Abstract
Introduction
One-size-fits-all chemotherapy does not improve survival in patients with small cell lung cancer (SCLC). Excision repair cross complementing group 1 (ERCC1), ribonucleotide reductase 1 (RRM1), thymidylate synthase (TS), and topoisomerase 2α (Topo2α) expression levels are predictive of chemotherapeutic efficacy in some malignancies. Our aim was to determine the expression levels of these proteins to assess their potential clinical utility in SCLC.
Methods
We used an immunofluorescence-based automated quantitative (AQUA) technique to score RRM1, ERCC1, TS, and Topo2α levels in tumor specimens from 100 patients with SCLC and immunohistochemistry (IHC) to semiquantitatively score levels of TS, 5-phosphoribosyl-glycinamide formyl-transferase, and folyl-polyglutamate synthase expression. Confocal microscopy was used for subcellular localization in SCLC cells.
Results
RRM1, ERCC1, and Topo2α staining was predominantly nuclear and TS mainly cytoplasmic. Using IHC, we found that TS (antibody 106) and TS (antibody 4H4) scores were strongly correlated (r=0.82, p<0.0001). By AQUA, RRM1 and Topo2α levels were highly correlated (r = .56, p <.0001). ERCC1 and TS levels had a narrow and low range of expression. There was no correlation between any of these biomarkers and patients’ age or sex.
Conclusion
Considering current clinical evidence, expression levels of RRM1 and Topo2α may have utility for chemotherapy customization. Clinical validation of their predictive power is desirable in a prospective clinical trial.
Keywords: Predictive biomarkers, Chemosensitivity, Small cell lung cancer (SCLC), immunofluorescence-based automated quantitative analysis (AQUA)
INTRODUCTION
Small cell lung cancer (SCLC) accounts for approximately 15% of all new lung cancer cases in the United States(1). SCLC is characterized by early dissemination, and 70%–80% of patients have widely metastatic disease at diagnosis (extensive stage, ES-SCLC). SCLC is considered a chemotherapy-responsive disease, and etoposide-platinum is the standard, first-line treatment in the United States. However, despite initial response rates of ≥ 60% and complete response rates of 20%–30%, the median survival time and 2-year survival rate range from 7–10 months and 10%–20%, respectively. No significant improvements in the efficacy of systemic chemotherapy have been achieved in the past 30 years(2, 3).
Newer cytotoxic drugs have been tested in patients with SCLC(4). Taxanes (paclitaxel(5) and docetaxel(6)), topoisomerase inhibitors (topotecan(7) and irinotecan(8)), and antimetabolites (pemetrexed(9) and gemcitabine(10)) have demonstrated acceptable activity as single agents and in combination with platinum agents. Unfortunately, in phase III trials, they have failed to produce a survival advantage(11–14). Novel approaches that focus on specific genomic characteristics of individual tumors may provide tools to overcome this stalemate.
Pharmacogenomics centers on the principle that molecular characteristics observed in tumors have the potential to affect therapeutic decisions and to improve patient outcomes. Specifically, pharmacogenomics has the potential to allow clinicians to select chemotherapy drugs that will give patients maximal benefit while simultaneously minimizing toxicity(15, 16). Specific gene products that may lend themselves to such an approach in SCLC include (1) RRM1, the regulatory subunit of ribonucleotide reductase, the enzyme responsible for the supply of deoxyribonucleotides for DNA synthesis, and the target of gemcitabine(17); (2) topoisomerase 2 (Topo2; subunits α and β), an enzyme that regulates the topological state of DNA, facilitating replication, mitosis, and chromatin condensation(18), and the target of etoposide; (3) excision repair cross complementing group 1 (ERCC1), a component of the 5’ endonuclease of the nucleotide excision repair complex, which is responsible for cisplatin-induced DNA damage repair(19); and (4) thymidylate synthase (TS), an enzyme responsible for thymidine production for DNA synthesis and one of the targets of pemetrexed(20).
Although not yet shown in SCLC, in different tumor types and using different methodologies, evidence from multiple preclinical and clinical studies has been accumulating to support the role of these proteins as predictive biomarkers of activity for their respective cytotoxic agents(21–32). Furthermore, their potential therapeutic advantage is highlighted by current prospective phase II(33) and III(34) clinical trials in non-SCLC (NSCLC), where these biomarkers have been used to select cytotoxic agents with promising clinical outcomes data.
In this study, our aim was to describe the pattern and range of expression levels as well as to find correlations in the expression levels among these biomarkers in a large cohort of patients with SCLC. Two biomarkers in particular, Topo2α and RRM1, showed promising patterns of expression that can be utilized to initiate a prospective clinical trial with individualized treatment combinations for patients with SCLC.
METHODS
Tissue Microarray Construction
A custom tissue microarray (TMA) was provided by Eli Lilly & Co. (Indianapolis, IN). The array (OD-CT-RsLug01) encompasses triplicate samples from 100 patients with SCLC. It was constructed by Shanghai Biochip (Shanghai, China), using 1.0-mm tissue cores from formalin-fixed and paraffin-embedded (FFPE) tissue blocks of patients with confirmed SCLC. Sections of 5 µm thickness were cut, transferred to 4x adhesive-coated slides using tape (Instrumedics, St. Louis, MO), and briefly exposed to ultraviolet light to enhance adherence. The surface of the TMA specimens was protected from light and environment by an opaque air-tight film. Clinical outcomes data are not available from patients who contributed specimens to this array (except age and gender). Investigational Review Board approval was obtained prior to performing the molecular analyses.
