Abstract
In zebrafish, TATA-box-binding protein (TBP)-related factor 3, Trf3, is required for early development and initiation of hematopoiesis, and functions by promoting expression of a single target gene, mespa. Recent studies have shown that in murine muscle cells, TRF3 interacts with the TBP-associated factor TAF3. Here we investigate the role of Taf3 in zebrafish embryogenesis. We find that like Trf3-depleted zebrafish embryos, Taf3-depleted embryos exhibit multiple developmental defects and fail to undergo hematopoiesis. Both Trf3 and Taf3 are selectively bound to the mespa promoter and are required for mespa expression. Significantly, Taf3 interacts with Trf3 but not Tbp, and a Trf3 mutant that disrupts this interaction fails to support mespa transcription, early development and hematopoiesis. Thus, a selective interaction between Trf3 and Taf3 is required for early zebrafish development and initiation of hematopoiesis. Finally, we provide evidence that TRF3 and TAF3 are also required for hematopoiesis initiation in the mouse.
Keywords: Taf3, Trf3, zebrafish, hematopoiesis, embryogenesis
INTRODUCTION
The faithful execution of many biological processes requires a precise and carefully orchestrated set of steps that depend on the proper spatial and temporal expression of genes. In many instances, this regulation is primarily imposed at the transcriptional level. Well-known examples include embryonic development and, in adults, commitment of multipotential cells, such as stem cells, to specific differentiation pathways (see, for example, Murre, 2005; Stennard and Harvey, 2005; Wegner and Stolt, 2005; Shi et al., 2006). Numerous studies have shown that these processes are regulated through specific networks of sequence-specific transcriptional activator proteins (activators).
In addition to activators, the general transcription machinery can also contribute to gene regulation. Although originally thought to be invariant, it is now clear that the general transcription machinery can also have substantial diversity (reviewed in Davidson, 2003; Hochheimer and Tjian, 2003). This notion is illustrated most clearly by the general transcription factor (GTF) TFIID, a multi-subunit complex consisting of the TATA-box-binding protein (TBP) and a set of tightly bound TBP-associated factors (TAFs) (Burley and Roeder, 1996; Albright and Tjian, 2000; Green, 2000). Distinct forms of TFIID have been described in different tissues and cell types, and are believed to direct specific transcriptional programs. For example, the presence of a gonad-specific form of TAF4, TAF4b, in mice (Freiman et al., 2001; Falender et al., 2005) and testis-specific forms of several TAFs in Drosophila melanogaster (Hiller et al., 2001; Hiller et al., 2004) enable the formation of tissue-specific TFIID complexes that direct gonad-specific transcriptional programs such as spermiogenesis.
TFIID diversity is also promoted through the existence of several TBP variants called TBP-related factors (TRFs). The most recently identified TRF, TRF3 (also called TBP2 or TBPL2), was originally found based upon a search of an initial draft of the human genome sequence (Tupler et al., 2001) and its existence subsequently confirmed by expression analyses (Persengiev et al., 2003). Unlike the two other known TRFs, TRF3 is present in vertebrates (from fish through humans) but not lower metazoans and has a conserved C-terminal region and DNA-binding domain virtually identical to that of TBP. However, despite the highly homologous TBP-like C-terminal core domain, gel filtration analysis indicates TRF3 is in a complex that is substantially smaller than TFIID (Persengiev et al., 2003; Deato and Tjian, 2007).
TRF3 is highly expressed during embryonic development, and studies in zebrafish and Xenopus have shown that TRF3 is required for normal embryogenesis (Bartfai et al., 2004; Jallow et al., 2004; Hart et al., 2007). We have found that Trf3-depleted zebrafish embryos exhibit multiple developmental defects and, in particular, fail to undergo hematopoiesis (Hart et al., 2007). Remarkably, in zebrafish embryos a single Trf3 target gene, mespa, is essential for embryogenesis and functions, in part, through a Trf3-Mespa-Cdx4 transcription factor pathway that commits mesoderm to the hematopoietic lineage (Hart et al., 2007).
Recent studies have demonstrated that in terminally differentiated murine muscle cells a complex containing TRF3 and TAF3 replaces TFIID (Deato and Tjian, 2007) and is recruited to the promoters of target genes through interaction with a muscle-specific activator (Deato et al., 2008). Here we investigate the role of Taf3 in early zebrafish development.
RESULTS
Taf3 is Required for Normal Development and Hematopoiesis in Early Zebrafish Embryos
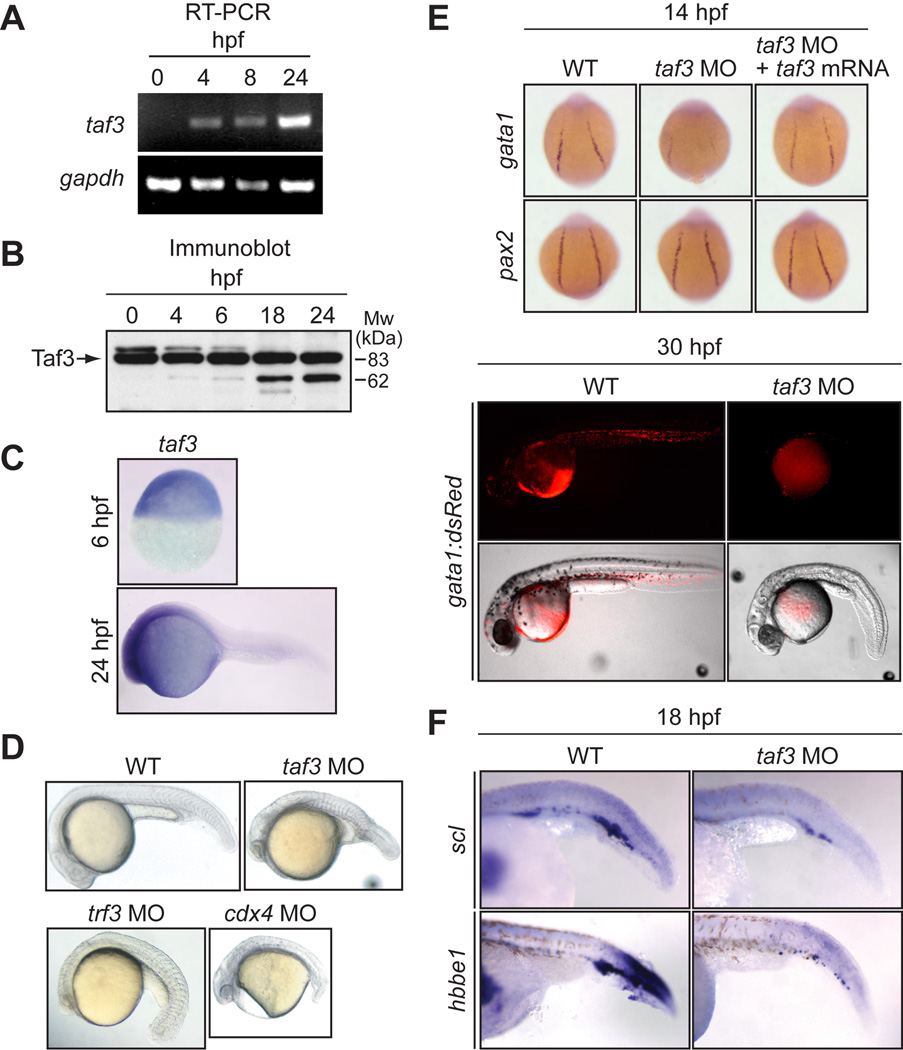
As a first step toward delineating the function of Taf3, we examined the expression profile of the taf3 gene in early zebrafish embryos. The RT-PCR analysis of Fig. 1A shows that the taf3 transcript was first detected at 4 hours post fertilization (hpf), coincident with the onset of zygotic transcription. By contrast, the Taf3 protein was detected as early as the one cell stage embryo (0 hpf; Fig. 1B), indicating that the protein is maternally deposited, similar to the Trf3 protein (Hart et al., 2007). Whole-mount in situ hybridization analysis revealed that taf3 mRNA was widely distributed throughout the zebrafish embryo with no discernible tissue-restriction during the first 24 hours of development (Fig. 1C).
