Abstract
A new test was recently developed that could use a high-density set of single nucleotide polymorphisms (SNPs) to determine whether a specific individual contributed to a mixture of DNA. The test statistic compared the genotype for the individual to the allele frequencies in the mixture and to the allele frequencies in a reference group. This test requires the ancestries of the reference group to be nearly identical to those of the contributors to the mixture. Here, we first quantify the bias, the increase in type I and type II error, when the ancestries are not well matched. Then, we show that the test can also be biased if the number of subjects in the two groups differ or if the platforms used to measure SNP intensities differ. We then introduce a new test statistic and a test that only requires the ancestries of the reference group to be similar to the individual of interest, and show that this test is not only robust to the number of subjects and platform, but also has increased power of detection. The two tests are compared on both HapMap and simulated data.
1. Introduction
Given a mixture of DNA samples from numerous individuals, it is often desirable to determine whether a specific individual contributes DNA to that mixture. Using forensics as an example, this mixture can be a specimen from a crime scene, and the goal can be determining whether a suspect’s DNA is included in that specimen. Many methods have been proposed to identify the presence of an individual within a mixture. Most of them focus on cases where only a few people contribute to the mixture. These methods usually compare short tandem repeats (STR) in the mixture to those in the individual (Fung and Hu, 2002; Balding, 2003; Foreman et al., 2003). When only interested in males, the comparison can be limited to STRs on the Y chromosome (Jobling and Gill, 2004). In cases where the DNA has degraded, a second approach, comparing Mitochondrial DNA (mtDNA), specifically the hypervariable region, between mixture and individual can be a better alternative (Stoneking et al., 1991). The limitations of each method were discussed in Homer et al. (2008), but in brief, methods based on STRs require the DNA to be in good condition and methods based on mtDNA, even when augmented by informative SNPs, can have limited discriminatory power (Homer et al., 2008).
In a ground-breaking paper, Homer et. al. propose a new method using a genome-wide set of SNPs (Homer et al., 2008) to identify an individual in a mixture. Their method compares , where YM j is the proportion or frequency of the “A” allele in the mixture at SNP j, with , where Y0 j is the proportion of the “A” allele in the individual. Clearly, the possible values of Y0 j are 0, 0.5, and 1. Note that we have assumed there are N SNPs and have labeled the two possible alleles at each SNP as A and B. Not only is their new method based on a more robust technique, it has higher resolution, able to identify an individual in a mixture containing thousands of subjects. This method also highlighted the potential for a new problem, important enough to be discussed in Science (Couzin, 2008) and require an immediate statement by the NIH. Their results proved that the practice of publicly releasing group-level results from case/control GWAS studies may jeopardize participants’ anonymity.
The test statistic, T, proposed by Homer et. al., compares the similarity between and to the similarity between and , where are the allele frequencies in a reference mixture. If T is large enough, the corresponding test will reject the null hypothesis that the individual of interest does not contribute DNA to the mixture. For this test to perform well, the subjects for the reference group must be carefully chosen so that their ancestral composition matches that of the subjects contributing to the mixture (e.g. if Caucasians contribute to the mixture, then the reference group must also be Caucasian), or only a select subgroup of ancestry independent SNPs can be used (Kidd et al., 2006). The need for similar reference groups is clear. Assume we fail to select a similar reference group, and the individual is more similar, in terms of ancestry, to those subjects contributing to the mixture. Then, even if the subject’s DNA is absent from the mixture, will be more similar to , T will be large, and the test will likely result in a false positive. Similarly, if the individual and the reference group are more similar, the test can lead to a false negative. The obvious problem is that the identities, and therefore the ancestries, of the individuals in the mixture are unknown, and this required matching can be very difficult.
