melody
When they called out ‘Change partners’
And you waltzed away from me
From ’Changing Partners’, Patti Page (1953); lyrics by Joe Darion.
An overview of centromere paring
Centromeres (see Glossary) are the regions of chromosomes that function as the interface between the chromosome and the microtubules and that mediate chromosome movement in the cell. Centromere pairing is the active association of chromosomes at their centromere regions. In some cases, centromere pairing represents the only point of contact between the chromosomes. In other cases, centromere pairing occurs in the context of end-to-end pairing of two homologous chromosomes. Although meiotic centromere pairing has been reported sporadically in the literature for >30 years [1–3], its role has been poorly understood. Recent studies point to the generality of centromere pairing across species and indicate that centromere pairing might be an important, but previously unappreciated, component of the complex cellular processes that determine the fidelity of meiotic chromosome segregation.
The meiosis-specific behavior of chromosomes is dictated by the developmental control of kinetochores and cohesins (Box 1). Kinetochores are centromeric-protein structures that attach to the microtubules, whereas cohesins are protein complexes that link sister chromatids together. Emerging data indicate that centromere pairing might be tied to kinetochore and cohesin function in meiosis.
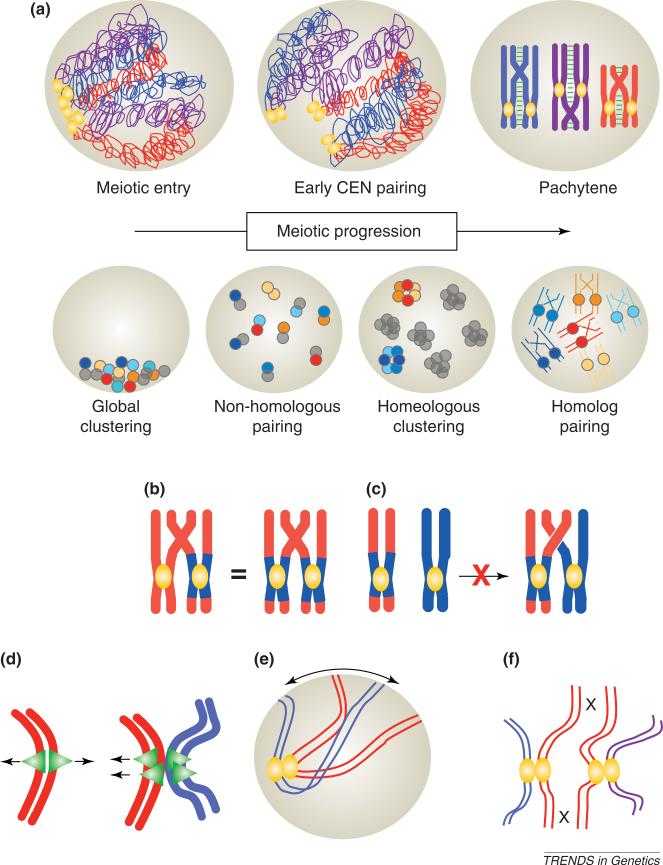
In many organisms, cells enter meiotic S phase with their centromeres clustered (Figure 1). As meiosis proceeds, chromosomes are replicated, homologous recombination is initiated and homologous chromosomes become synapsed from one end to the other, in a stage of meiotic prophase called pachytene [4] (Box 2). In pachytene, the centromeres of homologous chromosomes are in close apposition, but it is impossible to tell by simple observation whether they are actively paired or whether they are simply passengers in the synapsis process. Centromere pairing was first noted in early meiotic cells before chromosome arms were aligned, thus, making centromere pairing more noticeable [3] (Figure 1a). Meiotic centromere pairing has now been reported in a wide range of organisms, including mice [2,5], wheat [1], onion [3], rice [6], Drosophila melanogaster [7], the fission yeast, Schizosaccharomyses pombe [8] and the budding yeast, Saccharomyces cerevisiae [9,10]. Together, these observations indicate that centromere pairing is a common, potentially conserved event in the meiotic process. However, the roles of centromere pairing in meiotic chromosome dynamics remain unclear.
Figure 1.
Early centromere pairing. The arrangement of centromeres in pairs in early meiotic cells has been noted in many organisms. This pairing seems to be primarily between non-homologous centromeres. This non-homologous pairing gives way to the aligning of homologous centromeres as homologous chromosomes pair in later meiotic prophase. (a) Top row: in most organisms, cells enter meiosis with clustered centromeres (gold ovals) and decondensed chromosomes (homologous chromosomes are indicated by the same colors). It is not clear whether early centromere pairing occurs at exactly the same meiotic stage in all organisms in which it has been reported; furthermore, it has yet to be determined whether early pairing precedes or follows meiotic S phase and the early events of recombination initiation. In all cases, early pairing precedes alignment of homologous chromosomes and synaptonemal complex assembly. By pachytene, the stage of maximal homolog alignment and complete synaptonemal-complex formation (green), non-homologous centromere pairing has given way to the side-by-side arrangement of homologous centromeres. Bottom row: in wheat, early non-homologous pairing gives way to clustering of related (homeologous) centromeres (represented by dots of a similar shade). The organization of the chromosomes within the homeologous clusters is not known. By pachytene, centromeres have left the cluster and each is aligned with its true homologous partner. (b). Cytological experiments in wheat using wheat chromosomes (red) bearing rye centromere regions (blue) have addressed the roles of centromeres and chromosome arms in chromosome pairing [21]. Homologous chromosome pairs with either heterologous (one wheat centromere, one rye centromere) or homologous centromeres (both rye centromeres) show indistinguishable levels of pairing and chiasma formation, thereby demonstrating that centromere homology is not required for homologous chromosome pairing. (c) As a second test of the ability of homologous centromeres to drive chromosome pairing, heterologous chromosomes (one rye, blue; and one wheat, red) with homologous rye centromeres were examined. These showed no evidence of meiotic pairing or chiasma formation, demonstrating that homologous centromeres do not drive partner choice, a conclusion also supported by earlier genetic experiments in yeast [22]. (d) Mitotic kinetochores (green) are orientated towards opposite poles (red sister chromatids on the left). Early centromere pairing, shown for red and blue sister chromatid pairs on the right, might serve as a signal or provide a template for the assembly of sister kinetochores that orientate towards the same pole at meiosis I. (e) Experiments in budding yeast [45,46] and rat [47] show that meiotic telomeres move actively in early meiosis, probably to influence chromosome organization or interactions. Early centromere pairing might function as an anchor against telomere-led chromosome movements in early meiosis. (f) In most organisms tested, recombination is limited in centromere regions. Early non-homologous centromere pairing (different colored chromosomes are non-homologous) might sterically limit interactions of homologous centromere regions, thereby preventing homologous recombination near centromeres. (All chromosomes are drawn as condensed to simplify the illustration.)
