Abstract
Multiple testing has become an integral component in genomic analyses involving microarray experiments where a large number of hypotheses are tested simultaneously. However, before applying more computationally intensive methods, it is often desirable to complete an initial truncation of the variable set using a simpler and faster supervised method such as univariate regression. Once such a truncation is completed, multiple testing methods applied to any subsequent analysis no longer control the appropriate Type I error rates. Here we propose a modified marginal Benjamini & Hochberg step-up FDR controlling procedure for multi-stage analyses (FDR-MSA), which correctly controls Type I error in terms of the entire variable set when only a subset of the initial set of variables is tested. The method is presented with respect to a variable importance application. As the initial subset size increases, we observe convergence to the standard Benjamini & Hochberg step-up FDR controlling multiple testing procedures. We demonstrate the power and Type I error control through simulation and application to the Golub Leukemia data from 1999.
1. Introduction
Statistical analysis in genomics research often requires testing a large number of hypotheses simultaneously. This is especially true in microarray experiments where there are tens of thousands of variables and often less than 100 observations. A common approach to determine which genes are significant is to apply univariate regression to all variables and test the significance of the coefficient β using a standard t-statistic with the null hypothesis H0 : β = 0, and then adjust the marginal p-values for multiple testing.
When applying a more computationally intensive method such as targeted variable importance (Bembom et al., 2007; Tuglus and van der Laan, 2008) which requires data-adaptive estimation, one might want to initially reduce the dimensions of the data using a simpler method, restricting it to a conservative set of potentially relevant genes. One approach is to use an unsupervised method, for instance restricting the set to genes that have a variance higher than a specified threshold. However, when using these methods the potential of discounting relevant genes can be quite large since the threshold level is independent of the outcome. To alleviate this possibility simple supervised methods such as univariate regression or randomForest (Breiman et al., 1984; Breiman, 2001) can be used to prescreen the variables. Although, once the initial variable set is restricted with respect to the outcome, multiple testing procedures on the results of secondary analyses are biased and no longer control the Type I and Type II error appropriately. Any subsequent analysis must take this into account.
In this paper we propose a modification to marginal Benjamini & Hochberg step-up FDR controlling procedure for multi-stage analyses (FDR-MSA) that appropriately controls the False Discovery Rate (FDR) when applied to a reduced and data-adaptively selected set of null hypotheses. We also show that if the restricted set contains all relevant variables, this procedure has equivalent control of Type I error and equivalent power to applying the standard Benjamini & Hochberg step-up FDR controlling procedure (BH-FDR) to the entire variable set. We generalize this modification for any monotonic multiple testing adjustment to marginal p-values.
We introduce this method in terms of an application of targeted Maximum Likelihood (tMLE) variable importance methodology, which we introduce in section two. In section three, we present the FDR-MSA method in detail. In section four, we provide simulation results demonstrating the Type I and Type II error control. In section five, we present an application of the procedure to the commonly used Golub et al (1999) leukemia data (Golub et al., 1999), and we conclude with a discussion.
2. Variable Importance Application
As an example of the general application of FDR-MSA, we apply the proposed method in the context of variable importance analysis. The variable importance methodology is described thoroughly in van der Laan and Rubin (2006) and van der Laan (2005) and was previously applied in Bembom et al. (2007) and Tuglus and van der Laan (2008).
We observe data O = (W*, Y ∼ P) where P is the data generating distribution, the outcome Y is a vector of length n, and W* is an n by M matrix of covariates (i.e. genes). We define variable A as a single variable , in W*, where W* = { , j = 1, . . .M}. Note that in practice A can reflect a set of variables, but for this application is restricted to a single variable.
The objective is to identify variables in W* that are significantly associated with outcome Y. Therefore in this case, the parameter of interest will be a measure of the effect of variable A on Y controlling for possible confounders in .
We define our parameter of interest as the marginal variable importance of A at a particular value A = a as
where
satisfying m(0, W | β) = 0 for all β and W.