Fig. 1.
Taf3 is required for normal development and hematopoiesis in early zebrafish embryos. A: RT-PCR analysis monitoring expression of taf3 and, as a control, gapdh in zebrafish embryos at 0, 4, 8 and 24 hpf. B: Immunoblot analysis monitoring expression of Taf3 in zebrafish embryos at 0, 4, 6, 18 and 24 hpf. C: Whole-mount in situ hybridization with a riboprobe to taf3 at 6 and 24 hpf. D: Phenotypic analysis of embryos injected with a control, taf3, trf3 or cdx4 MO at 24 hpf. E: (Top) Whole-mount in situ hybridization with riboprobes to gata1 and pax2 (14 hpf). (Bottom) Fluorescence microscopy monitoring expression of an gata1:dsRed transgene at 30 hpf. F: Whole-mount in situ hybridization with riboprobes to scl and hbbe1 (18 hpf).
To investigate the role of Taf3 in zebrafish development, we used a morpholino oligonucleotide (MO) to ablate Taf3 function. We first performed immunoblot analysis to confirm that injection of the taf3 MO resulted in decreased levels of Taf3 (Supp. Fig. S1). Figure 1D shows that injection of the taf3 MO into one-cell stage fertilized zebrafish embryos resulted in a retardation of development and gross morphological defects in late somitogenesis. These developmental defects were strikingly similar to those observed in Trf3-depleted embryos (Hart et al., 2007), and bore a strong resemblance to Cdx4-depleted embryos (Fig. 1D and Davidson et al., 2003) and the Cdx4 mutant, kugelig (Hammerschmidt et al., 1996).
To determine whether Taf3-depleted embryos, like Trf3- and Cdx4-depleted embryos, failed to undergo hematopoiesis, we analyzed Taf3 morphants for expression of several hematopoietic markers. First, we analyzed expression of gata1, which is required for the expression of a variety of genes in the erythroid lineage. The whole-mount in situ hybridization experiment of Fig. 1E (top panel) shows that the characteristic expression pattern of gata1 in the lateral mesoderm was lost in Taf3-depleted embryos and restored by injection of taf3 mRNA bearing a silent mutation that prevented hybridization with the taf3 MO. Expression of a second lateral mesoderm marker, pax2, was unaffected by loss of Taf3, consistent with our previous finding that pax2 expression is not dependent upon Trf3 (Hart et al., 2007). We also examined expression of gata1 using a transgenic line in which the fluorescent protein dsRed is under control of the endogenous gata1 promoter (Traver et al., 2003). Figure 1E (bottom panel) shows that expression of the gata1:dsRed transgene was greatly reduced in Taf3-depleted embryos (n=168/255). In addition, we used in situ hybridization to monitor expression of two other hematopoietic markers, scl and hbbe1, which were both reduced in Taf3-depleted embryos (Fig. 1F). Expression of pu.1, a marker of myeloid cells that arise in the anterior lateral mesoderm, was also reduced in the absence of Taf3 (Supp. Fig. S2), consistent with our previous findings that pu.1 expression is diminished in Trf3- and Mespa-depleted embryos (Hart et al., 2007).
It has been previously shown that some MOs can non-specifically activate a p53 pathway resulting in apparent development defects (Robu et al., 2007). We therefore performed a series of experiments, the results of which conclusively demonstrate that the developmental and hematopoietic defects we observed in Taf3-depleted embryos were not due to non-specific p53 activation (Supp. Fig. S3). Collectively, the results of Fig. 1 indicate that, like Trf3, Taf3 is required for zebrafish hematopoiesis.
Taf3 is Selectively Recruited to the Promoter of the Trf3 Target Gene mespa in vivo and is Required for mespa Expression
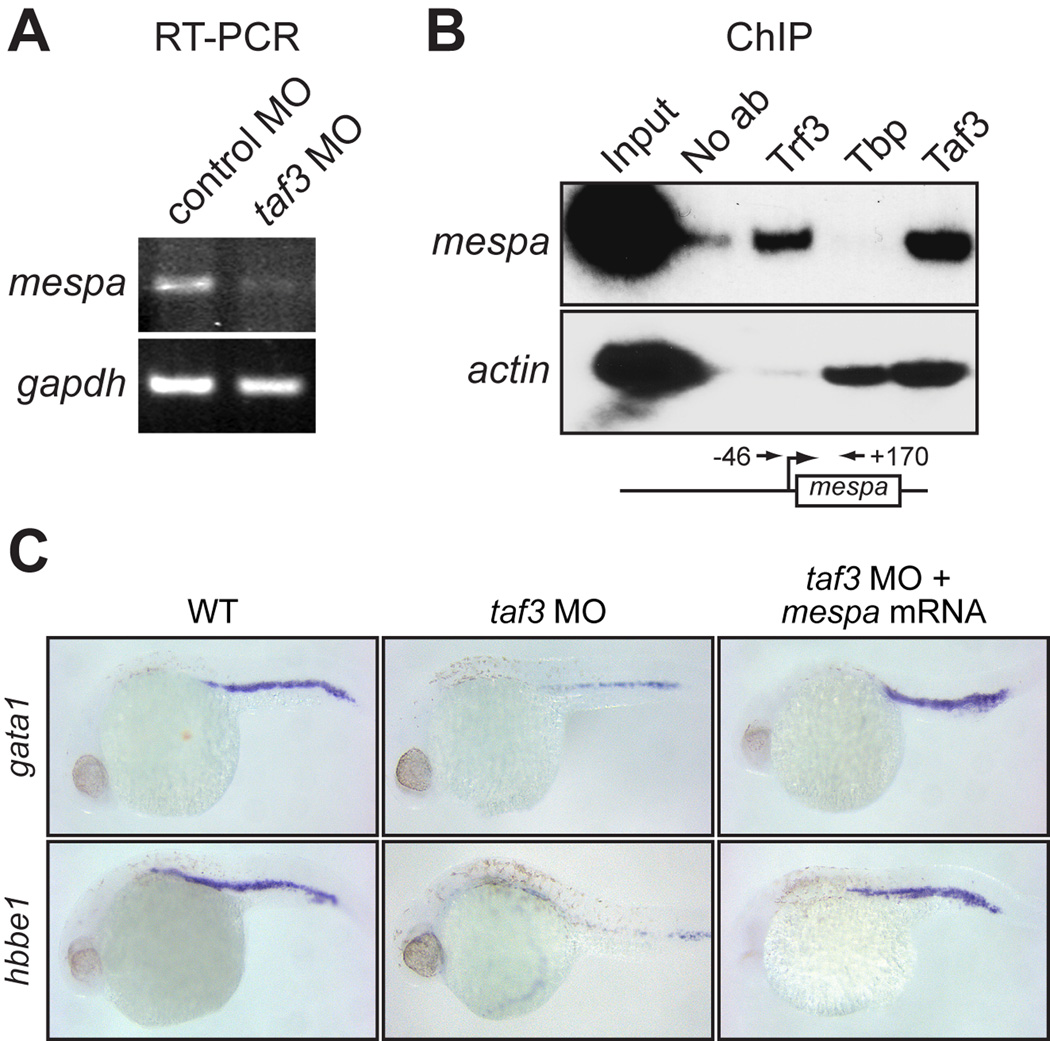
As stated above, we previously identified mespa as a direct Trf3 target gene; Trf3, but not Tbp, is recruited to the mespa promoter and is required for mespa expression (Hart et al., 2007). The RT-PCR analysis of Fig. 2A shows that mespa mRNA levels were significantly reduced in Taf3-depleted embryos, indicating that Taf3 is also required for mespa expression. Chromatin immunoprecipitation (ChIP) experiments in wild-type zebrafish embryos confirmed that Taf3 was bound to the mespa promoter (Fig. 2B). Collectively, these results indicate that Taf3 is an essential component of the pre-initiation complex involved in mespa expression.