Centromere pairing in early meiotic prophase
The first detailed descriptions of centromere pairing came from studies in plants (onions and wheat), in which astute investigators noted that the number of centromere structures in cells examined by electron microscopy was about half the total number of chromosomes [1,3]. The observation led to the conclusion that the centromeres must be arranged in pairs [1,3] (Figure 1a). Continued studies, many of which have been in wheat, have revealed details of early centromere pairing. An important aspect of the wheat experiments is that wheat is an allopolyploid: its genome comprises two (pasta wheat) or three (bread wheat) related, but distinct, diploid-chromosome sets. Thus, each chromosome has a true homolog but, also, other related chromosomes (referred to as homeologs) from the other chromosome sets. In wild-type wheat, each chromosome recombines only with its homologous partner in prophase and segregates from that partner at anaphase I, which is also true of chromosomes in diploid organisms (Box 2). The analysis of centromere pairing in wheat has been interwoven with questions of homology. Does the observed early centromere pairing occur between homologous chromosomes, homeologous chromosomes or non-homologous chromosomes? Are centromere behaviors in wheat related to the unique problems of chromosome-partner choice faced only by allopolyploids? The availability of multiple-pairing possibilities has confused the analysis (and, probably, those readers unfamiliar with wheat genetics) but has also provided unique opportunities to evaluate the relationships of early centromere pairing and its potential roles in meiotic chromosome-partner identification (for a review, see Refs [11,12]).
Wheat centromeres pass through four stages of interaction in the meiotic process: (i) clustering; (ii) non-homologous pairing; (iii) homeologous clustering; and (iv) homologous juxtaposition (Figure 1a). Wheat cells enter meiosis with clustered centromeres (as do many species); the centromeres subsequently become organized in pairs [13–15]. The exact stage at which this early pairing is initiated remains unclear, but it is possibly at, or just after, S phase [16]. Considerable effort has been invested in determining whether early meiotic centromere pairings occur between homologous or non-homologous chromosomes – a tricky endeavor owing to the absence of chromosome-specific probes in wheat. Most studies have examined meiosis in either hybrid plants (e.g. wheat-rye) or addition lines, which carry a complete wheat-chromosome set plus one or more chromosomes from another species. The introduction of non-wheat chromosomes has made it possible to use fluorescence in situ hybridization (FISH) probes to one specific chromosome or set (e.g. a rye chromosome pair in a field of wheat chromosomes) to examine pairing behaviors. These studies demonstrated, remarkably, that early centromere pairing occurs mainly between non-homologous centromeres [13,17,18] (Figure 1a). As meiosis continues, the centromeres cluster (about seven clusters per cell) [19]. This number is close to the number of chromosomes that compose each diploid set, indicating a model in which centromeres disengage from their early non-homologous pairing partners, then form seven multivalent clusters of related (homologous and homeologous) centromeres [20] (Figure 1a). The clusters dissolve as homologous partners become aligned (Figure 1a). How the congregation of homologous and homeologous centromeres to a single cluster relates to proper chromosome-partner choice in meiosis is still unclear and is the topic of active investigation [12,20].
The elucidation of these centromere behaviors has raised questions about the roles that centromeres have in meiotic chromosome-partner selection. Recently, Corredor et al. [21] tested how centromere homology dictates partner-choice in meiosis. These authors followed (via FISH) the meiotic behavior of wheat-homologous-chromosome pairs in which either one or both centromeres of the chromosome pair had been replaced with centromeres from rye chromosomes. In a cytological reprise of an elegant genetic experiment performed in yeast twenty-five years earlier [22], the authors tested whether centromere (or arm) homology drives homologous synapsis. First, they compared the pairing and recombination behaviors of homologous chromosomes that carried either homologous or heterologous (wheat-rye) centromeres and found no significant difference (Figure 1b). Second, they examined the pairing of heterologous chromosomes (wheat-rye) with homologous (rye) centromeres (Figure 1c) and found that centromere homology did not promote synapsis of the heterologs. Thus, congruent with the findings of Clarke and Carbon [22], homologous pairing and synapsis seems mainly to be driven by homology of the chromosome arms, not by the centromeres; at this stage of meiosis, centromeres seem to simply follow the course set by the arms. Thus, although centromere pairing has been suggested as a means by which to drive homologous-chromosome pairing, the reverse situation seems to be true; interactions of homologous-chromosome arms bring homologous centromeres together.