In Situ Detection and Quantification of Protein Expression
Immunofluorescence combined with automated quantitative analysis (AQUA) was used to assess in situ expression of the target molecules(35). Antigens were retrieved by incubating the tissue in a microwave oven for 15 minutes in 0.01 mol/L sodium citrate, Tris-HCl, or Tris-EDTA buffer at an optimized pH(36). The slides were blocked for 30 minutes with 0.3% bovine serum albumin and then incubated overnight at 4°C in optimal concentrations of antisera or antibodies to detect RRM1, ERCC1, TS, and Topo2α. The RRM1 antiserum was custom made in rabbits, affinity purified, and designated R1AS-6b(37). Commercial antibodies were used for the analysis of ERCC1 (clone SPM-243; Santa Cruz), TS (clone 106; Abcam), and Topo2α (clone Ki-S1; Dako Cytomation). For identification of carcinomatous cells, antibodies to cytokeratin were used (murine anti-human pancytokeratin AE1/AE3, 1:200, #M3515, Dako Cytomation; rabbit anti-human pancytokeratin AE1/AE3, 1:200, #Z0622, Dako Cytomation). Slides were washed and incubated with two different secondary antibodies for 1 hour: Envision™-labeled polymer horseradish peroxidase anti-rabbit (#K4011) or Envision-labeled polymer horseradish peroxidase anti-mouse (#K4007), specific to the primary antibody used for target protein detection (1:200; Alexa 555 goat anti-mouse (A21424) or goat anti-rabbit (A21429)), is based on the source of the anti-cytokeratin antibody (1:200) (Dako, Carpinteria, CA). For fluorescence amplification, slides were exposed to Cy5-tyramide (1:50) for 10 minutes at room temperature. Slides were mounted with Prolong Gold antifade reagent with 4’-6-diamidino-2-phenylindole (DAPI) solution. The final TMA slides were scanned with SpotGrabber, and image data were analyzed with AQUA (PM-2000, software version 1.6; HistoRx, New Haven, CT). For software version 1.6, the maximal range of scores is 0 to 33,333.
Immunohistochemistry (IHC) assays were developed, validated, and performed at a centralized laboratory (Ventana Medical Systems, Tucson, AZ) by Benchmark XT automated immunostainer. Commercial antibodies for TS were used (monoclonal TS106, Abcam, #AB3145 primary dilution 1:10; monoclonal 4H4B1, Zymed, #18–0399, primary dilution 1:20) with 1-hour primary incubation with CC1 standard (Ventana, #950-124) at room temperature and ultraView detection kit (Ventana, #760-500) for TS 4H4B1 or iViewPolymerDAB detection system (Ventana, #760-115) for TS106. Murine monoclonal antibodies for folyl-polyglutamate synthase (FPGS) and 5-phosphoribosyl-glycinamide formyl-transferase (GARFT) were developed by Integrated Biology/Translational Medicine at Eli Lilly (both primary dilution 1:160, with Ultraview detection). Anti-FPGS mAb was raised against purified recombinant N-terminal HIS-tagged full-length human FPGS protein. The mAb binds the full-length antigen with a dissociation constant of 2.58 × 10−12 M and stains a single protein at ~61 kDa in Western blots of whole cell extracts(38). Anti-GARFT mAb was raised against recombinant N-terminal HIS-tagged full-length human GARFT protein. The mAb binds the full-length antigen with a dissociation constant of 1.14 × 10−10 M and stains a single protein at ~110 kDa in Western blots of whole cell extracts(39). Expression was assessed semiquantitatively using the hybrid-score (H-score) method. For this, the percentage of tumor cells stained for a marker for each intensity category on a scale of 0 to 3 (for absent, slight, moderate, and marked staining, respectively) was enumerated. The percentage of cells in each category was then multiplied by its value, and the products were added. The maximal range of H-scores was 0 to 300(40).
Confocal Microscopy
Four different SCLC cell lines (H69, H82, H209, and H211) were grown in 75-cm2 flasks. Cells (5 × 104) were placed in a tube containing 250 µL of 20% fetal bovine serum-phosphate-buffered saline (PBS). Cell suspensions (250 µL) were added to each cytofunnel slot and spun at 570 rpm for 5 minutes. The cytofunnels were carefully removed from the slides. After the wet slides were air dried, the cells were fixed by incubation for 20 minutes in 4% paraformaldehyde in PBS and washed in PBS. They were permeabilized for 1 hour in 0.25% Triton X-100-PBS and washed in PBS. RRM1, ERCC1, TS, and Topo2α (1:100) antibodies were diluted in binding buffer (1% bovine serum albumin-0.1% Nonidet P40-PBS), added directly onto the cell spots (a Parafilm square covered the spots to prevent the cells from drying out), and incubated for 1 hour. After slides were washed in PBS, they were incubated for 45 minutes with 1:500 dilutions of Alexa Fluor 555 anti-mouse IgG (Molecular Probes-Invitrogen, Eugene, OR). The slides were washed with PBS and covered using ProLong Gold antifade reagent with DAPI (Molecular Probes-Invitrogen). As negative controls, the same procedure was performed without primary antibody. Samples were viewed with an inverted Zeiss LSM 510 confocal microscope with a 363/1.20NA water-immersion objective (Carl Zeiss, Oberkochen, Germany). Nuclei were observed with DAPI. Images were produced with dual photomultiplier detectors and the LSM 5 version 3.2.0.115 software suite.