The first goal of this paper is to quantify the magnitude of the bias, type I error, and type II error that can occur if the ancestries of the two groups are poorly matched. Then we identify two other possible sources of bias, the type of platform (e.g. Illumina, Affymetrix) and the number of subjects in the reference group. We use HapMap data to further quantify the extent of the bias in real samples (The International HapMap Consortium, 2003). After demonstrating the severity of the potential problems caused by using T, we propose a new statistic that only requires selecting a reference group with an ancestry that matches the ancestry of the individual of interest. This is a far simpler task, as the individual’s ancestry is usually known. Other benefits of the new statistic are that it is unbiased and has a known null distribution. We then compare the performance of the two statistics using simulated data. The paper is therefore ordered as follows. Section II starts by introducing notation and then continues with a discussion of the properties of both statistics and their associated tests. Section III demonstrates the performance of the statistics and their associated tests using both HapMap and simulated data. Finally, section IV contains our brief concluding remarks.
2. Methods
2.1. Notation
Let the individual of interest be indexed by 0, and let the nR subjects in the reference population and the nM subjects in the mixture be indexed by {1, ...,nR + nM}. For subject i, i ∈ {0, 1, ...,nR + nM}, let Ri = 1 (Mi = 1) if subject i is in the reference group (mixture), Ri = 0 (Mi = 0) otherwise. Unless explicitly stated otherwise, order the subjects so the first nM belong to the mixture (i.e. Mi = 1 for 1 ≤ i ≤ nM). Note that we use the shorthand that “subject i is in the mixture” or “subject i is in the mixture group” if subject i contributes DNA to the mixture. Let ei be the ancestry, or ethnicity, of subject i. We start by assuming homogenous groups and therefore ei ∈ {e0, eM, eR}, with ei = eR if Ri = 1, and ei = eM if Mi = 1.
Let there be N SNPs in the study. Let Qi j1 and Qi j2 indicate whether the minor allele (relative to ethnicity e0) is on chromosome 1 and 2, respectively, for subject i at SNP j. Let Yi j = 0.5 (Qi j1 + Qi j2) be the genotype for subject i at SNP j; Yi j ∈ {0, 0.5, 1}. We define the population allele frequencies by pM j ≡ P(Qi jk = 1 | Mi = 1, i > 0), pR j ≡ P(Qi jk = 1 | Ri = 1, i > 0), and p0 j ≡ P(Q0 jk = 1) where k ∈ {1, 2}. For calculating genotype probabilities, we assume Hardy-Weinberg Equilibrium.
Most genotyping platforms directly measure fluorescent intensity, which should be proportional to allele frequency. We shall label the intensity measures for allele A and allele B at SNP j from the mixture group as IAM j and IBM j. These intensities are measurements from a “pooled sample”. We label the ratio of intensities by γM j = IAM j / (IAM j + IBM j). If intensity measurements are also available for individuals, then we can calculate a constant kM j using known formulas (Pearson et al., 2007; Macgregor et al., 2008) and use a better proxy for the A allele frequency, γM j = IAM j / (IAM j + kM jIBM j). Similarly, define {IARj, IBRj, γR j, kR j} for the reference group. Therefore, the distance measure at each SNP from the Homer et. al. paper can be defined as
| (1) |
Their test statistic can then be defined as
| (2) |
where and μ0 = 0. In this manuscript, we will also discuss
| (3) |
and the corresponding values DL2 and TL2. The “L1” and “L2” subscripts emphasize the distance measure used for the statistic. We discuss the case when intensities perfectly reflect underlying allele frequencies and define X (j) ≡ | Y0 j – p̂R j | – | Y0 j – p̂M j|, where and .
In general, we assume that γM j and γR j can be described by the following statistical models:
| (4) |
where , , and {βM j, βR j} are SNP/platform specific biases. Therefore, the error is now linearly associated with the ratio of intensities, instead of the intensities themselves. As studies tend to exclude SNPs with rare minor alleles, we assume that 0 ≤ γM j, γR j ≤ 1 will generally hold. This description of γM j and γR j as normal variables is a simplification, but we use it only to show the existence of and then approximate expected biases. Note that the properties of our suggested test statistic do not depend on the validity of equation 4. We define , , , and . Finally, we use the abbreviation Θ to represent the set of all parameters, .