A progression from early non-homologous-centromere pairing to the alignment of homologous centromeres has also been demonstrated in budding yeast [10]. Here, the investigators discovered early pairing by observing centromeres in a spo11 (named for its role in sporulation)-mutant background. The spo11 mutation prevents the initiation of meiotic recombination and of homologous synapsis [23,24]. With no pairing of homologous chromosomes, centromeres were still able to pair; the 32 kinetochores became organized as 16 foci, as detected by antibodies against a kinetochore protein. This centromere pairing was shown to be homology-independent and was also found to occur in wild-type cells (SPO11) at a stage before homologous synapsis. Thus, in yeast too, an early stage of homology-independent centromere pairing gives way to the alignment of homologous centromeres, presumably, driven by recombination and synapsis of homologous chromosomes.
Pairs and clusters
In some early meiotic cells, centromeres have been observed to assemble in clusters and in pairs. In yeast [10], clusters are not observed but, in wheat and onion [1,25], investigators have noted examples of clustering. Do these observations reflect some mechanistic difference? They might simply reflect the fact that budding yeast has short centromere regions, whereas wheat and onion have much larger centromere regions. Perhaps the short centromere regions of yeast support only a single-pairing interaction, whereas longer centromeres can support multivalent associations. The centromere regions in wheat and onion are flanked by extensive heterochromatin [1,25]. The centromere pairs and clusters reported in these species might be related to the pairing, clustering and ‘stickiness’ of heterochromatin in meiotic cells, which has been noted by cytologists for >70 years [5].
The molecular basis of early centromere pairing
Little is known about proteins that mediate early centromere pairing. Studies in wheat have examined the role of the Ph1 (pairing homologous 1) locus in centromere behavior. Ph1 is not necessary for early non-homologous centromere pairing [14], but it is important for the transition to homologous-chromosome pairing. ph1 mutants exhibit promiscuous clustering of non-homologous centromeres (Figure 1a) and, later, pairing of non-homologous chromosomes. Because ph1 mutants have defects in the transition from non-homologous early centromere pairing to homologouschromosome pairing, it seems likely that Ph1 participates in promoting homologous interactions, blocking non-homologous interactions or dissolving early non-homologous centromere pairing. Recently, the Ph1 locus has been mapped to a chromosomal region that contains a cluster of genes with homology to human CDK2 (cyclin-dependent kinase 2), a regulator of cell-cycle progression [26,27], which thereby indicates that Ph1 might encode a regulator of early meiotic events.
The clearest example of a protein that is required for early meiotic centromere pairing is Zip1p (named for its role in zippering homologs together in meiosis) in budding yeast [10]. In the absence of Zip1p, instead of forming 16 non-homologous pairs, the 32 centromeres remain unpaired [10]. Zip1p is a major component of the central element of the synaptonemal complex (Figure 1a). There, Zip1p bridges the axial elements associated with the cores of homologous chromosomes [28]. How does Zip1p promote centromere pairing? The simplest possibility is that Zip1p bridges centromere pairs much like it bridges axial elements along chromosome arms in pachytene. This model predicts that Zip1p should localize to sites of centromere pairing in early meiosis and this is true in at least some situations [10,29]. If Zip1p does promote pairing directly at the centromeres, it will be of interest to determine whether this pairing involves the same proteins used to assemble the synaptonemal complex.
Clues regarding the molecular basis of early centromere pairing have come from the discoveries that Zip3p, a protein required for synaptonemal complex assembly, is a SUMO (small ubiquitin-related modifiers) ligase, and that Zip1p is a SUMO-binding protein [30]. Zip3p is required for Zip1p localization to axial elements and synaptonemal-complex formation; however, Zip3p is not required for the punctate foci of Zip1p and SUMO that are seen in early meiosis. The transition from punctuate-Zip1p localization in early meiosis to linear-synaptonemal-complex localization requires ubiquitin-like specific protease 2 (ULP2), which encodes a SUMO de-conjugating enzyme. These results have led to the model that localization of Zip1p to both early foci and the synaptonemal complex is mediated by interactions between Zip1p and SUMOylated binding partners [30]. By this model, Zip3p SUMOylates the Zip1p binding partners on the axial elements that enable synaptonemal-complex formation. By contrast, those binding partners that promote Zip1p localization in early meiosis are SUMOylated by a different SUMO ligase (there are at least three others in budding yeast [30]). In addition, the results indicate that the transition from non-homologous centromere pairing to homologous-chromosome alignment is driven not only by homologous-chromosome-arm interaction but also by the SUMOylation and de-SUMOylation of different Zip1p binding partners throughout the meiotic program. By analogy, the fact that ph1 mutants are compromised in the efficient transition from non-homologous centromere pairing to homologous-chromosome interactions raises the possibility that the Ph1 locus might regulate either of these processes.
Roles of early meiotic centromere pairing
Although the mechanics of early centromere pairing are beginning to be revealed, the role of early centromere pairing remains a mystery. In the absence of a means to block centromere pairing specifically, via mutations or inhibitors, current models are based on speculation. Below, three potential roles for early centromere pairing that will, probably, be addressed in future experimental work are described.
Directing the formation of meiosis-specific kinetochores?