Statistical Analysis/Methods
The primary objective was to describe the AQUA score and H-score expression distribution for each of the biomarkers. Descriptive statistics (mean, standard deviation, median, minimum, and maximum values) were calculated using the average values for the AQUA scores and H-scores from triplicate readings for each gene and treated as independent continuous variables. TS AQUA scores in the first three rows (13 values) of the TMA were excluded from analysis because the placement of the tissues on the slide precluded an accurate reading with the AQUA technology.
The secondary objective was to assess the correlations among the expression levels of the target proteins in their major cellular compartments, i.e., RRM1 nuclear versus ERCC1 nuclear, RRM1 nuclear versus Topo2α nuclear, RRM1 nuclear versus TS cytoplasmic, and so forth. Age and gender (the only clinical data available) were also correlated with protein expression. Spearman (rank) correlation was used to assess the associations between the different proteins and between proteins and age. Wilcoxon rank sum test was used to assess the association between proteins and gender. All P values were calculated with two-tailed significance levels. All statistical analyses were performed using SAS (version 9.1; SAS Institute; Cary, NC).
RESULTS
Patient Characteristics
Only gender and age were available for the patients contributing specimens to the studied TMA. Nineteen (19%) were women and 81 (81%) were men. The median age for all patients was 54 years (range 33–84).
Biomarker Expression Levels by AQUA and IHC
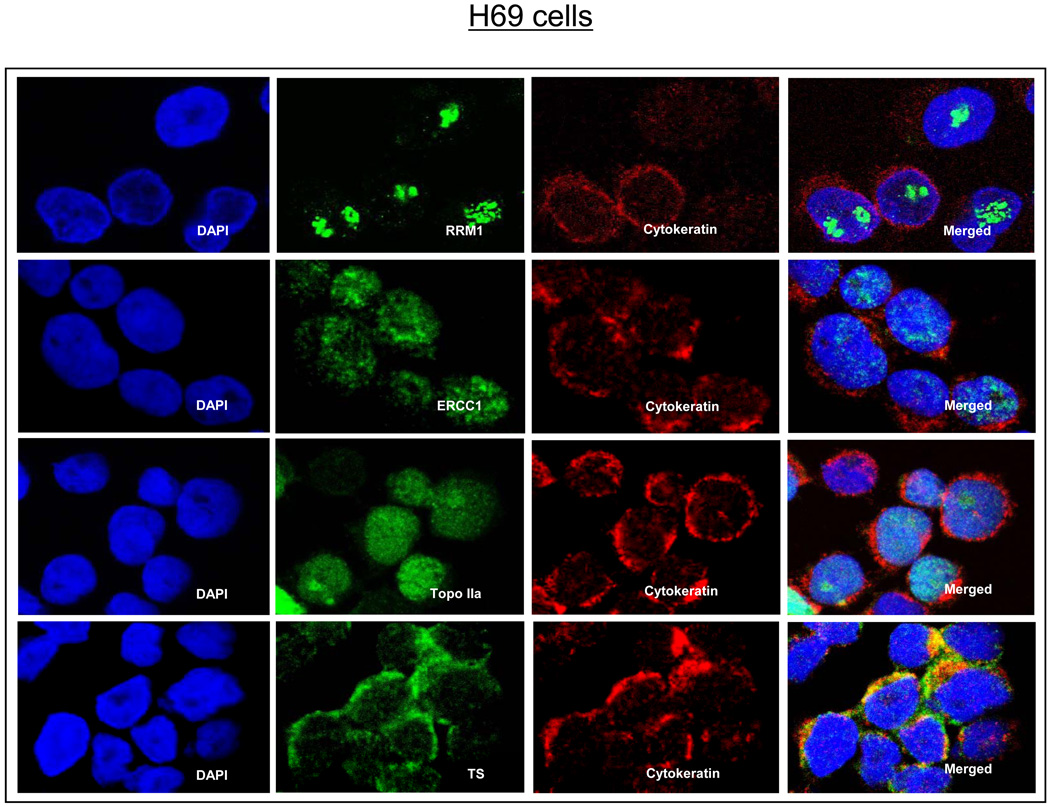
We used confocal microscopy to first determine the dominant cellular compartment of expression for each marker. As shown in Figure 1, RRM1, ERCC1, and Topo2α were predominantly nuclear in location, whereas TS was predominantly cytoplasmic. All scores were measured using the same scale.
Figure 1.