For this analysis we define our model m(A, W|β) = βA, where parameter β identifies the importance curve over the support of A. Inference is completed for β, but can be extended for any value A = a.
Estimation of β for each A ∈ W* is completed using tMLE variable importance (tVIM) methodology, which can require the estimation of E[Y|A, W] and E[A|W] for each individual variable, A. For optimal efficiency, we estimate E[Y|A, W] and E[A|W] using data-adaptive algorithms which are often computationally intensive, making pre-screening of the variable set a very attractive option. This is particularly true for microarray experiments where W* is very high dimensional. Possible prescreening methods include univariate regression, randomForest (Breiman et al., 1984; Breiman, 2001), and simple tVIM which uses univariate regression to estimate E[Y|A, W] and E[A|W]. In this paper, univariate regression will be used as the pre-screening method. Details on the tVIM method applied in this paper can be found in Tuglus and van der Laan (2008).
3. Methods
The modified marginal Benjamini & Hochberg step-up FDR controlling procedure for multi-stage analyses (FDR-MSA), is applied directly to the raw marginal p-values.
Given a multivariate parameter Ψ(P) = (ψ(m) : m = 1, ... M), we can define the null hypotheses and alternative hypotheses in terms of the parameter null value ψ0, which typically equals 0. For the two-sided hypothesis test, the null hypothesis is H0(m) = I(ψ(m) = ψ0(m)) and the alternative hypothesis is H1(m) = I(ψ(m) ≠ ψ0(m)).
Whether or not we reject the null hypothesis is determined by the value of the test statistic Tn = (Tn(m) : m = 1, ..., M). The parameter of interest can be tested using a standard t-statistic defined below
where ψn(m) is an asymptotically linear estimator of ψ(m) with specified influence curve ICm(P)(O), and (m) is an estimate of the variance of the influence curve.
Specifically for variable importance measures, ψ(m) = βn for a given A = , where W* = { , m = 1, . . .,= M,}. We test the null hypothesis H0 : ψ(m) = 0 using a standard t-test,
where σn(m) is the standard error estimated from the variance of the empirical influence curve (Tuglus and van der Laan, 2008).
3.1. Marginal Benjamini & Hochberg Step-up FDR Controlling Procedure
According to the standard marginal Benjamini & Hochberg step-up FDR controlling procedure (Benjamini and Hochberg, 1995) we define
where V = number of false positives and R= number of total rejections of the null.
For a set of variables W ={Wm, m=1, ... M} given a set of M test statistics Tn = {Tn(m) : m = 1, ..., M} and their associated p-values pn = {pn(m) : m = 1, ..., M}, define the ordered set of p-values as pn(1) ≤ pn(2) ≤ ... ≤ pn(M). According to Benjamini and Hochberg (1995), to control FDR at level α, find k̂ such that
and reject pn(1) ≤ ... ≤ pn(k̂). We define the set of rejected null hypotheses as .
3.2. Modified Marginal Benjamini & Hochberg Step-up FDR Controlling Procedure for Multi-Stage Analyses
FDR-MSA is applied using a simple modification of the standard marginal Benjamini & Hochberg step-up FDR (BH-FDR) controlling procedure (Benjamini and Hochberg, 1995). This method will be generalized to any monotonic multiple testing adjustment with marginal p-values.
3.2.1. Procedure
The BH-FDR adjusted p-values have the property such that if we replace pk by a qk for all k such that pk ≤qk, then the set of rejections of FDR applied to qk is included in the set of rejections of FDR applied to the original pk. Our proposed method is then applied by setting qk equal to pk for a supervised/data-adaptively selected subset of the null hypotheses and setting qk = 1 for all other null hypotheses. The procedure is outlined below.
Given set of M variables, select a subset of U variables based on an initial supervised analysis (i.e. univariate regression with a p-value cut-off)
Complete the desired test statistics for the null hypotheses of interest for these U variables only, and calculate their raw p-values.
-
Assign a value of one to the p-values of all M–U unselected variables.
Thus, with regard to construction of the ordered list of p-values, add to the end of the list of the U p-values K = (M–U) ones.