A key process that distinguishes meiotic from mitotic cells is the development of kinetochores that enable sister chromatids to segregate as a unit (rather than be pulled towards opposite poles) at meiosis I (Box 1). Kinetochore geometry differs in mitotic and meiotic cells. Whereas mitotic kinetochores are arranged in a back-to-back fashion, meiotic kinetochores are arranged side-by-side and, thus, are better able to attach to the same spindle pole at meiosis I [31,32]. Could early centromere pairing direct the assembly of properly orientated meiotic sister kinetochores? Recent work, primarily in nematode and yeast models, has identified several gene products, including kinetochore-associated proteins [33–35], cell-cycle kinases [33,36–39], proteins involved in chromatin condensation [33] and cohesins [40] that affect mitotic- and meiotic-kinetochore assembly. Notably, meiosis-specific kinetochore proteins Mam1p (monopolar microtubule attachment during meiosis I) in budding yeast [41] and Moa1p (monopolar attachment protein 1) in fission yeast [42], and the fission-yeast meiotic-cohesin component Rec8p [40] are required to assemble meiosis-specific kinetochores, implying that the mitotic kinetochore configuration is the default setting overridden in meiosis by the addition of these proteins. The timing of early centromere pairing could coincide with the assembly of meiosis-specific kinetochores. Experiments in mitotic cells demonstrate that some components of the outer kinetochore are added to kinetochores in pro-metaphase [43]. By analogy, key elements of the meiotic kinetochores are probably assembled in meiotic prophase. It is possible that early meiotic centromere pairing functions at that time to signal or act as a template for meiosis-specific-kinetochore assembly (Figure 1). In budding yeast, the Mam1p complex localizes to meiotic kinetochores and promotes meiosis-specific-centromere behavior [41]. Moa1p, Rec8p and the checkpoint kinase and kinetochore protein Bub1p have similar roles in fission yeast [34,40,42]. It will be of interest to determine at which point these centromere-associated proteins assemble on meiotic chromosomes. Those that assemble very early could contribute to early centromere pairing, whereas those that load later could be templated by early-pairing events to assemble in a meiosis-specific manner.
Anchoring the chromosomes against dynamic meiotic chromosome movements?
Like centromeres, telomeres also exhibit dynamic behaviors in meiosis. Telomeres cluster in early prophase (the bouquet stage) [44] and exhibit rapid movements (RPMs) in budding yeast [45,46] and, perhaps, also in mammals [47]. Could paired centromeres provide a counter-balance to these telomere-led movements (Figure 1e)? Early centromere pairing might function as an anchor to enable chromosome-arm extension as telomeres cluster, to provide a reference point for telomere migration or to enable chromosome oscillations that create tension along the chromosome arms [21,45]. These models predict that mutations disrupting early centromere pairing and bouquet formation will have overlapping phenotypes. Although several bouquet-formation and telomere-movement mutants have been identified [45,48–52], such comparisons will have to await the identification of mutations that cause specific centromere-pairing defects.
Preventing centromere proximal recombination?
In humans, Drosophila females and budding yeast, crossovers very near to centromeres predispose homologous chromosomes to segregation errors in meiosis I [53,54]. These crossovers probably occur within the region of centromeric cohesin, which is not removed until anaphase I (Box 1), thereby locking the homologs together in anaphase I and forcing them to migrate to the same pole. These deleterious crossovers are normally prevented by mechanisms that suppress the formation of crossovers near centromeres. Heterochromatin and an absence of recombination initiation sites near centromeres probably limit centromere proximal crossovers. The relative timing of early centromere pairing and recombination is unknown. If they are concurrent, non-homologous early pairing might sequester centromeres from their homologs at the time when recombination is initiated (Figure 1f). Consistent with this model, transplacing centromeres from their normal position increases recombination rates in the original region and reduces crossing-over in the new location [55,56].
Centromere pairing in later meiosis
It has not been easy to determine whether centromeres are actively paired in later prophase (pachytene) or simply placed side-by-side as the homologs become aligned. Compelling evidence for active centromere pairing in later meiosis came from observations of mouse spermatocytes [2]. As the spermatocytes transition from pachytene (complete synapsis) to diplotene (in which the synaptonemal complex is removed), the arms of the homologous chromosomes separate but homologs often remain linked – not only by chiasmata, but also by their centromeres (Figure 2a,b). Thus, in mouse spermatocytes, homologous-centromere pairing is mechanistically distinct from arm pairing. These results might be explained by any of several models. Most simply, centromeres could be paired by a structure distinct from the synaptonemal complex, such that removal of the synaptonemal complex releases arm, but not centromere, association. The presence of both meiosis-specific kinetochores and protected cohesins at the centromeres make it easy to imagine that there could be a molecular foundation for assembling specialized structures for centromere pairing. Alternatively, centromeres and arms might be associated by similar mechanisms, with the centromere regions able to resist removal of the pairing material (synaptonemal-complex elements), analogous to the removal of meiotic cohesins from the chromosome arms, but not the centromeres, at anaphase I [57] (Box 1).
Figure 2.
Centromere pairing in late prophase. In late meiotic prophase, homologous chromosomes align end-to-end. At pachytene, the stage of maximal alignment, a proteinaceous structure runs the length of the aligned homologs. This alignment juxtaposes homologous centromeres. This late centromere pairing has a role in the proper attachment of microtubules during meiosis I. (a) Evidence for active association of late prophase centromeres comes from studies of mouse spermatocytes. In pachytene, chromosomes are tightly synapsed along their length (synaptonemal complex is shown in green). As cells move into diplotene, the synaptonemal complex is lost and homologs remain tethered to one another by chiasmata and the centromeres (yellow). (b) Late centromere pairing is clearly revealed in this micrograph of mouse spermatocyte diplotene chromosomes, linked at chiasmata and centromeres (image provided by Huiling Xu, Peter MacCallum Cancer Centre; http://www.petermac.org/). The chromosome arms are stained with antibodies against SCP3 (synaptonemal-complex protein 3; green); each green line represents one chromosome (two tightly associated sister chromatids). Chiasmata are revealed as the points where the green lines come together. Centromeres are stained with CREST (a human autoimmune syndrome) antibodies against kinetochore components (red) [71]. (c) Centromere pairing of achiasmate chromosomes. In Drosophila, budding yeast and fission yeast, chromosomes that are not tethered by crossovers (red) pair at their centromere regions (but not their arms) in late meiotic prophase (pairing symbolized by blue rectangle), whereas exchange chromosomes (purple) are linked by chiasmata. Centromere pairing promotes disjunction at meiosis I. (d) Centromere pairing between achiasmate chromosomes seems to promote attachment of the chromosomes to opposite poles of the spindle. Centromere pairing could fulfill the same role for exchange chromosomes. Centromere pairing (blue rectangle) might secure the homologous kinetochores (green triangles) such that they face in opposite directions before microtubule attachment. In the absence of centromere pairing, kinetochores might have rotational freedom, thereby enabling attachment of the kinetochores to the same pole. For simplicity, the kinetochores of each sister chromatid pair is represented as a single triangle. (e) Does centromere pairing help orientate sister kinetochores towards the same pole? In mouse oocytes, chromosomes that have failed to pair with a partner (univalents) frequently segregate their sister chromatids to opposite poles at meiosis I. This indicates that partnerless sister chromatid pairs orientate their kinetochores to opposite poles (sister pair on left). Centromere pairing (blue rectangle) might help co-orientate sister kinetochores to the same pole (right).