Confocal microscopy images of the 4 biomarkers in the H69 small cell lung cancer cell line. DAPI, 4’,6-diamidino-2-phenylindole; RRM1, ribonucleotide reductase 1; ERCC1, excision repair cross complementing group 1; TS, thymidylate synthase; Topo2α, topoisomerase 2α.
AQUA
Most of the triplicate AQUA scores from each sample were very close in value, suggesting little expression heterogeneity for these proteins in SCLC (data not shown). Table 1 summarizes the key descriptive parameters for the average AQUA scores of each of the biomarkers in the TMA. Numerically, the scores followed the following order: ERCC1<TS<RRM1<Topo2α. The scores for Topo2α and RRM1 had a wide range. Conversely, the scores for ERCC1 and TS had a narrow range and low values.
Table 1.
Protein Expression Levels by AQUA and IHC in 100 SCLC Patients
| AQUA Score | ||||||
|---|---|---|---|---|---|---|
| AQUA (v 1.6) | N | Mean | SD of Mean | Median | Minimum | Maximum |
| RRM1, nuclear | 100 | 568.5 | 168.4 | 550.7 | 143.4 | 1311.6 |
| ERCC1, nuclear | 100 | 19.9 | 6.5 | 18.1 | 11.3 | 48.1 |
| Topo2α,, nuclear | 100 | 652.9 | 226.2 | 634.7 | 246.2 | 1481.8 |
| TS (antibody 106), cytoplasm | 87 | 45.1 | 27.5 | 36.6 | 12.0 | 166.8 |
| H-Score | ||||||
| IHC | N | Mean | SD of Mean | Median | Minimum | Maximum |
| TS (antibody 106), cytoplasm | 90 | 26.2 | 36.9 | 5.2 | 0.0 | 148.7 |
| TS (antibody 4H4), cytoplasm | 96 | 68.4 | 59.3 | 59.3 | 0.0 | 231.7 |
| GARFT, nuclear | 94 | 219.3 | 55.8 | 235.0 | 15.7 | 300.0 |
| FPGS, cytoplasm | 95 | 248.7 | 33.0 | 253.0 | 163.3 | 300.0 |
IHC
The descriptive parameters for the average H-scores for TS, GARFT, and FPGS are shown in Table 1. As with the AQUA scores, both TS H-scores tended to be low with narrow ranges, whereas the scores for GARFT and FPGS were higher and with wider ranges.
Associations Among Biomarker Expression Levels, Age, and Gender
H-scores for TS (106) and TS (4H4) were very strongly correlated (r = .82). More modest correlations were seen between all other pairs of these two measures and TS AQUA and GARFT H-scores (.22 < r < .29). ERCC1 was modestly correlated with all TS (AQUA and IHC) assessments (.27 < r < .34) and RRM1 (r=.20), as shown in Table 2. Topo2α levels were strongly and significantly correlated only with RRM1 (r=0.56, p<0.0001). Furthermore, the individual distribution of RRM1 and Topo2α AQUA scores (n=100) showed values above both median scores in 35 patients (35%) and below both median scores in 35 patients; in 15 patients, scores are only above one of each (i.e., among the 50 patients with high RRM1 values, 30% have low Topo2α levels and vice versa).
Table 2.
Correlations Among Biomarkers in 100 SCLC Patients
| ERCC1 AQUA nuclear |
Topo2α AQUA nuclear |
TS (106) AQUA cytoplasm |
TS (106) IHC cytoplasm |
TS (4H4) IHC cytoplasm |
GARFT IHC nuclear |
FPGS IHC cytoplasm |
|
|---|---|---|---|---|---|---|---|
| RRM1, AQUA Nuclear |
r = 0.20 p = 0.05 n = 100 |
r = 0.56 p < .0001 n = 100 |
r = 0.12 p = 0.25 n = 87 |
r = −0.02 p = 0.87 n = 90 |
r = 0.06 p = 0.58 n = 96 |
r = 0.01 p = 0.91 n = 94 |
r = 0.21 p = 0.04 n = 95 |
| ERCC1, AQUA Nuclear |
NA | r = 0.16 p = 0.10 n = 100 |
r = 0.34 p = 0.001 n = 87 |
r = 0.27 p = 0.01 n = 90 |
r = 0.27 p = 0.008 n = 96 |
r = 0.16 p = 0.12 n = 94 |
r = 0.10 p = 0.35 n = 95 |
| Topo2α, AQUA Nuclear |
NA | r = 0.08 p = 0.45 n = 87 |
r = 0.04 p = 0.71 n = 90 |
r = 0.11 p = 0.27 n = 96 |
r = −0.15 p = 0.14 n = 94 |
r = 0.20 p = 0.05 n = 95 |
|
| TS (106), AQUA cytoplasm |
NA | r = 0.25 p = 0.03 n = 78 |
r = 0.29 p = 0.009 n = 83 |
r = 0.22 p = 0.05 n = 82 |
r = 0.00 p = 0.97 n = 82 |
||
| TS (106), IHC Cytoplasm |
NA | r = 0.82 p <.0001 n = 90 |
r = 0.23 p = 0.03 n = 90 |
r = −0.21 p = 0.05 n = 89 |
|||
| TS (4H4), IHC Cytoplasm |
NA | r = 0.28 p = 0.007 n = 94 |
r = −0.16 p = 0.12 n = 95 |
||||
| GARFT, IHC Nuclear |
NA | r = −0.02 p = 0.89 n = 93 |
NOTE: r = Spearman correlation coefficient, p = p-value, n = number of observations.