Apply multiple testing procedure as usual, in this case standard marginal Benjamini & Hochberg step-up FDR controlling procedure (Benjamini and Hochberg, 1995).
Loss of power will only occur if the initial restriction excludes variables that would have been rejected by the BH-FDR procedure when applied to the original pk. In other words, the FDR-MSA procedure will maintain Type I and Type II error control equivalent to applying BH-FDR to all M variables given that the rank order of the p-values is maintained between the raw p-values of the pre-screening method and the adjusted p-values of the secondary analysis. The effectiveness of the FDR-MSA in preserving Type I and Type II error control is therefore contingent on the relationship between the pre-screening method and the test statistics of the secondary analysis.
For example, the supervised subset of the null hypotheses can be selected to be all variables for which their univariate regression p-value is smaller than 0.1. In the special case that our supervised subset does include the FDR selected set when applied to the p-values pk, then the two stage FDR procedure applied to qk is equivalent with FDR applied to pk. Thus, in this case the two stage FDR is equivalent to applying BH FDR to all variables.
3.2.2. Theorem
We propose the following theorem showing control of Type I error with the MSA method for any type I error which is a only a function of the number of false positives (i.e. FWER).
Theorem 1 Let be a set of rejections, i.e., a random subset of {1, . . ., M} consisting of the indicators of all null hypotheses H0(m), m = 1, . . ., M. If H0(m) is true we will denote that with H0(m) = 1. Let p(m), m = 1, . . ., M be the marginal p values for H0(m). Assume that is a deterministic function of these marginal p-values (p(m) : m = 1, . . ., M). Let be the number of false positives in . Let be the number of elements in .
Let g(V) be a function and let the type-I error rate of be defined as the expectation Eg(V). To express dependence on marginal p-values we use notation .
Assume the following monotonicity property in p-values: for any (for each component), we have
Then, for any random s.t. P( ) = 1, we have . As a consequence, if the procedure satisfies Eg(V) ≤ α, then the same applies to the modified procedure .
This theorem does not generalize for Type I errors that are a function of both the number of false positives and the total number of rejections (i.e. FDR) without the addition of constraints on the prescreening method. We investigate the control of FDR by FDR-MSA through simulation and we demonstrate that given a reasonable prescreening method, FDR-MSA will correctly control FDR.
The control of FDR under FDR-MSA is explored in simulation by applying two prescreening methods where the original list of M adjusted p-values is obtained from univariate regression. Method (1) is the ”worst possible” prescreening method in which p-values of true positives are set to one. In simulation this is obtained by setting the lowest U adjusted p-values to one. Method (2) sets a randomly selected set of U adjusted p-values to one. Results are summarized here, but are shown and discussed in more detail in Appendix A.
Results show that as expected by Theorem 1, the expected number of false positives (E[V]) is controlled correctly under both methods. In term of controlling the FDR, method (1) does not maintain control, but method (2) does and is actually conservative as U increases. However under method (1), the ”worst possible” prescreen, attempting to control FDR at 0.05 results in FDR control at ∼ 0.06. This increase is small and begins to decrease as U increases. As stated previously under method (2) where we are randomly selecting the p-values to set to one resulting in an arbitrary prescreen, E[V/R] ≤ α, becoming more conservative as the number of p-values set to one increases. Given these results, we are confident that in practice we can control FDR adequately.
In terms of selecting and applying a reasonable pre-screening method, we make the following points with regard to FDR-MSA.
The optimal pre-screening method has the property that given the original list of raw p-values, any variables selected to be removed will not have been found significant in the secondary analysis.
In practice, the goal is to conservatively approach the optimal pre-screen, reducing the variable set as much as possible without discounting potentially relevant variables
To define the reduced variable set, a p-value cut-off is recommended. P-values automatically scale with the sample size, adapting to the signal in the data and making the properties of the method invariant to sample size. Selecting the top k variables will not achieve this.
3.2.3. Generalization
It follows directly that the FDR-MSA method can be generalized to any multiple testing method with marginal p-values that have the monotonicity property, and that Theorem 1 holds for any Type I error that is a function of the number of false positives only.