Centromere pairing in later meiotic prophase is not unique to mouse spermatocytes [7,9]. In Drosophila oocytes and budding yeast, pairs of achiasmate chromosomes (chromosome pairs that are not tethered by a crossover) segregate from one another at meiosis I. There are important differences in the Drosophila and yeast achiasmate segregation processes but, in both, achiasmate partners pair at or near their centromeres (Figure 2c). In Drosophila, homologous achiasmate partners associate in regions of pericentric heterochromatin before segregating at anaphase I [7]. It is not yet known whether this pairing involves sequence-specific chromatin-associated proteins or, alternatively, interactions at the DNA level [58].
In yeast, late prophase pairing orientates the centromeres of achiasmate partners to opposite poles, thus, offering a chance for non-exchange partners to segregate properly [9]. This centromere pairing is homology-independent and it partitions homologous and non-homologous achiasmate pairs equally efficiently. Although late centromere pairing is homology-independent, the centromeres of exchange chromosomes always become aligned. Therefore, as in wheat, yeast centromeres pair non-homologously in early prophase, but homologous-arm interactions bring homologous centromeres into alignment in later meiosis. The proteins required for late centromere pairing have not been reported so, at this time, the molecular basis remains to be elucidated.
Non-homologous achiasmate partners have also been studied in Drosophila. In XXY flies (two X and one Y chromosome), when the X chromosomes recombine they segregate at meiosis I. By contrast, when they fail to recombine, the X chromosomes usually segregate to one pole of the spindle, away from the Y. Why? The phenomenon (confusingly referred to as secondary non-disjunction) has been considered to be a clue to the unanswered questions of meiotic-chromosome biology for nearly a century [59]. Recently, Xiang and Hawley [60] demonstrated that, in XXY flies, each X chromosome pairs with one arm of the Y chromosome via centromeric heterochromatin in early meiosis. If the X chromosomes recombine, they abandon the Y; alternatively, if they do not recombine, pairing persists, thereby orientating the X chromosomes to one spindle pole and the Y to the other. The work illustrates that pairing through centromeric heterochromatin can orientate meiotic partners and, strikingly, that homologous recombination can override non-homologous centromere associations.
The role of late prophase centromere pairing
A clear role for late centromere pairing has been established in the segregation of achiasmate chromosomes of budding yeast and Drosophila. Although these experiments might seem at first glance arcane, they provide informative clues to the roles that late centromere pairing might have in the segregation of typical exchange chromosomes.
Orientating centromeres at meiosis I
In budding yeast and Drosophila females, centromere pairing orientates the centromeres of achiasmate partners to opposite poles of the meiosis-I spindle, thereby promoting proper segregation (Figure 2c). In budding yeast, it remains unclear whether centromere pairing can fully mimic the role of a chiasma. The secure tether of a chiasma provides a means for a tension-sensing system to orientate homologous partners. When homologous chromosomes both attach to microtubules radiating from the same pole, they release then reattach until they have attached to microtubules from opposite poles, thereby generating tension at their kinetochores as they pull away from one another [61]. Achiasmate partners in yeast seem to dissociate early in metaphase; thus, centromere pairing might facilitate only one round of microtubule attachment [9]. Even so, the favorable bias provided by centromere pairing enables achiasmate partners to segregate properly in ~90% of meioses [9]. By contrast, in Drosophila females, centromere pairing maintains the association of partners as they proceed through the reorientation process [62]. This might, in part, be why achiasmate chromosomes in Drosophila achieve much higher fidelity (>99.9%) [63]. Exactly how paired centromeres participate in checkpoint signaling remains unclear, but achiasmate chromosomes in yeast and Drosophila both depend on the meiotic delays that are provided by the spindle checkpoint [62,64,65].
Does centromere pairing also promote the bi-orientation of exchange homologs? This notion was first suggested by Östergren >50 years ago [66]. He reasoned that, in the absence of an interaction between the centromeres, the two chromosomes of a homologous pair should just as often be directed to the same, as to opposite, poles (Figure 2). This is because, unlike mitotic centromeres, which are positioned directly back-to-back, centromeres of meiotic partners are linked only by crossovers. In the absence of centromere pairing, the centromeres should have rotational freedom and often point to the same pole (Figure 2). In some systems, such as Drosophila, yeast and mouse spermatocytes, centromere regions are in close proximity shortly before microtubules attach to kinetochores. In these organisms, centromere pairing might position the active faces of the homologous kinetochores away from one another (Figure 2), offering a good first chance for proper spindle attachment. This action would lessen the load on the spindle checkpoint that, in meiosis, sometimes offers only brief delays to correct potential errors [64,67].
Recently, crossovers near (but not at) centromeres were found to reduce the need for the spindle checkpoint in yeast [68]. Unlike the deleterious crossovers in centromere regions described earlier, these beneficial crossovers probably lie beyond the centromeric cohesins that are not removed until anaphase II. An artificial tether (a tetramerizing lacI protein that bridges the chromosomes at lac-operator arrays positioned near the centromeres) had the same favorable effect on chromosomes as a centromere proximal crossover. These findings indicate that centromere proximal crossovers might promote a kinetochore topology that increases the likelihood of bipolar attachment. Another possibility is that centromere proximal crossovers increase the efficiency of centromere pairing (for example by bringing the centromeres closer together), which in turn could orientate the kinetochores.