Fig. 2.
Taf3 is selectively recruited to the promoter of the Trf3 target gene mespa in vivo and is required for mespa expression. A: RT-PCR analysis monitoring expression of mespa in control and taf3 MO-treated embryos at 6 hpf. B: ChIP analysis monitoring occupancy of Trf3, Tbp and Taf3 at the mespa and actin promoters in embryos at 6 hpf. Control ChIP experiments were performed in the absence of antibody (No ab). A schematic representation of the mespa promoter showing the location of the primer pairs used for ChIP analysis is shown (bottom). C: Whole-mount in situ hybridization with riboprobes to gata1 and hbbe1 (28 hpf) in embryos injected with a control MO or taf3 MO, or in taf3 MO-treated embryos injected with a mespa mRNA.
We have previously shown that the developmental and hematopoietic defects of Trf3-depleted embryos can be rescued by ectopic expression of mespa (Hart et al., 2007). We therefore examined whether mespa expression could similarly rescue the defects of Taf3-depleted embryos. Figure 2C shows that injection of mespa mRNA restored normal expression of gata1 and hbbe1 to taf3 MO-injected embryos, consistent with our conclusion that mespa is a downstream target of Taf3.
Taf3 Selectively Interacts with the C-terminal Domain of Trf3 but not Tbp
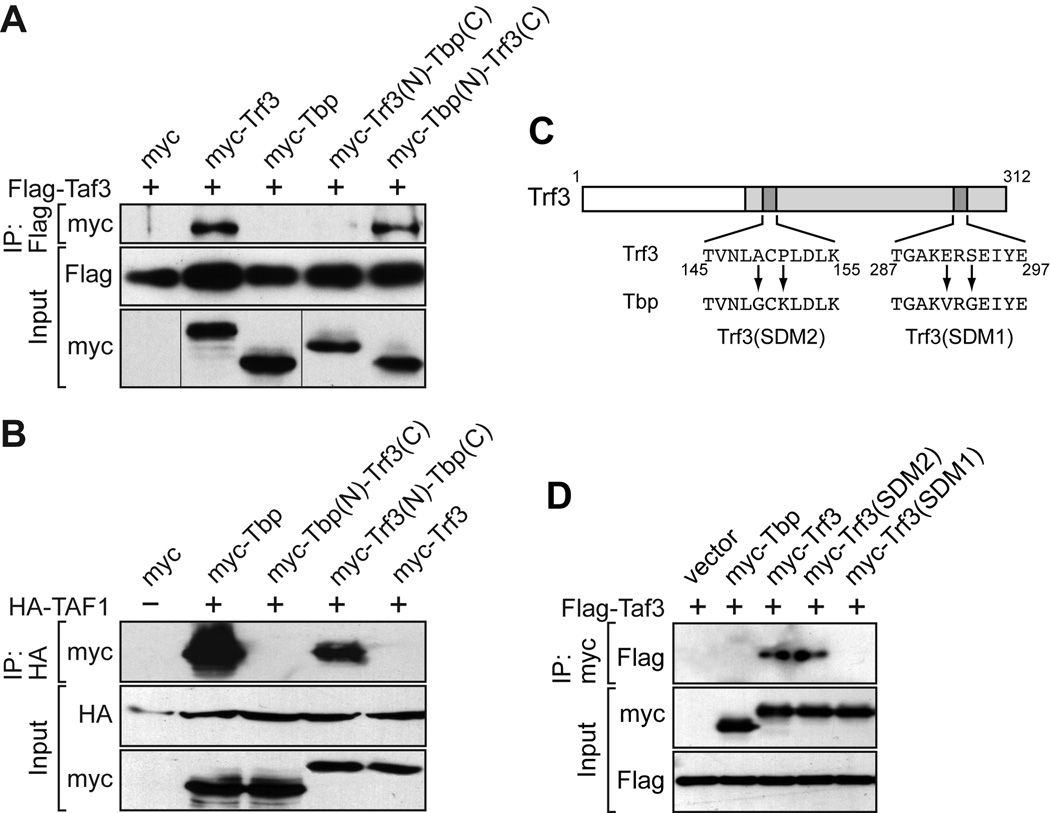
We next investigated whether, as in mouse cell lines (Deato and Tjian, 2007), zebrafish Trf3 and Taf3 physically interact. Plasmids expressing Flag-tagged Taf3 and either myc-tagged Trf3 or Tbp were co-transfected into COS-7 cells. After 48 hours, extracts were prepared, Taf3 was immunoprecipitated with an anti-Flag antibody, and the immunoprecipitates were analyzed for the presence of Trf3 or Tbp by immunoblotting with an anti-myc antibody. The results of Fig. 3A show that Taf3 interacted with Trf3 but not Tbp.
Fig. 3.
Taf3 selectively interacts with the C-terminal domain of Trf3 but not Tbp. A: Co-immunoprecipitation assays. Plasmids expressing Flag-tagged Taf3 and either myc-tagged Trf3, Tbp or Trf3/Tbp chimeric protein were co-transfected into COS-7 cells. Taf3 was immunoprecipitated with an anti-Flag antibody, and the immunoprecipitates were analyzed for the presence of Trf3, Tbp or Trf3/Tbp chimera by immunoblotting with an anti-myc antibody. B: Co-immunoprecipitation assays as described in (A) using HA-tagged TAF1. C: Schematic diagram of the zebrafish Trf3 protein, showing the substitution mutations. D: Co-immunoprecipitation assays. Plasmids expressing Flag-tagged Taf3 and either myc-tagged Tbp, wild-type Trf3 or Trf3 mutant were co-transfected into COS-7 cells. Tbp or Trf3 was immunoprecipitated with an anti-myc antibody, and the immunoprecipitates were analyzed for the presence of Taf3 by immunoblotting with an anti-Flag antibody.