2.2. DL1 : The Original Test
The definition of any test requires a statement of the null hypothesis and the list of outcomes that would lead to the rejection of that null. Therefore, the desired null hypothesis is that the individual of interest is not in the mixture, M0 = 0, and the original test is to reject this null when TL1, with μ0 = 0, is large. The next goal is to calculate the appropriate threshold, type I error rate, and power. Unfortunately, TL1 is not a pivotal statistic, in that its distribution still depends on the parameter set Θ. Stating M0 = 0 does not lead to a single distribution of TL1. Therefore, for the original paper to discuss the type I error rate and power, there needed to be three additional assumptions
Identical Ethnicities: e0 = eM = eR
Identical Sample Sizes: nM = nR
Identical Platforms: βM j = βR j and
With these added assumptions, TL1 no longer depends on Θ and they could choose a threshold tα so that the type I error of the test would be
| (5) |
The power was then calculated as
| (6) |
Unfortunately, if one or more of these three assumptions are false, the type I error rate and power can differ from α and βPOW. Here, we consider a test to be biased for a given Θ if P(TL1 > tα |M0 = 0, Θ) ≠ α or P(TL1 > tα |M0 = 1, Θ) ≠ βPOW. In the following sections, we show the magnitude of the bias when each of the assumptions is violated.
2.2.1. DL1 : Bias from Ancestry
In this section, we show that unless the ancestries of the reference and mixture groups are extremely well matched, the false positive rate can easily be near 1, as opposed to the predicted α. Ignoring the sample size and platform parameters, there are seven possible scenarios (Table 1). The original paper thoroughly discussed scenarios H0 and H1 with assumptions II and III holding, but only warned of potential bias in the other cases (i.e P(TL1 > tα |M0 = 0, eM, eR, e0) ≠ α and P(TL1 > tα |M0 = 1, eM, eR, e0) ≠ βPOW for all combinations of e0, eM, and eR). Here, we want to estimate the magnitude of the type I error for the scenario Hf p where the result is likely to be a false positive (Table 1). Under Hf p, the individual is not in the mixture, but only the ancestries of the individual of interest and the mixture are the same (e0 = eM ≠ eR).
Table 1:
There are seven scenarios depending on whether the individual of interest is in the mixture (column 2) and depending on whether the ethnicity of the individual of interest (e0), the ethnicity of the mixture group (eM), and the ethnicity of the reference group (eR) are the same (columns 3,4,5). ‘Yes’ indicates the statement at the top of the column is true and a blank indicates the statement is false. For example, in scenario 1, which we label as “H0”, the individual of interest is not in the mixture (the statement M0 = 1 is false), the ethnicities of the individual and mixture are the same (e0 = eM), the ethnicities of the individual and reference are the same (e0 = eR), and the ethnicities of the mixture and reference are the same (eR = eM). In this case, all individuals share a common ethnicity. Scenarios 1 and 2 were studied by Homer et al. (2008). Here, we also study scenario 3, which we label as Hf p because it is likely to lead to a false positive result.
| Ethnicities | ||||
|---|---|---|---|---|
| M0 = 1 | e0 = eM | e0 = eR | eR = eM | |
| 1 (H0) | Yes | Yes | Yes | |
| 2 (H1) | Yes | Yes | Yes | Yes |
| 3 (Hf p) | Yes | |||
| 4 | Yes | |||
| 5 | Yes | |||
| 6 | ||||
| 7 | Yes | Yes | ||
Estimating the true type I error rate requires understanding the distribution of TL1, or equivalently, DL1 (j), for the three scenarios, H0, H1, and Hf p. As the allele frequencies at many of the SNPs will likely be the same for all populations, p0 j = pR j = pM j, the type I error rate will depend on s, defined to be the proportion of all SNPs that are ancestry independent, and the differences between pR j and pM j for the other (1 – s)N SNPs. To start, we calculate the asymptotic distribution of DL1 (j) (see appendix 5.1), the fundamental component of TL1, in all three scenarios. When n is large, n ≡ nM = nR, and βR j = βM j ∀ j,