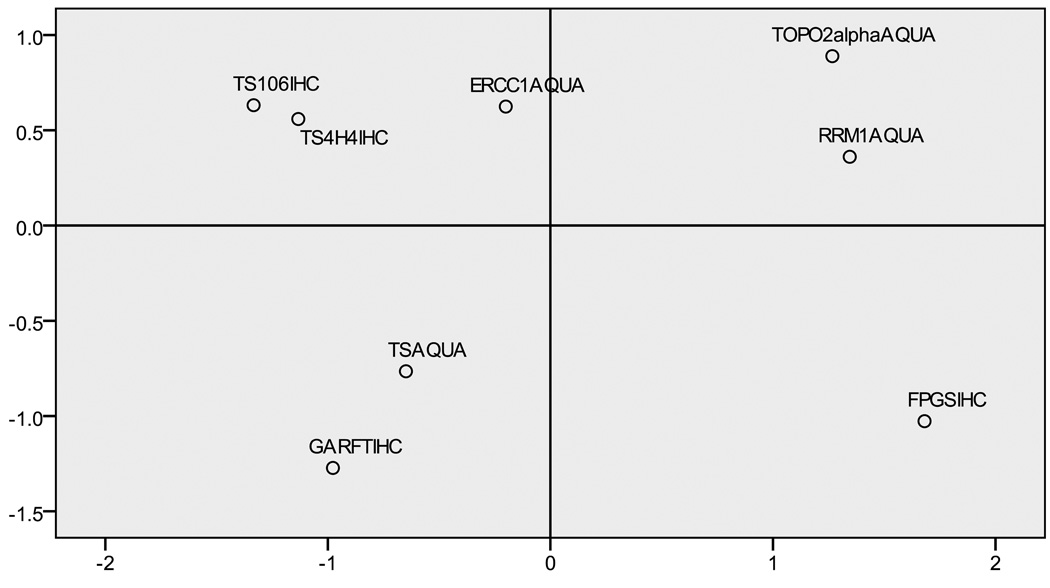
We used a two-dimensional spatial representation (Figure 2) to further examine the correlations between proteins. The distance between the protein locations represents the strength of the correlation (large distance represents low or negative correlation). Two correlative groups are shown (Figure 2, left and right). ERCC1 (Figure 2, middle) played a more central role, but with slightly stronger correlations with the TS biomarkers.
Figure 2.
Multidimensional scaling plot of IHC and AQUA scores.
We found no significant correlations between expression levels for any of these biomarkers and the patients’ age or gender.
DISCUSSION
Cancer develops as a result of gradual accumulation of genetic alterations that can affect normal cellular processes and function. The role that these “modified” gene products play, when involved in the actions of cytotoxic agents, can be used to tailor therapeutic choices; i.e., observed patterns of sensitivity or resistance could be used to customize treatments. A “customized” strategy has several advantages: (1) patients are more likely to be treated with agents that they will respond to, (2) patients can be spared the toxicity of agents that they are resistant to, and (3) effective treatment can be delivered early in the course of the disease.
Customized therapy has been previously investigated in SCLC using in vitro drug sensitivity testing(41, 42). In two trials, SCLC cell suspensions were incubated with various concentrations of individual cytotoxic drugs (cisplatin, etoposide, adriamycin, cyclophosphamide, vincristine, methotrexate, and carmustine). The dye exclusion assay(43) was used for all drug sensitivity tests to rank these 7 drugs and to select the in vitro best regimen. Although in both studies treatment with individualized chemotherapy was feasible and associated with numerically favorable outcomes (compared to non-randomized controls), the studies were small and the in vitro best regimen selection proved labor intensive and time consuming(44).
These trials highlight the biggest obstacles that customization therapy for SCLC currently faces: (1) lack of defined predictive biomarkers and (2) lack of reliable and reproducible technology to timely quantification. Multiple preclinical and clinical studies have confirmed the role of ERCC1, RRM1, Topo2α, and TS as predictive biomarkers for their respective cytotoxic agents,(21–32) thus providing a solution to the first obstacle. Solutions to the second obstacle have proved to be more elusive.
For example, RT-PCR analysis on FFPE tissue has been questioned due to cross-linking and degradation of mRNA by formalin fixation(45). Furthermore, mRNA expression may not accurately reflect protein expression and/or function. IHC provides information on protein expression and localization; thus, it is the standard protein in situ assay. However, IHC scoring is semiquantitative, subjective, and highly dependent on poorly controlled variables(46). TMAs reduce the variables in IHC scoring and provide a high-throughput method to analyze potential biomarkers on multiple samples but are limited by the pathologist’s ability to reproducibly score on a continuous scale, discriminate between subtle low-level staining differences, and accurately score expression within subcellular components. In addition, TMAs cannot be used for real-time patient specimen assessments.