As stated above, TRF3 has a C-terminal DNA-binding domain nearly identical to that of TBP, whereas the N-terminal domain is highly divergent. It therefore seemed likely that residues in the N-terminus would provide specificity for Taf3 association, and that a Trf3 derivative lacking the N-terminus would be unable to interact with Taf3. To test this hypothesis, we performed a domain-swapping experiment. We constructed a set of myc-tagged chimeric proteins consisting of the N-terminus of Trf3 fused to the C-terminus of Tbp [Trf3(N)-Tbp(C)] or the N-terminus of Tbp fused to the C-terminus of Trf3 [Tbp(N)-Trf3(C)], and tested the ability of these chimeric proteins to co-immunoprecipitate with Flag-tagged Taf3. Unexpectedly, we found that the Tbp(N)-Trf3(C) but not the Trf3(N)-Tbp(C) chimeric protein interacted with Taf3 (Fig. 3A), suggesting that the determinants required for interaction with Taf3 reside within the C-terminus of Trf3.
As a control, we next performed a similar set of experiments with TAF1, which directly interacts with TBP and is believed to function as a scaffold of the TFIID complex (Chen et al., 1994). Because the zebrafish Taf1 had not yet been cloned, we used human TAF1, which is 76% identical to the zebrafish protein. Co-immunoprecipitation analysis showed that TAF1 interacted with zebrafish Tbp but not Trf3 (Fig. 3B). Domain-swapping experiments using the chimeric proteins described above revealed that the interaction between Tbp and TAF1 was mediated by the C-terminus of Tbp. Collectively the results of Figs. 3A and B suggest that residues within the C-terminal conserved domains of Tbp and Trf3 are responsible for selective interaction with Taf1 and Taf3, respectively.
To confirm this conclusion we attempted to isolate Trf3 mutants that were defective for interaction with Taf3. Of the 192 amino acids in the C-terminal domain of Trf3, only 14 differ from the corresponding residue in the C-terminal domain of Tbp (Supp. Fig. S4). Within these 14 amino acids we generated a series of site-directed double amino acid substitutions that changed Trf3 residues to the corresponding Tbp sequence (Fig. 3C). This approach enabled us to identify a pair of amino acid substitutions, E291V and S293G (hereafter called site-directed mutant 1 or SDM1), which abolished the ability of Trf3 to interact with Taf3 (Fig. 3D). Notably, these two amino acids also differ between the human TRF3 and TBP proteins (Persengiev et al., 2003).
Interaction with Taf3 is Required for Trf3 Function in Zebrafish Development
We used the Trf3(SDM1) mutant to ask whether the Trf3-Taf3 interaction was required for Trf3 function in vivo. Synthetic mRNAs encoding Trf3, Tbp, Trf3(SDM1) and as a control another Trf3 mutant harboring a pair of amino acid substitutions, A149G and P151K (hereafter referred to as SDM2), which retains interaction with Taf3 (see Fig. 3D), were injected into one cell stage Trf3 morphants and analyzed for their ability to rescue morphological development. As expected, injection of a trf3 mRNA bearing a silent mutation that prevented hybridization with the trf3 MO restored normal morphological development whereas injection of a tbp mRNA did not (Fig. 4A). Injection of an mRNA encoding the Trf3(SDM2) mutant was also able to rescue normal development. By contrast, injection of an mRNA encoding the Trf3(SDM1) mutant failed to rescue development.
Fig. 4.
Interaction with Taf3 is required for Trf3 function in zebrafish development. A: Whole-mount in situ hybridization with a riboprobe to hbae1 (22 hpf) in trf3 MO-treated embryos injected with mRNAs expressing tbp, trf3 or a trf3 mutant. Arrowheads denote staining in the intermediate cell mass (ICM). B: Whole-mount in situ hybridization with riboprobes to gata1 or pax2 (14 hpf). C: RT-PCR analysis of scl1, lmo2 or, as a control, gapdh expression at 18 hpf.
To assess the completeness of rescue in greater detail, we analyzed expression of several hematopoietic markers. The in situ hybridization results of Fig. 4A show that expression of hbae1, a terminal marker of erythroid cell fate, was rescued by injection of an mRNA encoding the Trf3(SDM2) mutant but not the Trf3(SDM1) mutant. Similar results were obtained for all other hematopoietic markers analyzed, including gata1 (Fig. 4B) and earlier markers of hematopoietic precursors, scl1 and lmo2 (Fig. 4C). These results indicate that interaction with Taf3 is required for Trf3 function in zebrafish development and hematopoiesis.
TRF3 and TAF3 are also Required for Initiation of Hematopoiesis in the Mouse
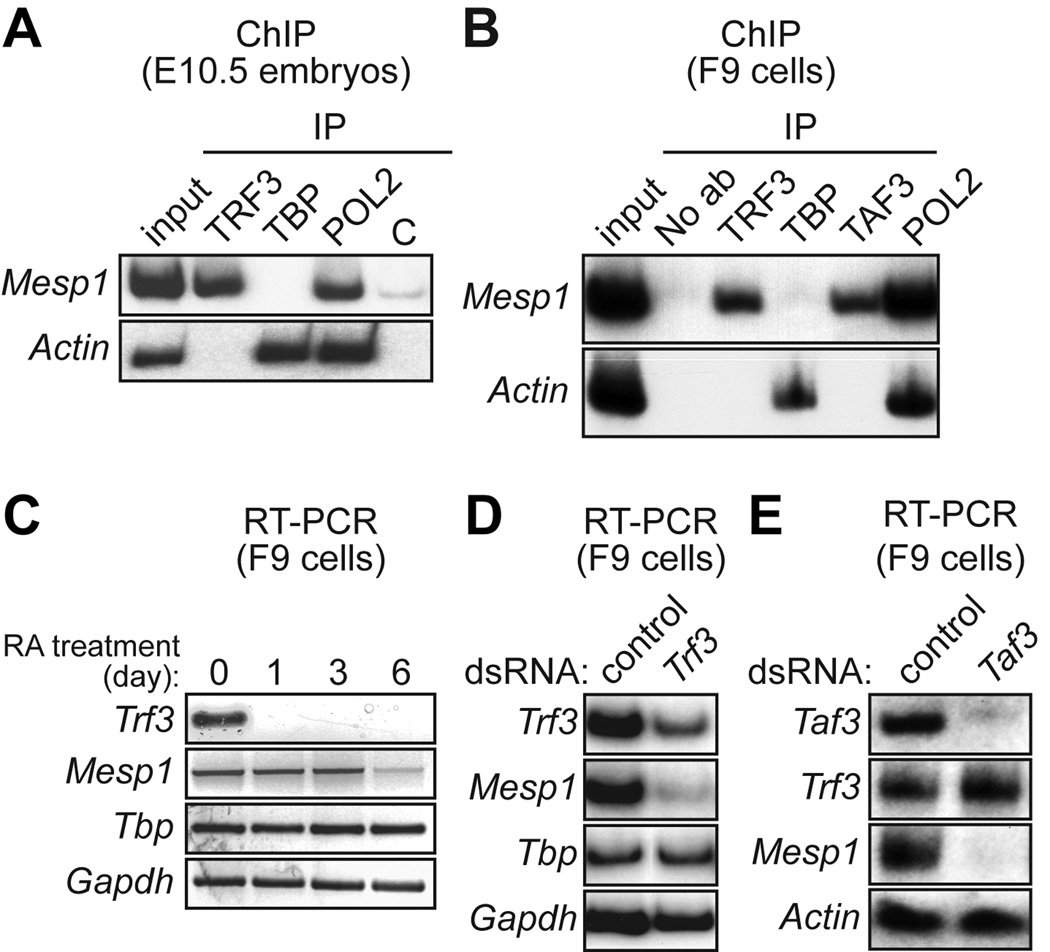
The pathways required for hematopoietic development are in general highly conserved throughout vertebrates, although some notable differences have been documented (reviewed in Fossett and Schulz, 2001). We therefore asked whether TRF3 and TAF3 had roles in initiation of hematopoiesis in the mouse similar to those in zebrafish embryos. We first analyzed whether, as in zebrafish (Hart et al., 2007), there was a TRF3-MESP1-CDX4 transcription factor pathway in mouse (MESP1 is the murine orthologue of zebrafish Mespa). In the initial experiments we tested whether mouse Mesp1 was, like zebrafish mespa, a Trf3 target gene. Figure 5A shows, identical to the results obtained in zebrafish embryos, that in embryonic day 10.5 (E10.5) mouse embryos TRF3 but not TBP was bound selectively to the Mesp1 promoter.