4. Simulations
We compare the performance of FDR-MSA under different levels of initial data screening. In this case the initial screen is determined by ranked p-values from univariate linear regression results. The different levels of screening correspond to different (increased) p-value cut-off values. The comparison is completed in terms of Type I error and power.
4.1. Simulated Data
Covariate matrix W consists of 100 independent variables, each with 100 observations simulated from a multivariate normal distribution with variance 1 and mean vector created by randomly sampling mean values from {0.1,0.2, . . .,50}.
Outcome Y is simulated from a main effect linear model using ten variables each with coefficient 4. These ten variables are designated as “true effects.” A normal error with mean zero and a standard deviation σY = 10 is added as noise. We use σY = 10 to simulate a realistic noise scenario and provide enough variation to motivate false positive findings. Power and Type I error is calculated over 500 samples.
4.2. Analysis
Univariate linear regressions are applied to all 100 independent variables. Of these 100 variables we define subsets according to their ranked raw p-values from the univariate tests. We compare five different levels of screening corresponding to p-value cut-off values of ks = {0.05, 0.1, 0.2, 0.3, and 1}, where a subset is defined as all variables with raw univariate p-value less than or equal to a specific cut-off. A p-value cut-off of 1 corresponds to no initial screening of the data at which point FDR-MSA is equivalent to standard BH-FDR.
For each subset, we apply tMLE variable importance methodology and obtain measures β and associated inference under a null hypothesis H0 : β = 0. Initial density estimate for and are estimated using lasso regression (Tibshirani, 1996), applied by the lars() R package (Efron and Hastie). FDR-MSA is applied to each set of variable importance p-values.
We compare the performance of FDR-MSA under different levels of screening in terms of Type I error and power. Type I error (or 1-Specificity) is defined as the probability of rejecting the null hypothesis (β = 0) when the null hypothesis is true and power (or Sensitivity) is defined as the probability of rejecting the null hypothesis (β = 0) when the alternative hypothesis (β ≠= 0) is true.
Results are compared using plots representing levels of power and Type I error. We use a p-value cut-off to define the reduced variable sets. We select all variables with p-value less than the specified cut-off (α) and assess the power and Type I error among the variables in that group. Results are shown using the following plots.
Sensitivity (Power) versus p-value cut-off (α)
Type I error (1-Specificity) versus p-value cut-off (α)
4.3. Results
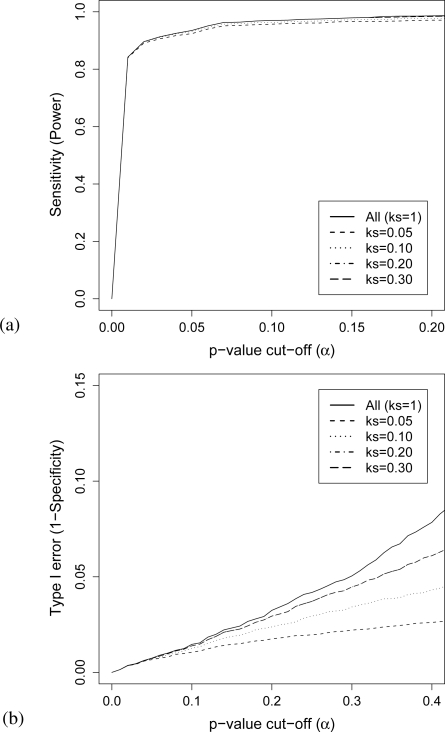
Overall, as the size of the initial subset of variables increases (ks increases), the performance of FDR-MSA in terms of both power and Type I error converge to standard BH-FDR (under no truncation).
From the slight loss of power in Figure 1a, it is evident that initially truncating the set of variables according to ks = 0.05 was too harsh and did not allow all truly significant variables into the subset. As ks is increased we see power converging to the power of BH-FDR applied to the full data (under no truncation).