AQUA is an immunofluorescence-based technique that allows for rapid, automated, and quantitative analysis of proteins(35), thus reducing the human variability occurring with IHC scoring. AQUA automatically measures protein expression in subcellular compartments (i.e., nuclear versus cytoplasmic), providing a continuous score in an accurate, reliable, and reproducible way. Reported results in NSCLC(37, 47) not only confirm ERCC1, RRM1, and TS as important biomarkers but also support AQUA as a valid methodology for use.
The future use of biomarkers to customize chemotherapy in patients with SCLC will first require a description of the biomarker expression levels to identify the cut-off points needed for dichotomization. Second, the biomarkers’ predictive powers will need to be validated. In this report, we have characterized the expression levels of 4 different proteins in SCLC (Table 1) using AQUA. Furthermore, with the help of confocal microscopy, we have also identified the appropriate subcellular compartment where each of these proteins should be measured (nucleus for RRM1, ERCC1, and Topo2α and cytoplasm for TS; Figure 1).
Not surprisingly, the AQUA scores for ERCC1 in our investigation were universally and homogeneously low and had a narrow distribution range (Table 1). This suggests a uniform level of sensitivity to cisplatin, in accordance with the well-known natural history of SCLC (typically sensitive to cisplatin and radiation), and thus renders the ERCC1-cisplatin biomarker-drug combination of low interest for SCLC chemotherapy customization.
Although numerically higher than ERCC1 scores, TS AQUA scores appear similarly low and narrow in range. However, unlike cisplatin, pemetrexed has shown substandard antitumor activity in recent SCLC trials(13). This is likely related to the differential TS expression observed in different lung cancer histologies. In NSCLC, TS expression (AQUA or RT-PCR) is lower in adenocarcinomas than in squamous cell carcinomas(47), correlating with the inferior antitumor activity recently described for pemetrexed in the latter(48). Furthermore, SCLC TS mRNA expression appears to be even higher than that of any NSCLC histology(49), thus putatively explaining the resistance to pemetrexed. Contrary to this observation however, our SCLC TS AQUA scores appeared numerically lower than those of both NSCLC histologies(47), but a direct comparison is unfortunately precluded by the different technology versions utilized (different ranges, accuracy, and variability).
In contrast, the distributions observed for the Topo2α and RRM1 AQUA scores had much wider ranges (Table 1), making them amenable to separation by a meaningful cut-off point. Additionally, an analysis of the individual distribution of Topo2α and RRM1 AQUA scores indicated that, in 30% of patients, these scores localized at opposite sides of their respective medians, suggesting the existence of “different populations” identifiable by their Topo2α and RRM1 AQUA scores, raising our enthusiasm for a possible chemotherapy customization using Topo2α and RRM1 to select etoposide and gemcitabine containing platinum regimens, respectively. Of course, unlike pemetrexed, the known activity of etoposide and gemcitabine regimens in SCLC strengthens the potential usefulness of these biomarkers as chemosensitivity predictors.
The predictive and prognostic values of some of these enzymes in SCLC have been recently reported(50). With the use of mRNA extracted from microdissected sections of FFPE biopsies from 103 LS- and ES-SCLC patients, ERCC1, RRM1, and Topo2α were quantified by RT-PCR. In LS-SCLC, only Topo2α mRNA predicted for better response (especially a complete response), and ERCC1 mRNA was the only independent prognostic factor for survival. No prognostic or predictive role was detected for any of these genes in ES-SCLC. Given the small numbers and the limitations inherent to the use of mRNA, reevaluation of these data, using AQUA, is justified.
The ultimate goal is the design of a prospective clinical trial in SCLC where the predictive powers of these biomarker expression levels are used to select the “most effective” cytotoxic agents for an “individualized” combination. Based on our present results, we hypothesize that selecting etoposide and gemcitabine as part of a platinum-doublet combination, based on Topo2α and RRM1 protein expression, is feasible and may improve the response and survival rates of patients with SCLC. A clinical trial to test this hypothesis is in development.
ACKNOWLEDGEMENTS
We thank Victor Chen and Steve Zuckerman with Eli Lilly Integrated Biology and Greg Stella and Anne Lodge with Ventana Medical Systems for their contributions in the IHC assay development and Rasa Hamilton with the Moffitt Cancer Center for her contribution editing this manuscript.