Fig. 5.
Requirement of TRF3 and TAF3 for expression of mouse Mesp1. A: ChIP analysis monitoring occupancy of TRF3, TBP, RNA polymerase II (POL2) or, as a negative control (C), an irrelevant protein (yeast Gal4) on the Mesp1 and Actin promoters in mouse E10.5 embryos. B: ChIP analysis monitoring promoter occupancy of TRF3, TBP, TAF3 and POL2 in mouse F9 embryonal teratocarcinoma cells. C: RT-PCR analysis of Trf3 and Mesp1 gene expression in F9 cells at 0, 1, 3 or 6 days after addition of retinoic acid (RA). D: RNAi. Trf3 double-stranded RNA (dsRNA) or, as a control, GFP dsRNA, was transfected into F9 cells and 48 h later Trf3, Mesp1, Tbp and Gapdh expression were monitored by RT-PCR analysis. E:Taf3 dsRNA or control (GFP) dsRNA was transfected into F9 cells and 48 h later Taf3, Mesp1, and Actin expression were analyzed by RT-PCR.
We also performed ChIP analysis in mouse F9 embryonic teratocarcinoma cells, a well-established model system for investigating early events in mouse development (Strickland and Mahdavi, 1978; Fischer et al., 2000; Kubota et al., 2001). Figure 5B shows that in F9 cells the Mesp1 promoter was also selectively bound by TRF3 but not TBP. Significantly, the Mesp1 promoter in F9 cells was also bound by TAF3. When F9 cells were differentiated by treatment with retinoic acid, Trf3 expression was rapidly extinguished followed by loss of Mesp1 expression (Fig. 5C), raising the possibility that in F9 cells TRF3 was required for Mesp1 expression.
To confirm this conclusion we performed an RNA interference (RNAi) experiment. Figure 5D shows that following transfection of Trf3 double-stranded RNA (dsRNA) into F9 cells, Trf3 expression levels were, as expected, substantially reduced. By contrast, expression of Tbp and Gapdh were unaffected, confirming the specificity of RNAi-mediated TRF3 depletion. Significantly, expression of Mesp1 was also dramatically reduced, indicating that TRF3 is required for Mesp1 expression. RNAi-mediated depletion of TAF3 also resulted in loss of Mesp1 expression (Fig. 5E). Thus, analogous to the results in zebrafish embryos, Mesp1 is a TRF3 target gene in mouse embryos and F9 teratocarcinoma cells. In addition, as in zebrafish embryos, in F9 teratocarcinoma cells TAF3 is an essential TRF3 co-factor.
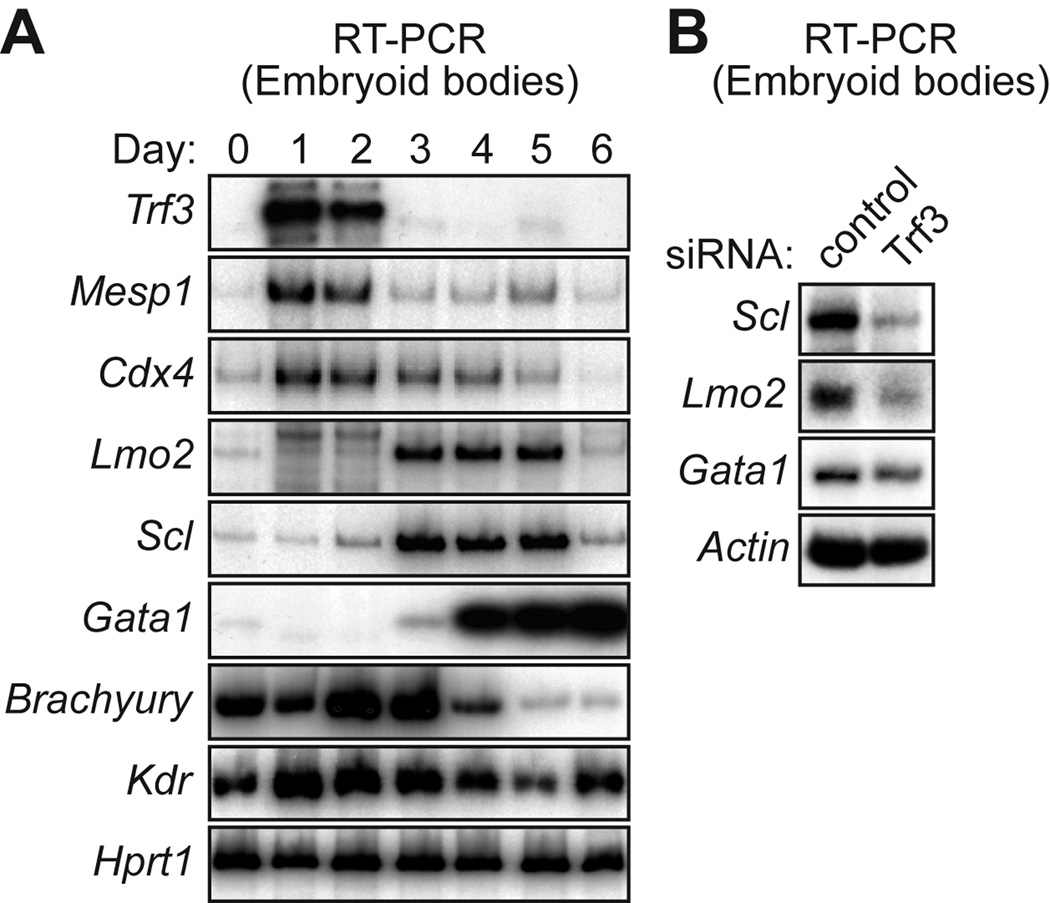
Mouse embryonic stem (ES) cells, when differentiated as suspension aggregates called embryoid bodies, give rise to differentiated hematopoietic cells (reviewed in Choi et al., 2005). We analyzed the expression profile of Trf3, Taf3, Mesp1 and Cdx4 in mouse embryoid bodies. The RT-PCR results of Fig. 6A show that Trf3, Mesp1 and Cdx4 were first detected at very early times following differentiation (day 1). Trf3 expression was substantially decreased by day 3 followed by the loss of Mesp1 and Cdx4 expression by day 4. As expected, Taf3, which is also a component of the GTF TFIID, was present at a relatively constant level throughout the 6-day time course (Supp. Fig. S5).
Fig. 6.