| (7) |
where
| (8) |
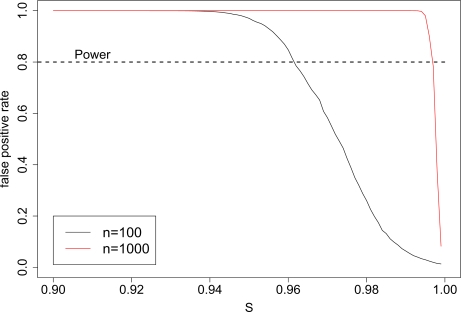
Figure 1 shows the type I error rate as a function of s assuming that pM j is uniformly distributed over the interval (0, 0.5), pR j = pM j at SNPs 1,...,sN, and pR j is uniformly distributed over the interval (0,0.5) for the remaining (1-s)N SNPs independent of pM j. This will overestimate the type I error rate for a given s, because, in truth, pR j will often be relatively close to pM j. Nevertheless, this simplified example demonstrates the magnitude of the type I error rate. Figure 1 shows that when nM is large (i.e. nM = 1000), over 99% of all SNPs would need to be ancestry independent to prevent the false positive rate from being near 1. Therefore, the false positive rate can easily equal or exceed the power of the test. Next, we can calculate the value of s needed for the false positive rate to equal the power, which is essentially when μ1 =μ̄f p, where μ1 ≡ E[μ1 (j)] and μ̄f p ≡ (1 – s) E[μf p (j)]. In appendix 5.2, assuming the allele frequencies have the above uniform distributions, we show that μ1 = 0.23/n and μ̄f p = 0.062(1 – s). Therefore, if 1 – s = 3.7/n, μ1 = μ̄f p. If our mixture contained 1000 subjects, the reference and mixture groups would only need to differ at 0.37% of all SNPs for the false positive rate to exceed the power. Had we allowed the allele frequencies for those (1-s)N SNPs to be randomly distributed over the interval [0, 1], even fewer SNPs would need to differ. Here, μ1 = μ̄f p if 1 – s = 1.2/n, and only 0.12% of all SNPs would need to differ.
Figure 1:
The false positive rate can easily exceed power when tests are based on TL1. Using the normal approximations given in equations 7 and 8, we plot the false positive rate when Hf p is true, showing its decrease with the proportion, s, of SNPs that are ancestry independent. Under H1, the power was set to be 0.8. Calculations assume N=50,000 and n ∈ {100, 1000}.
2.2.2. DL1 : Bias from Sample Size
In this section, we show that unless the number of subjects in the mixture and reference groups are equal, either type I or type II error can be higher than expected. Here, we keep assumptions I and III, identical ancestries and platforms, and examine the test’s bias if nM and nR are finite and unequal. Before discussing the origin and extent of this bias, we state a useful fact, which is proven in appendix 5.3.
Inequality of Absolute Values:
Let p < 0.5, , , and p1 ⊥ p2.
If , then E [|0.5 – p1|] > E [|0.5– p2|]
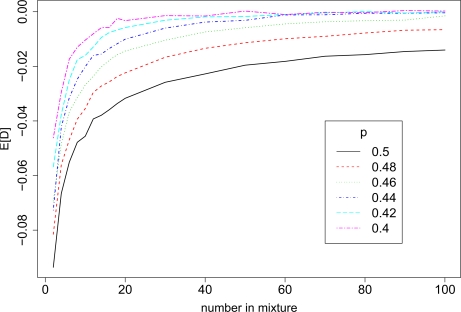
Our goal is to show that if nM ≠ nR, then E[DL1 (j)] ≠ 0 when M0 = 0, even under otherwise ideal conditions. This is demonstrated empirically by figure 2. A more complete discussion follows. Without loss of generality, assume that we have a large reference sample and nR > nM. Then var(p̂M j) > var (p̂R j). Because p̂R j is a more precise estimate of p, the expected L1 distance between Y0 j and p̂R j tends to be smaller than the expected distance between Y0 j and p̂M j, or, equivalently, E[DL1 (j)] < 0. This would be a trivial statement if distance were measured in L2. For L1, we will show that E[Xj] < 0, which is essentially equivalent to E[DL1 (j)] < 0. Recall, X (j) ≡ | Y0 j – p̂R j | – | Y0 j – p̂M j |. To use the Inequality of Absolute Values, we start by assuming and , and decompose X (j) as
| (9) |
where
| (10) |
It is immediately clear that when the individual of interest is not in the mixture, E[Xj1] = E[p̂R j] – E[p̂M j] = 0 and E[Xj0] = E[p̂R j] – E[p̂M j] = 0. With our assumptions of normality and the above inequality, E[Xj0.5] < 0. Consequently, E[Xj] < 0. For an even more intuitive understanding, let nR ≈ ∞ and p̂R j = pR j ≡ 0.5 Then, clearly |0.5 – p̂R j | = 0, and Xj0.5 < 0.