Research Support: This work was partially funded by NIH-R01-CA129343 to Gerold Bepler
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Janne PA, Freidlin B, Saxman S, et al. Twenty-five years of clinical research for patients with limited-stage small cell lung carcinoma in North America. Cancer. 2002;95:1528–1538. doi: 10.1002/cncr.10841. [DOI] [PubMed] [Google Scholar]
- 2.Simon GR, Wagner H. Small cell lung cancer. Chest. 2003;123:259S–271S. doi: 10.1378/chest.123.1_suppl.259s. [DOI] [PubMed] [Google Scholar]
- 3.Davies AM, Lara PN, Lau DH, Gandara DR. Treatment of extensive small cell lung cancer. Hematol Oncol Clin North Am. 2004;18:373–385. doi: 10.1016/j.hoc.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Ettinger DS. New drugs for chemotherapy-naive patients with extensive-disease small cell lung cancer. Semin Oncol. 2001;28:27–29. [PubMed] [Google Scholar]
- 5.Ettinger DS. Single-agent paclitaxel in the treatment of small cell lung cancer. Semin Oncol. 1996;23:16–17. [PubMed] [Google Scholar]
- 6.Hesketh PJ, Crowley JJ, Burris HA, 3rd, et al. Evaluation of docetaxel in previously untreated extensive-stage small cell lung cancer: a Southwest Oncology Group phase II trial. Cancer J Sci Am. 1999;5:237–241. [PubMed] [Google Scholar]
- 7.Schiller JH, Kim K, Hutson P, et al. Phase II study of topotecan in patients with extensive-stage small-cell carcinoma of the lung: an Eastern Cooperative Oncology Group Trial. J Clin Oncol. 1996;14:2345–2352. doi: 10.1200/JCO.1996.14.8.2345. [DOI] [PubMed] [Google Scholar]
- 8.Masuda N, Fukuoka M, Kusunoki Y, et al. CPT-11: a new derivative of camptothecin for the treatment of refractory or relapsed small-cell lung cancer. J Clin Oncol. 1992;10:1225–1229. doi: 10.1200/JCO.1992.10.8.1225. [DOI] [PubMed] [Google Scholar]
- 9.Socinski MA, Weissman C, Hart LL, et al. Randomized phase II trial of pemetrexed combined with either cisplatin or carboplatin in untreated extensive-stage small-cell lung cancer. J Clin Oncol. 2006;24:4840–4847. doi: 10.1200/JCO.2006.07.7016. [DOI] [PubMed] [Google Scholar]
- 10.Chiappori AA, Rocha-Lima CM. New agents in the treatment of small-cell lung cancer: focus on gemcitabine. Clin Lung Cancer. 2003;4 Suppl 2:S56–S63. doi: 10.3816/clc.2003.s.005. [DOI] [PubMed] [Google Scholar]
- 11.Hanna N, Bunn PA, Jr, Langer C, et al. Randomized phase III trial comparing irinotecan/cisplatin with etoposide/cisplatin in patients with previously untreated extensive-stage disease small-cell lung cancer. J Clin Oncol. 2006;24:2038–2043. doi: 10.1200/JCO.2005.04.8595. [DOI] [PubMed] [Google Scholar]
- 12.Lara PN, Jr, Gandara DR, Natale RB. Randomized phase III trial of cisplatin/irinotecan versus cisplatin/etoposide in patients with extensive-stage small-cell lung cancer. Clin Lung Cancer. 2006;7:353–356. doi: 10.3816/CLC.2006.n.019. [DOI] [PubMed] [Google Scholar]
- 13.Socinski MA, Smit EF, Lorigan P, et al. Phase III study of pemetrexed plus carboplatin compared with etoposide plus carboplatin in chemotherapy-naive patients with extensive-stage small-cell lung cancer. J Clin Oncol. 2009;27:4787–4792. doi: 10.1200/JCO.2009.23.1548. [DOI] [PubMed] [Google Scholar]
- 14.Heigener DF, Freitag L, Eschbach C, et al. Topotecan/cisplatin compared to cisplatin/etoposide for patients with extensive stage small cell lung cancer. Final results of a phase III randomised study. J Clin Oncol (abst # 7513) 2008;26(15S):400s. [Google Scholar]
- 15.Bepler G. Using translational research to tailor the use of chemotherapy in the treatment of NSCLC. Lung Cancer. 2005;50 Suppl 1:S13–S14. doi: 10.1016/s0169-5002(05)81553-3. [DOI] [PubMed] [Google Scholar]
- 16.Bepler G. Pharmacogenomics: a reality or still a promise? Lung Cancer. 2006;54 Suppl 2:S3–S7. doi: 10.1016/j.lungcan.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plunkett W, Huang P, Searcy CE, Gandhi V. Gemcitabine: preclinical pharmacology and mechanisms of action. Semin Oncol. 1996;23:3–15. [PubMed] [Google Scholar]
- 18.Larsen AK, Skladanowski A, Bojanowski K. The roles of DNA topoisomerase II during the cell cycle. Prog Cell Cycle Res. 1996;2:229–239. doi: 10.1007/978-1-4615-5873-6_22. [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Yu JJ, Mu C, et al. Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer Res. 2000;20:645–652. [PubMed] [Google Scholar]
- 20.Rose MG, Farrell MP, Schmitz JC. Thymidylate synthase: a critical target for cancer chemotherapy. Clin Colorectal Cancer. 2002;1:220–229. doi: 10.3816/CCC.2002.n.003. [DOI] [PubMed] [Google Scholar]
- 21.Uesaka T, Shono T, Kuga D, et al. Enhanced expression of DNA topoisomerase II genes in human medulloblastoma and its possible association with etoposide sensitivity. J Neurooncol. 2007;84:119–129. doi: 10.1007/s11060-007-9360-0. [DOI] [PubMed] [Google Scholar]