Promoting proper sister kinetochore behavior
Just as early centromere pairing might impact sister-kinetochore development, so too might late centromere pairing. In mouse oocytes, chromosomes that fail to associate with a partner in late prophase often separate their sister chromatids at meiosis I [67,69] (Figure 2e). Thus, it might be that late centromere pairing promotes or maintains the orientation of sister kinetochores toward a common pole in meiosis.
Concluding remarks
Meiotic centromere pairing has been reported for decades, but has only recently become a topic of focused investigation. In a variety of experimental systems, centromere pairing occurs at two stages of meiosis. In early meiosis, centromeres pair in a non-homologous fashion. Later in meiosis, centromeres change partners, leaving their non-homologous partners and aligning with their homologous partners while homologous chromosomes pair. Late pairing can orientate the centromeres, but the mechanism of late centromere pairing remains a mystery. Questions abound. What is the relationship between early and late centromere pairing? What mechanisms mediate each process? Will genes that are required for one also be required for the other? How do early and late centromere pairing contribute to meiotic segregation fidelity? These are fundamental questions, to which the answers are beginning to be revealed.
In humans, aneuploidy (an incorrect chromosome number) occurs in up to 30% of zygotes, making aneuploidy the leading cause of both failed pregnancy and mental retardation [70]. These aneuploidies are usually the result of either a failure of homologs to segregate to opposite poles at meiosis I (Figure 2) or precocious separation of sister chromatids at meiosis I (Figure 2e). Both errors are directly related to the known and proposed roles of centromere pairing in meiosis. Elucidating the roles and mechanisms of centromere pairing will not only shed light on the workings of fundamental meiotic processes but, also, will probably provide clues to the molecular basis of human meiotic errors.
Box 1. Kinetochores and cohesins dictate mitotic and meiotic chromosome-segregation behaviors.
Mitosis is the process of nuclear division that results in identical daughter cells, whereas meiosis produces cells with half the genetic material of the parent, a process necessary for sexual reproduction. A key difference between mitotic and meiotic cell divisions is that in mitosis, the sister chromatids separate, whereas in meiosis I, homologous chromosomes separate. This differential behavior is accomplished through the regulation of kinetochores and cohesins. During S phase of the mitotic cell cycle, a protein complex called cohesin secures the newly replicated sister chromatids together. Mitotic kinetochores are assembled in a back-to-back fashion that predisposes them to attach to microtubules from opposite spindle poles [Figure Ia (i)]. At metaphase of mitosis, the kinetochores of sister chromatids attach to microtubules from opposite poles of the spindle. In early anaphase, the cohesins that hold chromatids together are removed from the chromosome [Figure Ib (i)]. These two properties, cohesin regulation and outwardly orientated sister kinetochores, together enable the sister chromatids to remain linked through metaphase and then to segregate to opposite poles at mitotic anaphase. In meiosis I, the assembly of sister kinetochores is altered, such that both sister chromatids attach to the same spindle pole at meiosis I [72] [Figure Ia (ii)]. If meiosis-specific kinetochores are not assembled properly, microtubules from opposite poles can attach to sister kinetochores, as in mitosis, causing premature separation of sister chromatids at meiosis I [Figure Ia (iii)]. In meiosis, cohesins are removed in a step-wise manner [72]. Arm cohesin is removed at anaphase I, whereas centromeric cohesin is protected, enabling the sisters to segregate as a unit in meiosis I [Figure Ib( ii)]. Finally, at anaphase II, centromere cohesin is removed, enabling sister chromatids to segregate to opposite poles, as in mitosis [Figure Ib (ii)]. Proper orientation of the meiosis I kinetochores and the step-wise removal of cohesins enable homologs to segregate to opposite poles at meiosis I, followed by the proper segregation of sister chromatids at meiosis II.0
Box 2. Meiotic progression and potential errors.
In meiosis, chromosomes complete two rounds of segregation after a single round of replication to yield gametes with half the number of chromosomes of somatic cells (Figure I). DNA is replicated in a specialized S phase as cells enter meiosis; each homolog is now made up of two sister chromatids. After S phase, there are five substages that constitute prophase I. During leptotene, chromosomes begin to condense and search for their homologous partner. Homologous recombination probably initiates in leptotene [73]. Zygotene is the stage at which homologous chromosomes become aligned and begin to assemble the synaptonemal complex. Chromosomes are fully condensed and synapsed at pachytene. In diplotene and diakinesis, the synaptonemal complex is removed and recombination is completed to produce mature crossovers (viewed as chiasmata cytologically). At metaphase I, the homologs of each pair (called a bivalent) attach to microtubules that are opposite spindle poles. At anaphase I, homologs segregate away from each other, whereas sister chromatids remain paired. Finally, at anaphase II, sister chromatids segregate, thereby producing four haploid cells.
Two common errors occur at anaphase I. Meiosis I non-disjunction is the segregation of both homologs to one pole. This error yields two nuclei with zero copies of the errant chromosome (nullisomic) and two nuclei with two copies (disomic). The second error, precocious separation of sister chromatids, occurs when a pair of sister chromatids migrates to opposite poles at meiosis I. This error yields two normal nuclei and one each that is nullisomic and disomic for the errant chromosome. Fertilization of disomic and nullisomic gametes yields trisomic and monosomic zygotes, respectively.
In humans, the segregation errors described result in aneuplodies, which occur in 5–30% of clinically recognized pregnancies [70]. In most cases, aneuploidy will result in death but, among live births, it is also the most common cause of mental retardation, the best known example being Down's Syndrome, which is caused by trisomy of chromosome 21 [70].