Figure 2:
E[DL1 (j)] ≠ 0 even when pR j = pM j ≡ p. In this example, where we let nR = ∞ (p̂R j = p), we find that E[DL1 (j)] decreases as the number of individuals in the mixture shrinks toward 0. We plot E[DL1 (j)] (y-axis) vs nM (x-axis) for multiple values of p.
2.2.3. DL1: Bias from Platform
In this section, we show that unless the allele intensities for the mixture and reference group are measured on the same platform, the test may be biased, often leading to an increase in type II error. Normally, we need not concern ourselves with platform bias, as we can ensure that both samples will be measured on the same platform. Unfortunately, this is not the case here. For the mixture, we have a sample of DNA and we can choose our preferred platform. For the reference group, we often do not have access to actual samples of DNA, and instead base our estimates of allele frequencies on previously recorded genotypes for a group of individuals. Comparing γM j and is equivalent to comparing allele frequencies measured on two different platforms. Also, even if the reference sample were an actual mixture of DNA, that sample might no longer be available for analysis.
First, if βM ≠ βR, then E[DL1] ≠ 0 when M0 = 0, even under otherwise ideal conditions. Simple algebra shows that if model (4) were true,
| (11) |
Here, we presume E[DL1 (j)] > 0 implies E[DL1] > 0, or that . Fortunately, this bias can be easily removed by substituting and into the equations for DL1 (j), where {β̂M j, β̂R j} is an unbiased approximation of {βM j, βR j}, or by using the appropriate constants kM j and kR j for calculating γM j and γR j.
However, this does not eliminate the potential bias caused by platform. If , then E[| Y0 j – γR j | – | Y0 j – γM j |] ≠ 0. The origin of this confounding is identical to that for the nM ≠ nR case. Here, we would assume that and . Next, note that even if E[β̂M j] = βM j, E[β̂R j] = βR j, and , there can still be bias if var (β̂M j) ≠ var (β̂R j), as . The origin of the confounding is easily identified if we assume that and . The main importance of these statements is that the reference sample cannot simply be the average of known genotypes. In this case and βR j = 0 (i.e. var (β̂R j) = 0). This would create a considerable test bias.
2.3. DL2 : Switching from L1 to L2
Using the L2 distance (recall DL2 (j) = (Y0 j – γR j)2 – (Y0 j – γM j)2) greatly simplifies calculations. Under the null hypothesis H0, if we keep assumptions II and III, the mean and variance of DL2 (j) can be easily calculated as (see appendex 5.4):
| (12) |
With the same assumptions, under the alternative hypothesis, H1, the mean of DL2 (j) is
| (13) |
and the variance can be approximated by σ2 (j). In contrast to DL1, we have estimated the parameters for the normal approximation of DL2 without assuming large n. Equivalently, the approximation of requires only DL2 (j1) ⊥ DL2 (j2) ∀ j1 ≠ j2 and N being large. In addition to the ease of L2, we found the statistic TL2 to outperform TL1. This can be verified by simulation (data not shown) or by comparing the two normal approximations for TL1 and TL2. In the online supplementary material, we compare the power of the statistics TL1 and TL2 for specific values of n, N, , and σ2, assuming independent SNPs and pM j ∼ uniform[0,0.5]. The plots suggest a mild improvement using TL2.