Figure 1:
(a) Sensitivity (power) and (b) Type I error versus p-value cut-off α
In Figure 1b, Type I error is compared with respect to p-value cut-off. We see that when controlling at a level of α=0.05 or below, the methods are equivalent for ks values as high as 0.2. Above α = 0.05, the FDR-MSA method slowly converges to the Type I error of BH-FDR on the entire data as the raw p-value cut-off is increased, becoming equivalent at higher α values.
5. Application - Leukemia
To illustrate its application in practice, the FDR-MSA method is applied to the Golub et al (1999) leukemia data in conjunction with targeted variable importance (Tuglus and van der Laan, 2008). Targeted variable importance is applied to the full data and subsets of the data defined by an initial univariate raw p-value cut-off. The resulting p-values from each case will be adjusted with FDR-MSA. The resulting ranked lists will be compared.
5.1. Data
Variable importance methods (Bembom et al., 2007; Tuglus and van der Laan, 2008) are used to identify genes which distinguish patients with acute lymphoblastic leukemia (ALL) from patients with acute myeloid leukemia (AML). For the study presented in Golub et al (1999), the gene expression levels were measured using Affymetric oligonucleotide arrays with 6,817 human genes for n=38 patients (27 ALL, 11 AML). The gene expression set was pre-processed using unsupervised methods and reduced to 3,051 genes according to methods described in Dudoit et al. (2002). This dataset was obtained from the R package multtest, dataset golub (Pollard et al., 2005).
5.2. Analysis
Univariate logistic regressions are applied to all 3,051 variables. Of these variables we defined subsets according to their raw p-values from the univariate regressions. We restrict the data to all variables with raw p-values less than 0.01, 0.025, 0.05, 0.1, 0.2, 0.3, and 1. We obtain tMLE variable importance measures and associated p-values for all subsets. The initial density estimate for is estimated using polymars regression, applied by the polspline() R package (O’Connor). To estimate Q(A, W) = E[Y|A, W], we use lasso regression using the lars R package (Efron and Hastie). There are more powerful methods to data-adaptively select Q(A, W), such as DSA (Sinisi and van der Laan, March 2004), and super Learner (van der Laan et al., 2007). Using a less powerful method does cost us consistency and efficiency with respect to our variable importance estimate. However lars provides a quick implementation of lasso regression making it convenient for this particular demonstration. Future work on the variable importance method will use more data-adaptive estimates for Q(A, W) estimates.
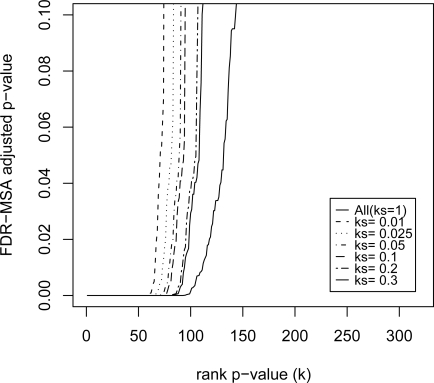
We apply FDR-MSA to the resulting sets of variable importance p-values. Results are compared plotting the rank of the p-value versus its value.
5.3. Results
As we weaken the restriction on the initial cut and become more generous, we find that the results for FDR-MSA converge to the results when BH-FDR is applied to all the data.
The apparent loss in power from truncating the data is due to the initial screening process discounting important and significant variables. Ideally screening the data would result in an initial subset of variables that contain all true variables. The fact that univariate regression does not accomplish this suggests that more sophisticated screens are necessary.
6. Discussion
Pre-screening of a set of variables is not uncommon and is very attractive for microarray data where the number of genes can be over 20,000. Ideally prescreening is done in a supervised fashion, with respect to the outcome. However, once applied, any inference on secondary analyses must account for the initial set of tests. The FDR-MSA method presented here does this in a simple straightforward fashion. The procedure allows for proper control of Type I and Type II error when pre-screening occurs assuming prescreening is applied conservatively and no significant variables are removed in the process.