Figure I.
Kinetochore orientation and cohesin removal during mitosis and meiosis. (a) (i) Mitotic kinetochores (green triangles) are assembled in a back-to-back in a fashion. (ii) In meiosis, sister chromatids have kinetochores that function as a unit. (iii) Failure to adopt a meiosis-specific kinetochore assembly would result in precocious separation of the sister chromatids in meiosis I, rather than meiosis II. (b) (i) In mitosis, cohesins (orange lines) are all removed by anaphase, facilitating sister chromatid separation. In meiosis, cohesins are removed in two steps. (ii) Before anaphase I, cohesins distal to the chiasmata are removed, enabling homologs to separate. (iii) Before anaphase II, the remaining cohesins are removed, thereby facilitating separation of sister centromeres.
Figure I.
The stages of meiosis. Depicted is a diploid cell with only one chromosome; one homolog in blue and the other in purple. To simplify the illustration of meiotic events, the chromosomes are represented as condensed throughout the process (which is not actually the case). The time at which early centromere pairing initiates has not been determined, but could be before or during meiotic S phase (red dashed line). Early pairing continues into leptotene (red line). Known examples of late centromere pairing initiate in leptotene or zygotene and continue until chromosomes separate at metaphase or anaphase I.
Acknowledgements
We apologize to our colleagues whose work could not be cited owing to space limitations. We thank Michael Dresser, Scott Hawley and David Obeso for their comments on the manuscript. Huiling Xu (Peter MacCallum Cancer Centre; http://www.petermac.org/) generously provided the image of mouse diplotene chromosomes. D.D. is supported by grant R01 GM075363.
Glossary
Achiasmate chromosome: a chromosome that has failed to become linked to its meiotic partner by a chiasma at meiosis I.
Centromere: the region of the chromosome involved in attaching to the microtubules that direct chromosome segregation at anaphase.
Chiasma: the point of contact between homologous chromosomes, at the site of a crossover or exchange between the homologs that links them together until anaphase I.
Cohesin: a particular protein complex deposited along the arms of sister chromatids that links them together until its removal at anaphase.
Exchange chromosomes: homologous chromosomes that are linked by a chiasma in meiosis I.
Kinetochore: the proteinaceous structure assembled at centromeres that attaches to the microtubules.
Non-disjunction: the errant segregation of chromosome partners towards the same pole of the spindle at anaphase.
Precocious separation of sister chromatids: the errant segregation of sister chromatids to opposite rather than the same pole of the spindle at meiosis I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bennett MD. Centromere arrangements in Triticum aestivum and their relationship to synapsis. Heredity. 1979;43:157. [Google Scholar]
- 2.Brinkley BR, et al. Arrangements of kinetochores in mouse cells during meiosis and spermiogenesis. Chromosoma. 1986;94:309–317. doi: 10.1007/BF00290861. [DOI] [PubMed] [Google Scholar]
- 3.Church K, Moens PB. Centromere behavior during interphase and meiotic prophase in Allium fistulosum from 3-D, E.M. reconstruction. Chromosoma. 1976;56:249–263. [Google Scholar]
- 4.Kleckner N. Meiosis: how could it work? Proc. Natl. Acad. Sci. U. S. A. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yunis JJ, Yasmineh WG. Model for mammalian constitutive heterocromatin. In: DuPraw FJ, editor. Advances in Cell and Molecular Biology. Academic Press; 1972. pp. 1–46. [Google Scholar]
- 6.Prieto P, et al. Chromosomes associate premeiotically and in xylem vessel cells via their telomeres and centromeres in diploid rice (Oryza sativa). Chromosoma. 2004;112:300–307. doi: 10.1007/s00412-004-0274-8. [DOI] [PubMed] [Google Scholar]
- 7.Dernburg AF, et al. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 8.Ding DQ, et al. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 9.Kemp B, et al. A role for centromere pairing in meiotic chromosome segregation. Genes Dev. 2004;18:1946–1951. doi: 10.1101/gad.1227304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsubouchi T, Roeder GS. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308:870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- 11.Moore G. Meiosis in allopolyploids–the importance of ‘Teflon’ chromosomes. Trends Genet. 2002;18:456–463. doi: 10.1016/s0168-9525(02)02730-0. [DOI] [PubMed] [Google Scholar]
- 12.Naranjo T, Corredor E. Nuclear architecture and chromosome dynamics in the search of the pairing partner in meiosis in plants. Cytogenet. Genome Res. 2008;120:320–330. doi: 10.1159/000121081. [DOI] [PubMed] [Google Scholar]
- 13.Maestra B, et al. Chromosome arrangement and behaviour of two rye homologous telosomes at the onset of meiosis in disomic wheat-5RL addition lines with and without the Ph1 locus. Chromosome Res. 2002;10:655–667. doi: 10.1023/a:1021564327226. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Perez E, et al. Homologous chromosome pairing in wheat. J. Cell Sci. 1999;112:1761–1769. doi: 10.1242/jcs.112.11.1761. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Perez E, et al. Polyploidy induces centromere association. J. Cell Biol. 2000;148:233–238. doi: 10.1083/jcb.148.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jasencakova Z, et al. Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma. 2001;110:83–92. doi: 10.1007/s004120100132. [DOI] [PubMed] [Google Scholar]
- 17.Aragon-Alcaide L, et al. Association of homologous chromosomes during floral development. Curr. Biol. 1997;7:905–908. doi: 10.1016/s0960-9822(06)00383-6. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Perez E, et al. The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature. 2001;411:204–207. doi: 10.1038/35075597. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Perez E, et al. Chromosomes form into seven groups in hexaploid and tetraploid wheat as a prelude to meiosis. Plant J. 2003;36:21–29. doi: 10.1046/j.1365-313x.2003.01853.x. [DOI] [PubMed] [Google Scholar]
- 20.Prieto P, et al. Homologue recognition during meiosis is associated with a change in chromatin conformation. Nat. Cell Biol. 2004;6:906–908. doi: 10.1038/ncb1168. [DOI] [PubMed] [Google Scholar]
- 21.Corredor E, et al. Terminal regions of wheat chromosomes select their pairing partners in meiosis. Genetics. 2007;177:699–706. doi: 10.1534/genetics.107.078121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke L, Carbon J. Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature. 1983;305:23–28. doi: 10.1038/305023a0. [DOI] [PubMed] [Google Scholar]