2.4. : A New Statistic
Our ideal goal would be to develop a pivotal statistic that, when M0 = 0, is a) independent of the individual of interest’s ethnicity; b) independent of the number of individuals and the ethnicity of those individuals in the mixture; and c) independent of the platform chosen to analyze the mixture. Although we could find no such statistic, we can take advantage of being able to easily identify an individuals’ ethnicity by genotyping a small set of SNPs (< 0.01N). The previous requirement of identifying the ethnic composition of the mixture is a far more difficult task. Therefore, we introduce a new statistic that, given e0, will have a N(0,1) distribution when M0 = 0 regardless of the remaining parameters. The key difference in deriving this statistic is that the individuals in the reference group will be selected to have the same ancestry as the individual of interest, as opposed to the same ancestral composition as the mixture. Note that the small set of SNPs used to identify the individuals’ ancestries can be removed from the later analyses without greatly diminishing power. A suggested list of SNPs will be made available by the authors in the near future. Moreover, we found that the suggested statistic will lead to a test with increased power. The details of the statistic follow.
To describe the new statistic, we change the notation slightly as we no longer have Yi j values for subjects in the mixture. Therefore, in addition to having γM j and Y0 j, 1 ≤ j ≤ N, choose nR subjects for a reference group and let Yi j be the genotype for subject i, 1 ≤ i ≤ nR at SNP j. Note that i is bounded by nR instead of nR + nM. Select the reference subjects so their ancestry is similar to that of the individual of interest (i.e. ei = e0 if Ri = 1).
Step 1:
Create nR + 1 reference samples, , , where , k ∈ {0, ...,nR} and
| (14) |
Here, in model (4). We have immediately removed a source of variation.
Step 2:
Measure the distance between each individual and the appropriate reference group, (Yi j – γRi j)2, and compare the resulting value to the distance between that individual and the mixture
| (15) |
Because the term is absent from , . In fact,
| (16) |
All variance and covariance calculations will assume n ≡ nR = nM and pj ≡ pR j = p0 j = pM j.
Step 3:
We then average those differences over all reference subjects,
| (17) |
to obtain an expected difference between the distance to the reference sample and the distance to the mixture, under the null hypothesis. The covariance between two terms in the sum is (see appendix 5.4)
| (18) |
Each term on the right side of equation 18 must be positive, so the covariance must be positive (i.e. ).
Step 4:
Compare to this averaged value
| (19) |
By noting the exchangeability between any two terms and , we can calculate the variance of
| (20) |
The key result is that, except for those values of where the power is essentially 1 regardless of the chosen statistic, . As the means for the L2 statistics are the same, the smaller variance suggests that our proposed statistic , based on , will usually have higher power than the original TL2. This improvement is demonstrated in our simulations. Moreover, the statistic was designed so if the ancestries of the reference group and individual of interest are matched correctly, regardless of platform and sample size.
Step 5:
Average over all SNPs to get
| (21) |
Because of the large number of SNPs, the CLT suggests that under the null hypothesis. When allowing for dependency,
| (22) |
Therefore, can be estimated by
| (23) |
In practice, we restrict the double sum to j1 ≠ j2 and | j1 – j2 | ≤ 500. In the on-line supplementary material, we use the HapMap samples to show . We now define our new statistic,
| (24) |
and note that . For purposes of future comparisons, we also define , , and by replacing the L2 distance with the L1 distance in , , and .
Although appears to be one of the better performing statistics, it is not necessarily the most intuitive. We first tried a simpler alternative, , but as we discuss here, this statistic proved to have extremely low power. Let
| (25) |
where
| (26) |
This alternative also has the desirable characteristics that neither platform nor sample size can invalidate the equalities E[(Y0 j – γM j)2] = E[(Yi j – γM j)2] and , and that the ancestry of the reference group need only match that of the individual of interest. The problem with such a simple approach is that the . To understand the origin of this vast difference between variances, we turn to a simple example, where Yi j ∈ {0, 1} and P(Yi j = 1) = pjM. Here, , and the var(DL2 (j)), under H0, asymptotically approaches 0. In contrast, , where is the appropriate function of the reference group. Without the cancelation of the Y0 j terms in the statistic, asymptotically, . Therefore, we designed to not only avoid potential bias, but retain the advantage gained by the cancelation of Y0 js in DL1. Here we also note that the variance of DL2 (j) or any similar statistic will be minimized when E[γM j] = E[γR j].