- 22.Tinari N, Lattanzio R, Natoli C, et al. Changes of topoisomerase IIalpha expression in breast tumors after neoadjuvant chemotherapy predicts relapse-free survival. Clin Cancer Res. 2006;12:1501–1506. doi: 10.1158/1078-0432.CCR-05-0978. [DOI] [PubMed] [Google Scholar]
- 23.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 24.Simon GR, Sharma S, Cantor A, Smith P, Bepler G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest. 2005;127:978–983. doi: 10.1378/chest.127.3.978. [DOI] [PubMed] [Google Scholar]
- 25.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 26.Bepler G, Sommers KE, Cantor A, et al. Clinical efficacy and predictive molecular markers of neoadjuvant gemcitabine and pemetrexed in resectable non-small cell lung cancer. J Thorac Oncol. 2008;3:1112–1118. doi: 10.1097/JTO.0b013e3181874936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu JM, Wang LS, Huang MH, et al. Topoisomerase 2alpha plays a pivotal role in the tumor biology of stage IV thymic neoplasia. Cancer. 2007;109:502–509. doi: 10.1002/cncr.22404. [DOI] [PubMed] [Google Scholar]
- 28.Reed E. ERCC1 and clinical resistance to platinum-based therapy. Clin Cancer Res. 2005;11:6100–6102. doi: 10.1158/1078-0432.CCR-05-1083. [DOI] [PubMed] [Google Scholar]
- 29.Lord RV, Brabender J, Gandara D, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 30.Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64:3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- 31.Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 32.Rosell R, Felip E, Taron M, et al. Gene expression as a predictive marker of outcome in stage IIB-IIIA-IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clin Cancer Res. 2004;10:4215s–4219s. doi: 10.1158/1078-0432.CCR-040006. [DOI] [PubMed] [Google Scholar]
- 33.Simon G, Sharma A, Li X, et al. Feasibility and efficacy of molecular analysis-directed individualized therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2007;25:2741–2746. doi: 10.1200/JCO.2006.08.2099. [DOI] [PubMed] [Google Scholar]
- 34.Rosell R, Cobo M, Isla D, et al. ERCC1 mRNA-based randomized Phase III trial of docetaxel (doc) doublets with cisplatin (cis) or gemcitabine (gem)in stage IV non-small clell lung cancer (NSCLC) patients (p) Proc ASCO. 2005;23:621s. [Google Scholar]
- 35.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 36.Shi SR, Cote RJ, Taylor CR. Antigen retrieval immunohistochemistry: past, present, and future. J Histochem Cytochem. 1997;45:327–343. doi: 10.1177/002215549704500301. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 38.Dotzlaf J, Carpenter J, Luo S, et al. Derivation and characterization of monoclonal antibodies against human folypolyglutamate synthetase. Hybridoma (Larchmt) 2007;26:155–161. doi: 10.1089/hyb.2007.004. [DOI] [PubMed] [Google Scholar]
- 39.Dotzlaf J, Carpenter J, Luo S, et al. Derivation and characterization of a monoclonal antibody against human glycinamide ribonucleotide formyltransferase. Hybridoma (Larchmt) 2006;25:139–144. doi: 10.1089/hyb.2006.25.139. [DOI] [PubMed] [Google Scholar]
- 40.Detre S, Saclani Jotti G, Dowsett M. A "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gazdar AF, Steinberg SM, Russell EK, et al. Correlation of in vitro drug-sensitivity testing results with response to chemotherapy and survival in extensive-stage small cell lung cancer: a prospective clinical trial. J Natl Cancer Inst. 1990;82:117–124. doi: 10.1093/jnci/82.2.117. [DOI] [PubMed] [Google Scholar]
- 42.Cortazar P, Gazdar AF, Woods E, et al. Survival of patients with limited-stage small cell lung cancer treated with individualized chemotherapy selected by in vitro drug sensitivity testing. Clin Cancer Res. 1997;3:741–747. [PubMed] [Google Scholar]
- 43.Weisenthal LM, Marsden JA, Dill PL, Macaluso CK. A novel dye exclusion method for testing in vitro chemosensitivity of human tumors. Cancer Res. 1983;43:749–757. [PubMed] [Google Scholar]
- 44.Cortazar P, Johnson BE. Review of the efficacy of individualized chemotherapy selected by in vitro drug sensitivity testing for patients with cancer. J Clin Oncol. 1999;17:1625–1631. doi: 10.1200/JCO.1999.17.5.1625. [DOI] [PubMed] [Google Scholar]
- 45.Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol. 2001;195:72–79. doi: 10.1002/path.893. [DOI] [PubMed] [Google Scholar]
- 46.McCabe A, Dolled-Filhart M, Camp RL, Rimm DL. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97:1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 47.Zheng Z, Li X, Schell MJ, et al. Thymidylate synthase in situ protein expression and survival in stage I nonsmall-cell lung cancer. Cancer. 2008;112:2765–2773. doi: 10.1002/cncr.23491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 49.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ceppi P, Longo M, Volante M, et al. Excision repair cross complementing-1 and topoisomerase IIalpha gene expression in small-cell lung cancer patients treated with platinum and etoposide: a retrospective study. J Thorac Oncol. 2008;3:583–589. doi: 10.1097/JTO.0b013e3181734f24. [DOI] [PubMed] [Google Scholar]