Requirement of TRF3 for initiation of hematopoiesis in mouse embryoid bodies. A: RT-PCR analysis monitoring expression of various genes in mouse embryoid bodies at the indicated times (days) following induction of differentiation. B: Mouse embryonic stem cells were transfected with a Trf3 or control (luciferase) siRNA, differentiated into embryoid bodies, and 4 days later expression of various markers were analyzed by RT-PCR.
Definitive markers of hematopoiesis, such as Lmo2 and Scl, first appeared on day 3, and Gata1 was first detected on day 3 and subsequently increased markedly. By contrast, expression of mesoderm/endothelial markers, such as Brachyury and Kdr, first appeared on day 0, prior to expression of Trf3, Mesp1 or Cdx4. Thus, the temporal expression pattern in mouse embryoid bodies also supports the involvement of a TRF3-MESP1-CDX4 pathway in hematopoiesis.
Finally, to provide functional evidence for the role of TRF3 in hematopoietic development in mouse embryoid bodies we performed an RNAi experiment. A small interfering RNA (siRNA) was used to knockdown TRF3 in mouse ES cells (Supp. Fig. S6), followed by differentiation and analysis of hematopoietic markers. The results of Fig. 6B show that TRF3 knockdown resulted in markedly decreased levels of Scl, Lmo2 and Gata1, indicative of decreased initiation of hematopoiesis. Thus, as in zebrafish embryos, TRF3 is required to initiate hematopoiesis in mouse embryoid bodies.
DISCUSSION
In this report we have studied the most recently identified member of the Tbp family, Trf3. Previous studies have shown that Trf3 is required for normal vertebrate embryogenesis and commitment of mesoderm to the hematopoietic lineage (Hart et al., 2007). Remarkably, Trf3 promotes early development and hematopoiesis through a single target gene, mespa. Here we show that Taf3 is a Trf3 co-factor required for both transcriptional activity and developmental function during early zebrafish embryogenesis.
We find that Taf3 selectively interacts with Trf3 and not Tbp, and that this interaction is critical for Trf3 function. Like other members of the Tbp family, the C-terminal region of Trf3 is highly conserved whereas the N-terminal region of Tbp and Trf3 are divergent. It was therefore surprising that the determinants for the selective interaction of Taf3 with Trf3 reside within the conserved C-terminal domain of Trf3. However, consistent with this finding, TAF1, which interacts with Tbp but not Trf3, also makes contact through the conserved C-terminal region. TAF3 was originally identified as a component of TFIID (Dynlacht et al., 1991; Tanese et al., 1991). Because TBP and TAF3 do not interact, incorporation of TAF3 into TFIID must occur through an interaction(s) with one or more TFIID TAFs.
It is remarkable that the TRF3–TAF3 complex stimulates the expression of target genes that are required for both early embryonic development (this study) and terminal differentiation (Deato and Tjian, 2007; Deato et al., 2008). In both instances the TRF3 target genes are involved in developmental programs involving mesoderm specification. However, a notable difference is that in the early embryo, TRF3 must select its target genes in the presence of the complete repertoire of GTFs and, in particular, TFIID. By contrast, in terminally differentiated muscle cells, many TFIID components, as well as other GTFs, are absent. Thus, in terminally differentiated muscle selective recruitment of TRF3 is likely due, at least in part, to elimination of the potentially competing TFIID complex.
The results presented here and in our previous study (Hart et al., 2007) indicate that protein-coding genes, such as mespa and actin, are differentially bound by TBP and TRF3. However, TRF3 binds to a canonical TATA box and all of the residues involved in the TBP–TATA box interaction are conserved in TRF3 (Persengiev et al., 2003). Thus, the basis by which TRF3 binds selectively to certain promoters remained to be determined. The results presented here suggest that interaction with TAF3 provides the specificity required for selective TRF3 recruitment.
EXPERIMENTAL PROCEDURES
Zebrafish Maintenance, Embryo Production and Microinjection
Zebrafish (Danio rerio) were maintained under standard conditions (Westerfield, 1993), and embryos were produced and staged as described (Kimmel et al., 1995). Antisense morpholino oligonucleotides (MOs) were designed against the start codon/5′UTR to block translation and their sequences are as follows: control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; taf3 MO, 5′-GCGCGAAGCTCTCACACATCTCTCC-3′; and trf3 MO, 5'-GATGCCTCCTCATCC ATGTTCAT-3'. The cdx4 MO (5'-CTCCAAAAGGTATCCAACGTACATG-3') was purchased from Open Biosystems. Control and trf3 MOs were injected at a concentration of 5 mg/ml, the taf3 MO was injected at 0.1 or 0.3 mg/ml, and the cdx4 MO was injected at 0.2 mg/ml. Embryos were analyzed morphologically up to 24 h post injection.
To generate mRNAs for phenotypic rescue, full-length taf3, trf3 and mespa genes were amplified from shield-stage cDNA by RT-PCR and subcloned into pCS2 (Turner and Weintraub, 1994) for mRNA synthesis as previously described (Hart et al., 2007). To generate the mespa mRNA containing the silent mutation, the forward primer 5′-GAATTCAATGTGCGAAAGTTTTGCACGCTCT-3′ was used, which incorporates five nucleotide changes (represented by underlined nucleotides) that do not result in a change in amino acid sequence but are sufficient to protect the resulting mRNA from MO binding. Trf3 mutants were generated by PCR amplification from pCS3-trf3 plasmid DNA (described below). The mRNAs were then co-injected into MO-treated embryos at the 1–2 cell stage at concentrations of 100 ng/µl.
Mouse F9 Cells, ES Cells and Embryoid Bodies
Mouse F9 embryonal teratocarcinoma cells (ATCC CRL 1720) were cultured in DMEM supplemented with 10% fetal bovine serum. The 129/Sv-derived ES cell line CCE (Keller et al., 1993) was obtained from StemCell Technologies and grown in the presence of leukemia-inhibitory factor (LIF) on 0.1% gelatin coated tissue culture dishes without feeder cells. For in vitro differentiation, cells were trypsinized and resuspended in IMDM culture medium without LIF, supplemented with 15% fetal calf serum, 0.2 mM L-glutamine, 0.1 mM β-mercaptoethanol, and 0.1 mM nonessential amino acid stock. Cells were cultured into embryoid bodies by the hanging drop method (Keller, 2005).
RT-PCR
RT-PCR analysis was performed according to standard protocols (Ausubel et al., 2001). For zebrafish embryos, total RNA was prepared using TRIzol reagent (Invitrogen) and analyzed using the following primers: taf3 forward 5′-AAGGAACGGGAGAAGGAACG-3′ and reverse 5′-GGGCAGGTGGAGGAAGAACT-3′; mespa forward 5′-GCTGTTA TCAGACTCAGAATCCAGT-3′ and reverse 5′-CCGGAAAGATGTAGCAAGTATAAAT-3′; scl forward 5′-TCGTCTTGTGTTTCATTGTTTATTG-3′ and reverse 5′-A TAGTCACACCTTCCTCATCACTGT-3′; lmo2 forward 5′-CGAGGACTGTCTGAGCTGT G-3′ and reverse 5′-GTCCGAGTTGATGAGCAGGT-3′; and gapdh forward 5′-GATACACGGAGCACCAGGTT-3′ and reverse 5′-CGTTGAGAGCAATACCAGCA-3′.