We have shown that the FDR-MSA multiple testing procedure applied to a restricted subset of variables has equivalent power and Type I error control to FDR applied to all variables when all BH-FDR-significant variables are present in the restricted set. Thus it conservatively controls FDR while only requiring calculation of the test statistics for the restricted and data adaptively selected set of null hypotheses.
Generally pre-screening can be beneficial and allow for more in depth analysis while conserving computational time. However if p-values from the prescreening method and those of the secondary analysis are expected to be far apart or nonmonotonic then pre-screening can be dangerous. Choosing an appropriate cut-off to define the reduced variable set becomes very important. We must first carefully consider the relationship between the prescreening method and testing from the secondary analysis. If we believe that our prescreening method is reasonable and wish to detect significance at the 0.05 level, we can simply use a raw p-value cut-off of 0.05 on the ranked list from the prescreening results. For example if we believe tVIM will never find anything more significant than univariate regression, the using a p-value cut-off of 0.05 on the raw univariate p-values will not discount any variables that tVIM would have identified as significant had it been applied to the entire variable set, and it is guaranteed to be conservative. However, if we do not believe our prescreening method is reasonable and for instance think that the significance of variables can improve, then we must be extremely careful on how we define our reduced variable set. This is often the case when dealing with independent SNP data or a randomized trial, where you might expect to gain efficiency through adjustment (i.e. the p-values of tVIM might be much smaller than those of univariate regression).
We see this effect both in simulation and in practice, where restricting solely based on raw p-values from univariate regression did not necessarily provide a conservative reduction of the variable set. Therefore, in most cases, we recommend to be thoroughly generous on how the cut-off is defined. Another possibility is to apply multiple supervised learning methods and take the union of the selected variables as the restricted set. For instance applying randomForest to the full variable set and taking all variables with non-zero importance or univariate regression p-values less than a particular cut-off (0.1 for instance). Alternatively, one can apply the tMLE-variable importance analysis with a simple and less computationally intensive initial regression estimator as a first stage analysis, selecting the restricted set based on a p-value cut-off. Approaches like this one will be investigated in more detail in order to improve the power of the FDR-MSA method while still maintaining the reduction in computation time.
Finally, we reiterate again that the MSA method for multiple testing applies to any multiple testing procedure based on marginal p-values that has the monotonicity property in the p-values. For example, one can carry out a MSA modified method for controlling the generalized family-wise error (FWER-MSA) based on the appropriate multiple testing procedure controlling this Type I error.
A Type I Error Control of FDR-MSA
We investigate the ability of FDR-MSA to control the desired FDR level under two prescreening methods. The first method is what we consider to be the “worst possible” prescreening method, in which p-values for true positives are set to one. In practice/simulation is can be obtained most effectively by setting the k lowest BHFDR adjusted p-values to one. The second method randomly selects k BH-FDR adjusted p-values to set to one. We assess the ability of each to control both the expected number of false positive (E[V], where V is the number of false positives) and the expected FDR (E[V/R], where R is the number of rejections).
The simulation was set up as follows. Covariate matrix W consists of 100 independent variables, each with 500 observations simulated from a multivariate normal distribution with variance 1 and mean vector created by randomly sampling mean values within the range [0.17,0.83]. The means are decreased from the simulation presented in section 4.1 to encourage a greater number of false positives.
Outcome Y is simulated from a main effect linear model using ten variables each with coefficient 2. These ten variables are designated as “true effects.” A normal error with mean zero and variance σY = 5 is added as noise. We use σY = 5 to simulate a realistic noise scenario and provide enough variation to motivate false positive findings.
On each sample prescreening is completed using BH-FDR adjusted univariate p-values and tVIM is applied. Under α = 0.05, the expected number of false positives and the expected FDR are calculated from 500 samples. The results are shown in the plots below.