- 23.Loidl J, et al. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J. Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 25.Moens PB, Church K. Centromere sizes, positions, and movements in the interphase nucleus. Chromosoma. 1977;61:41–48. [Google Scholar]
- 26.Al-Kaff N, et al. Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: with deletion mutants and expression profiling. Ann. Bot. (Lond.) 2008;101:863–872. doi: 10.1093/aob/mcm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths S, et al. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- 28.Sym M, et al. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 29.Henderson KA, Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng CH, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldstein LS. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell. 1981;25:591–602. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- 32.Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore LL, et al. HCP-4/CENP-C promotes the prophase timing of centromere resolution by enabling the centromere association of HCP-6 in Caenorhabditis elegans. Mol. Cell. Biol. 2005;25:2583–2592. doi: 10.1128/MCB.25.7.2583-2592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernard P, et al. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat. Cell Biol. 2001;3:522–526. doi: 10.1038/35074598. [DOI] [PubMed] [Google Scholar]
- 35.Kiburz BM, et al. Shugoshin promotes sister kinetochore biorientation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:1199–1209. doi: 10.1091/mbc.E07-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maure JF, et al. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 2007;17:2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauf S, et al. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 2007;26:4475–4486. doi: 10.1038/sj.emboj.7601880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monje-Casas F, et al. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petronczki M, et al. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 40.Yokobayashi S, et al. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol. Cell. Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toth A, et al. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- 42.Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 43.McCleland ML, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- 45.Conrad MN, et al. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3 and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 46.Koszul R, et al. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parvinen M, Soderstrom KO. Chromosome rotation and formation of synapsis. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- 48.Bass HW, et al. The desynaptic (dy) and desynaptic1 (dsy1) mutations in maize (Zea mays L) cause distinct telomere-misplacement phenotypes during meiotic prophase. J. Exp. Bot. 2003;54:39–46. doi: 10.1093/jxb/erg032. [DOI] [PubMed] [Google Scholar]
- 49.Chikashige Y, et al. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 50.Conrad MN, et al. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 51.Jin Y, et al. Fission yeast mutants affecting telomere clustering and meiosis-specific spindle pole body integrity. Genetics. 2002;160:861–876. doi: 10.1093/genetics/160.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nimmo ER, et al. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- 53.Koehler KE, et al. Recombination and nondisjunction in humans and flies. Hum. Mol. Genet. 1996;5:1495–1504. doi: 10.1093/hmg/5.supplement_1.1495. Spec No. [DOI] [PubMed] [Google Scholar]
- 54.Rockmill B, et al. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beadle GW. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 1932;18:160–165. doi: 10.1073/pnas.18.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lambie EJ, Roeder GS. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986;114:769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe Y. Shugoshin: guardian spirit at the centromere. Curr. Opin. Cell Biol. 2005;17:590–595. doi: 10.1016/j.ceb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Karpen GH, et al. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- 59.Bridges CB. Non-disjunction as proof of the chromosome theory of heredity. Genetics. 1916;1:1–52. doi: 10.1093/genetics/1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiang Y, Hawley RS. The mechanism of secondary nondisjunction in Drosophila melanogaster females. Genetics. 2006;174:67–78. doi: 10.1534/genetics.106.061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 62.Gilliland WD, et al. The multiple roles of mps1 in Drosophila female meiosis. PLoS Genet. 2007;3:e113. doi: 10.1371/journal.pgen.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hawley RS, Theurkauf WE. Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet. 1993;9:310–317. doi: 10.1016/0168-9525(93)90249-h. [DOI] [PubMed] [Google Scholar]
- 64.Cheslock PS, et al. The roles of MAD1, MAD2 and MAD3 in meiotic progression and the segregation of nonexchange chromosomes. Nat. Genet. 2005;37:756–760. doi: 10.1038/ng1588. [DOI] [PubMed] [Google Scholar]
- 65.Gilliland WD, et al. The meiotic defects of mutants in the Drosophila mps1 gene reveal a critical role of Mps1 in the segregation of achiasmate homologs. Curr. Biol. 2005;15:672–677. doi: 10.1016/j.cub.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 66.Östergren G. The mechanism of co-orientation in bivalents and multivalents. Hereditas. 1951;37:85–156. [Google Scholar]
- 67.LeMaire-Adkins R, et al. Lack of checkpoint control at the metaphase/anaphase transition: a mechanism of meiotic nondisjunction in mammalian females. J. Cell Biol. 1997;139:1611–1619. doi: 10.1083/jcb.139.7.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lacefield S, Murray AW. The spindle checkpoint rescues the meiotic segregation of chromosomes whose crossovers are far from the centromere. Nat. Genet. 2007;39:1273–1277. doi: 10.1038/ng2120. [DOI] [PubMed] [Google Scholar]
- 69.Kouznetsova A, et al. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat. Genet. 2007;39:966–968. doi: 10.1038/ng2065. [DOI] [PubMed] [Google Scholar]
- 70.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 71.Xu H, et al. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep. 2004;5:378–384. doi: 10.1038/sj.embor.7400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 73.Padmore R, et al. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]