For F9 cells, total RNA was prepared using TRIzol reagent and analyzed using the following primers: Trf3 forward 5′-ATGGCAGTGAGCTGAACTTGAATAG-3′ and reverse 5′-TAAAGATGAGAGCTGTTGTCCTTGG-3′; Mesp1 forward 5′-GTTCCTGTACGCAGAAACAGCAT-3′ and reverse 5′-GCCTGCTTCATCTTTAGAGCGTGTA-3′; Tbp forward 5′-CTTCGTGCAAGAAATGCTGA-3′ and reverse 5′-ACAAGGCCTTCCAGCCTTA-3′; Taf3 forward 5′-CATCCCAAAAGAAAAGAAGTCAC-3′ and reverse 5′-AGAGATATATGGGAAGGTGGGAG-3′; Gapdh forward 5′-ATCAACGACCCCTTCATTGACC-3′ and reverse 5′-AGATGATGACCGTTTGGCTC-3′; and Actin forward 5′-GACGGCCAGGTCATCACTAT-3′and reverse 5′-ACATCTGCTGGAAGGTGGAC-3′.
For embryoid bodies, RT-PCR was performed using the following primers: Cdx4 forward 5′-TATTCAGAAACCATCCCCAGAC-3′ and reverse 5′-AATCACCTCCTGATGCTGTTTT-3′; Lmo2 forward 5′-ACCATGTCCT CGGCCATCGAAAGGA-3′ and reverse 5′-TAGATGATCCCATTGATCTTGGT-3′; Scl forward 5′-ATGGAGATTTCTGATGGTCCTCAC-3′ and reverse 5′-AAGTGT GCTTGGGTGTTGGCTC-3′; Gata1 forward 5′-GGAATTCGGGCCCCTTGTGAGGCCAG AGAG-3′ and reverse 5′-CGGGGTACCTCACGCTCCAGCCAGATTCGACCC-3′; Brachyury forward 5′-ATCAAGGAAGGCTTTAGCAAATGGG-3′ and reverse 5′-GAACCTCGGATTCACATCGTGAGA-3′; Kdr forward 5′-CTGTGTCCCGCAGCCGG ATA-3′ and reverse 5′-AAGTCACAGAGGCGGTATGG-3′ (Shalaby et al., 1995); and Hprt1 forward 5′-GTTCTTTGCTGACCTGCTGGATTAC-3′ and reverse 5′-GTCAAGGGCATATCCAACAACAAAC-3′; Trf3, Mesp1 and Actin primers are listed above. RT-PCR products were separated by agarose gel electrophoresis and visualized by autoradiography.
Immunoblot Analysis
Protein extracts were prepared from zebrafish embryos as previously described (Westerfield, 1993). Blots were probed with an α-beta tubulin (Sigma) antibody or an α-zebrafish Taf3 polyclonal antibody raised to a mixture of two peptides, C+PPTPGHKPKLPSPSPARQKNKSPKRGGA-NH2 and C+TPPKTNQKAGKRSPGMPKSPRSPRPSSA-NH2 (corresponding to amino acids 248–275 and 288–315, respectively) that was used to immunize rabbits (AnaSpec) followed by affinity purification.
For immunoblot analysis in mouse ES cells, cells were washed twice with phosphate-buffered saline (PBS) pH 7.2 and lysed in a solution of 25 nM NaCl, 1 mM EDTA and 1% NP40. Protein concentrations were determined by Bradford assay, and equal amounts were subjected to SDS-PAGE and subsequent transfer to nitrocellulose membranes, and blots were probed with an α-human TAF3 polyclonal (Persengiev et al., 2003) or α-Actin monoclonal (A-5316; Sigma) antibody.
In situ Hybridization
In situ hybridization was performed on whole-mount zebrafish embryos as described (Hauptmann and Gerster, 1994). Digoxygenin-labeled antisense probes were synthesized in vitro and obtained as follows: gata1 (Nathan Lawson), pax2 (Charles Sagerstrom) and scl (NIH Zebrafish Gene Collection). For the hbbe1 and hbae1 probes, PCR-amplified cDNA fragments of each were first subcloned into the vector pGEM-T (Promega). Full-length taf3 was PCR amplified from shield stage (6 hpf) cDNA and cloned into the EcoRI and XhoI sites of the pCS3 vector. Riboprobes were synthesized from the corresponding constructs as T7 or SP6 transcripts using the DIG RNA labeling mix (Roche). Riboprobe primers are as follows: hbbe1 forward 5′-ATCTTCGCCAAGGCTGACTA-3′ and reverse 5′-CAGCGGCATTGTAGAGGTTT-3′; hbae1 forward 5′-GGACACGATGCTCTCTCCAG-3′ and reverse 5′-GAGGATGT TGTGGGACAGAA-3′; taf3 forward 5′-GAATTCAATGTGTGAGAGCTTCGCGCGCTCT-3′ and reverse 5′-CTCGAGTCAGTGCGCTTTGCGTTTCC-3′.
Fluorescence Microscopy
Live dechorionated embryos (30 hpf) expressing the gata1:dsRed transgene [TG(gata1:dsRed)y1; (Traver et al., 2003)] were treated with a solution of 0.016% tricaine and monitored for gata1 expression in control and MO-injected embryos using the dsRed filter on a Leica MZFLIII epifluorescent microscope.
Chromatin Immunoprecipitation
ChIP analysis on zebrafish embryos was performed as previously described (Hart et al., 2007) using α-Trf3 polyclonal (Hart et al., 2007), α-Tbp monoclonal (3G3; Eurogentec), or α-Taf3 polyclonal (described above) antibodies. ChIP samples were analyzed by PCR using the following primers: mespa forward 5′-CGCACCTCTCAGAGCACATA-3′ and reverse 5′-AAAACTGCAGGAATCGATGG-3′ (which amplified −46 bp to +170 bp with respect to the mespa transcription start site and encompassed the putative TATA-box); and actin forward 5′-CGAAAGTTTACCTTATATGGAAGGAG-3′ and reverse 5′-TTATTGTGAACAAGGGGAGTGAAG-3′.
For mouse experiments, each set of ChIPs used 15 mg mouse embryo tissue (E10.5; C57/BL6), or 107 F9 cells. Formaldehyde was added directly to the cell culture to a final concentration of 1%, and samples were processed as previously described (Hart et al., 2007) using the following antibodies: α-human TRF3 polyclonal (Persengiev et al., 2003); α-TBP monoclonal (3G3; Eurogentec); α-RNA Pol II monoclonal (8WG16; Covance Research Products); α-yeast Gal4 polyclonal (sc577; Santa Cruz Biotechnology); and α-human TAF3 monoclonal (1C7, a kind gift from Irwin Davidson).