This same simple example, where Yi j ∈ {0, 1} and P(Yi j = 1) = p jM, illustrates why the original statistic, DL1 (j) = | Y0 j – γM j | – | Y0 j – γR j |, or any of our adaptations, needs to include the reference term, | Y0 j – γR j |. Under the null hypothesis, var(| Y0 j – γR j | – |Y0 j – γM j |) = 2p jM (1 – p jM) /n → 0. In contrast, without the cancelation of Y0 j terms, . Even if the ethnicities were well matched, the variance of the genome-wide statistic would be proportional to the sum of these variances over the entire genome, not just over the subset of informative SNPs. For choosing the best statistic, the variance will be the deciding value. The additional term | Y0 j – γR j | does not change the expected difference between the test statistic under the null and alternative hypotheses.
2.5. Data and Simulations
The power and type I error for the test based on TL1 has already been thoroughly described when e0 = eR = eM. Here, we use simulations to accomplish two goals. First, we show that tests based on are more powerful than tests based on TL1. Second, we show that if the threshold tα is chosen so that the type I error rate is α when e0 = eR = eM (equation 5), but, in fact, assumption I is not true, then the true type I error rate can greatly exceed α.
The simulations generated distributions of TL1, TL2, , and for each of the three scenarios, H0, H1, and Hf p. Recall that the denominator for TL1 was , which assumes independence across SNPs. Therefore, in order to provide a fair comparison, we let the genotypes be independently distributed and let the denominator for be .
First, we simulated 10,000 datasets (and 10,000 values of each test statistic) under H0, with e0 = eM = eR, nM = nR, and βM = βR = 0. Each dataset is simulated as follows. For each of 5000 SNPs (when nM = 100) or 50,000 SNPs (nM = 1000), we randomly generated p0 j, the frequency of allele ‘A’ for the individual of interest, from a uniform(0.1,0.5) distribution. Then, we set pM j = pR0j = pRM j = p0 j, where pM j, pR0 j, and pRM j are the ‘A’ allele frequencies for subjects in the mixture, reference group R0 (selected to match the individual of interest), and reference group RM (selected to match the mixture). Given the allele frequencies and assuming Hardy-Weinberg Equilibrium, we then generated the genotypes for the individual of interest ( ), the nm individuals in the mixture (YM), and the nR individuals in each reference group (YR0, YRM). Given knowledge of the individuals in the mixture and reference groups, we generated the microarray intensities at each SNP as and , where . Simulations were performed for a range of values. The results are a single dataset and a set of values . After simulating all 10,000 datasets under H0, we denote the 99th percentile of the four test statistics by t̂αL1, t̂αL2, , and .
Next, we simulated 1000 datasets under H1. To create the datasets, we repeated the above steps, but let . Then the power for the original test was the propotion of these 1000 TL1 values exceeding t̂αL1. A similar estimation of power was performed for TL2, , and . Finally, we simulated datasets under Hf p. The type I error rate will depend on how the allele frequencies differ between the two ancestries. In one case, we assumed that a proportion, s, of the SNPs to be ancestry independent, (i.e. pM j = pR0j = pRM j = p0 j). For j > s × 5000 or j > s × 50, 000, we still enforced pM j = pR0j = p0 j, but generated pRM j from a uniform(0.1,0.5) distribution. In the second case, we assumed that the allele frequencies differed at all SNPs and let pRM j = (1 + kj)±1 p0 j, where {k1, ..., kN} was a vector of equally spaced points over the range 0 to 0.15, 0.20, and 0.25 when nM = 100 and 0 to 0.03, 0.05, and 0.07 when nM = 1000. The superscript ± 1 indicates that the exponent was randomly generated to be −1 or 1 with equal probability. Here, we can quantify the extent of the difference between the two ethnicities by the measure λ, the variance inflation factor typical in case/control studies. For nM = 100, the three ranges correspond to a λ of 1.16, 1.36, and 1.65, and for nM = 1000 the three ranges correspond to 1.09, 1.32, and 1.67. For each of these cases, we simulated 1000 datasets and defined the type I error rate to be the percentage of these 1000 TL1 (TL2) values exceeding t̂αL1 (t̂αL2).