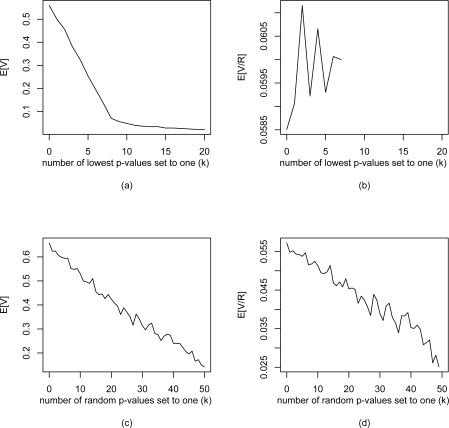
We show that under both prescreening methods, E[V] is appropriately controlled as expected from the results of Theorem 1 in section 3.1.2. Under the worst prescreening method, FDR is not controlled, but is never greater than 0.06 and decreases as k increases. When selecting a random set of adjusted p-values and setting them to one, the FDR-MSA does control FDR at the desired level. We propose that, in practice, using reasonable prescreening method with FDR-MSA will control FDR.
Figure 2:
(a) FDR adjusted p-value versus p-value rank for FDR-MSA for raw p-value cut-offs ks = {0.01, 0.025, 0.05, 0.1, 0.2, 0.3, and 1}
Figure 3:
Results of simulations assessing performance of FDR-MSA. Shown above are (a, c) expected number of false positives (E[V]) and (b, d) expected False Discovery rate (E[V/R] versus the number of original adjusted p-values (k) set to one. The k p-values are selected by (a, b) taking the k lowest adjusted p-values, and by (c, d) selecting k random p-values
Acknowledgments
This work was done under the grant for Targeted Empirical Super Learning in HIV Research, funding through NIH National Institute of Allergy and Infectious Diseases; Award number R01 A1074345-01.
References
- Bembom O, Petersen ML, Rhee S, Fessel WJ, Shafer RW, Sinisi SE, van der Laan MJ.Biomarker discovery using targeted maximum likelihood estimation: Application to the treatment of antiretroviral resistant hiv infection U.C. Berkeley Division of Biostatistics Working Paper Series, Working Paper 221, 2007. URL http://www.bepress.com/ucbbiostat/paper221 [DOI] [PMC free article] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Breiman L, Freidman JH, Olshen RA, Stone CJ. Classification and Regression Trees. Wadsworth; 1984. [Google Scholar]
- Breiman L.Random forests Machine Learning 4515–32.2001. URL http://dx.doi.org/10.1023/A:1010933404324 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. Journal of the American Statistical Association. 2002;97(457):77–87. doi: 10.1198/016214502753479248. [DOI] [Google Scholar]
- B. Efron and T. Hastie. lars. R package.
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- M. O’Connor. polymars. R package polspline.
- Pollard KS, Dudoit S, van der Laan MJ.Multiple Testing Procedures: R multtest Package and Applications to Genomics in Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Number 209–229Springer; (Statistics for Biology and Health Series), 2005 [Google Scholar]
- Sinisi SE, van der Laan MJ.Loss-based cross-validated deletion/substitution/addition algorithms in estimationWorking paper 143, U.C. Berkeley Division of Biostatistics Working Paper Series, March 2004. URL http://www.bepress.com/ucbbiostat/paper143
- Tibshirani R. Regression shrinkage and selection via the lasso. J. Royal Statist. Soc B. 1996;58(1):267–288. [Google Scholar]
- Tuglus C, van der Laan MJ. UC Berkeley Division of Biostatistics Working Paper Series, submitted. 2008. Targeted methods for biomarker discovery: The search for a standard. [Google Scholar]
- van der Laan MJ.Statistical inference for variable importanceTechnical Report Working Paper 188, U.C. Berkeley Division of Biostatistics Working Paper Series, 2005. URL http://www.bepress.com/ucbbiostat/paper188
- van der Laan MJ, Rubin D.Targeted maximum likelihood learningWorking paper 213, U.C. Berkeley Division of Biostatistics Working Paper Series, 2006. URL http://www.bepress.com/ucbbiostat/paper213
- van der Laan MJ, Polley EC, Hubbard AE.Super learner U.C. Berkeley Division of Biostatistics Working Paper Series, (Working Paper 222), 2007. URL http://www.bepress.com/ucbbiostat/paper222