Construction of Trf3 and Tbp Derivatives
Zebrafish taf3 was cloned by PCR amplification from shield-stage cDNA using primers 5′-GAATTCAATGTGTGAGAGCTTCGCGCGCTCT-3′ and 5′-CTCGAGTCAGTGCGCTTTGCGTTTCC-3′. The PCR product was digested with EcoRI and XhoI and cloned into the expression vector p3xFLAG-myc-CMV (Sigma) to generate Flag-Taf3. Chimeric cDNAs of zebrafish Trf3(N)-Tbp(C) and Tbp(N)-Trf3(C) were prepared by first PCR amplifying fragments corresponding to the N- and C-termini of Trf3 and Tbp using the following primers: Trf3(N) forward 5′-GAATTCCATGAACATGGATGAG-3′ and reverse 5′-GATATCTGAACT CTCTGCCAC-3′; Trf3(C) forward 5′-GATATCGGCATCATCCCGCA-3′ and reverse 5′-CTCGAGCTATTGCTTCCTAAATCCTT-3′; Tbp(N) forward 5′-GAAT TCCATGGAGCAGAACAAC-3′ and reverse 5′-GATATCAGAGCTCTCGGAGG-3′; and Tbp(C) forward 5′-GATATCGGCATCGTCCCGCA-3′ and reverse 5′-CTC GAGTTACGAGGTCTTCCT-3′. The primers were designed to tag the N-terminal fragments with 5′ EcoRI sites and 3′ EcoRV sites, and C-terminal fragments with 5′ EcoRV sites, and 3′ XhoI sites. These fragments were then digested with EcoRV and ligated at their junction. The resulting linear fragments were then digested with EcoRI and XhoI and cloned into EcoRI/XhoI-digested pCS3 vector. The resulting Trf3(N)-Tbp(C) chimeric protein fused residues 1–133 of Trf3 to 123–302 of Tbp, and the Tbp(N)-Trf3(C) protein fused residues 1–122 or Tbp to 134–312 of Trf3.
Site-directed Trf3 mutants were generated using a Stratagene kit, according to the manufacturer’s instructions, using the following primers: SDM1 forward 5′-CACAGGTGCAAAAGTGCGGGGCGAGATCTATGAAGC-3′ and reverse 5′-GCTTCATAG ATCTCGCCCCGCACTTTTGCACCTGTG-3′, and SDM2 forward 5′-CAACTGTGAATC TTGGGTGCAAACTGGATCTCAAATCC-3′ and reverse 5′-GGATTTGAGATCCAGTTTG CACCCAAGATTCACAGTTG-3′ (where the underlined sequences represent the mutated amino acids).
Co-immunoprecipitations
COS-7 cells were co-transfected with plasmids expressing myc-tagged Trf3, Tbp or chimeric protein (described above), and Flag-Taf3 (described above) or HA-TAF1 (a gift from Edith Wang), and lysed 48 h later. Protein extracts were prepared by lysis in a buffer containing 50 mM Tris-HCl, pH 7.4, 0.1% Triton X-100, 5 mM EDTA, 250 mM NaCl, 50 mM NaF, 0.1 mM Na3VO4 and protease inhibitors. Protein concentration was measured by Bradford method. Protein (600 µg/IP) was pre-cleared with rProtein G-agarose beads (Invitrogen) for 1 h and then immunoprecipitated using 2 µg antibody overnight at 4°C on a rocking platform. rProtein G-agarose beads (Invitrogen) were then added and incubated for 1 h on a rocking platform at 4 °C. Samples were briefly centrifuged and the beads washed 3 times with buffer. The protein complex was eluted by boiling with Laemmli buffer. Blots were probed with α-Myc monoclonal (9E10; Pierce), α-Flag monoclonal (Sigma) or α-HA monoclonal (Sigma).
RNA Interference
Double strand RNA interference in F9 cells was performed essentially as previously described (Billy et al., 2001). To generate templates for production of double-stranded RNA, fragments of Trf3 (nt 222–513) or Taf3 (nt 1128–1698) were PCR amplified from murine F9 cDNA (using primers Taf3 forward 5′-CAT CCCAAAAGAAAAGAAGTCAC-3′ and reverse 5′-AGAGATATATGGGAAGGTGGGAG-3′; Trf3 forward 5′-TCCACATCTTGGTGGAGTCA-3′ and reverse 5′-GGCAT TGGAGTTTGGTGTTT-3′; and GFP forward 5′-ATGGTGAGCAAGGGCGAGGAG-3′ and reverse 5′-GAAGTTCACCTTGATGCCGT-3′), and cloned into the pGEM-T TA cloning vector (Promega). The inserts were then PCR amplified using SP6 and T7 primers, and double-stranded RNAs were generated from the PCR products by in vitro transcription using opposing T7 and SP6 promoters. Double-stranded RNA was transfected into mouse F9 teratocarcinoma cells (ATCC) using Effectene (Qiagen) according to the manufacturer’s protocols. F9 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS on 0.1% gelatin-coated plates. Cells were passaged every 2 days, and split 24 h before transfection.
For siRNA-based experiments, embryonic stem cells were transfected with 100 nM siRNA duplexes using Lipofectamine 2000 according to the manufacturer’s instructions. Anti-sense siRNA sequences are as follows: Trf3 5′-CAUUUCUUAAUCAUGGCUATT-′3 and luciferase 5'-UCGAAGUAUUCCGCGUACGdTdT-3′ (Elbashir et al., 2001).
Supplementary Material
ACKNOWLEDGMENTS
We thank Nathan Lawson, Charles Sagerstrom, Irwin Davidson and Edith Wang for reagents, and Sara Evans for editorial assistance. This work was supported in part by a grant from the National Institutes of Health to M.R.G. M.R.G. is an Investigator of the Howard Hughes Medical Institute.
Grant sponsor: National Institutes of Health; Grant number: GM33977
REFERENCES
- Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York, NY: John Wiley and Sons; 2001. [Google Scholar]
- Bartfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orban L, Muller F. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- Choi K, Chung YS, Zhang WJ. Hematopoietic and endothelial development of mouse embryonic stem cells in culture. Methods Mol Med. 2005;105:359–368. doi: 10.1385/1-59259-826-9:359. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–306. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- Davidson I. The genetics of TBP and TBP-related factors. Trends Biochem Sci. 2003;28:391–398. doi: 10.1016/S0968-0004(03)00117-8. [DOI] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate Myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht BD, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer HS, Berti I, Schatz DS, Humpel C, Saria A. Retinoic acid treatment enhances the acetylcholine contents in the human teratocarcinoma cell line NTera-2. Regul Pept. 2000;96:59–63. doi: 10.1016/s0167-0115(00)00201-9. [DOI] [PubMed] [Google Scholar]
- Fossett N, Schulz RA. Functional conservation of hematopoietic factors in Drosophila and vertebrates. Differentiation. 2001;69:83–90. doi: 10.1046/j.1432-0436.2001.690202.x. [DOI] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- Green MR. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nusslein-Volhard C. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development. 1996;123:143–151. doi: 10.1242/dev.123.1.143. [DOI] [PubMed] [Google Scholar]
- Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci USA. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Keller G, Kennedy M, Papayannopoulou T, Wiles MV. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kubota H, Chiba H, Takakuwa Y, Osanai M, Tobioka H, Kohama G, Mori M, Sawada N. Retinoid X receptor alpha and retinoic acid receptor gamma mediate expression of genes encoding tight-junction proteins and barrier function in F9 cells during visceral endodermal differentiation. Exp Cell Res. 2001;263:163–172. doi: 10.1006/excr.2000.5113. [DOI] [PubMed] [Google Scholar]
- Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Dixit BL, Maston GA, Kittler EL, Green MR. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci USA. 2003;100:14887–14891. doi: 10.1073/pnas.2036440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sun G, Stewart R. Nuclear receptors in stem cell biology. Crit Rev Eukaryot Gene Expr. 2006;16:171–181. doi: 10.1615/critreveukargeneexpr.v16.i2.50. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Strickland S, Mahdavi V. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978;15:393–403. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- Tanese N, Pugh BF, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A guide for the laboratory use of zebrafish (Danio rerio) Eugene: University of Oregon Press; 1993. The zebrafish book. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.