To better quantify the type I error rate possible in real experiments, we used the 90 CEU and 45 Japanese individuals from the HapMap samples. For each CEU individual i, we selected 9 unrelated CEU individuals to create a positive mixture, , or 10 unrelated individuals to create a negative mixture, , where , εij ∼ N(0,0.012) and εi j1 ⊥ εi j2 if j1 ≠ j2. To achieve meaningful levels of power, we chose N = 1000. For each CEU individual, 11 reference groups were similarly created where γRt included t Japanese individuals and 10 – t CEU individuals. We calculated the distribution of TL1 (36000 simulations, 90 subjects × (400,000 SNPs/1000)=400 sets of SNPs) under H0, H1, and Hf p. We chose the threshold t̂αL1 so that α = 0.05 if H0 were true and then calculated the probability of rejection under H1 (i.e. power) and under Hf p (i.e. type I error).
3. Results
3.1. Comparison of Power
Our first goal is to show that tests based on are more powerful than tests based on TL1. As the results for the two sets of simulations, nM = nR = 1000 and nM = nR = 100 were similar, we limit our discussion to the latter. Recall, we chose to empirically estimate the 99th percentile of the test statistics under H0. This was a necessity because our reference mixture is an average of known genotypes, , and as discussed in section 2.2.3, TL1 cannot be expected to follow a N(0, 1) distribution. Under H0, when σM = 0.03, 0.05, 0.07, and 0.09, the averages of TL1 over the 10,000 simulations were −0.76, −1.71, −2.77, and −3.91 respectively. Similarly, the averages of TL2 were −1.85, −4.13, −6.46, and −8.68 for those same values of σM. The empirical variances were close to 1. In contrast, followed a N(0, 1) distribution, as promised, and this empirical step was superfluous. The empirical means for and were near 0, –0.015 ≤ ≤ –0.010 and –0.024 ≤ ≤ –0.015, regardless of σM. Moreover, not only were the empirical variances near 1, the empirical 99th percentiles were approximately 2.326 (2.29 ≤ ≤ 2.32 and 2.31 ≤ ≤ 2.32).
For each value of σM, we then calculated the power for the four tests, based on TL1, TL2, , and . The alternative hypothesis was H1 with the other parameters unchanged. These values are plotted against signal noise or σM in Figure 3. Clearly, power is larger when the statistics are based on L2 and when the reference group is matched to the individual of interest. Note, had we included an error term in the reference mixture (and possibly attained TL1, TL2 ∼ N (0, 1)), the benefit of using the new statistic would have been larger.
Figure 3:
Tests based on have the highest power. Power for tests based on the four statistics, TL1, TL2, , and , are plotted for multiple values of or noise. Other parameters in the simulation were nM = nR = 100, N = 5000, s = 1, and βM = βR = 0.
3.2. Examination of False Positive Rate
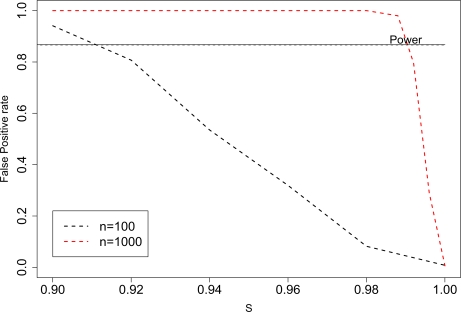
Our second goal is to show that when assumption I is violated, the type I error rate can be much larger than α. Again, we presume tα was selected so equation 5 holds. Simulations showed the quick increase in the false positive rate, P(TL1 > tα | Hf p), as the proportion of ancestry-independent SNPs decreased in those tests based on TL1. Parameter values were chosen so that the ideal test (e0 = eM = eR), for both nM = 100 and nM = 1000, would have had power = 0.87 to detect an individual’s presence. Clearly, unless s is close to 1, the false positive rate exceeds this estimated power. Because we have finite sample sizes and , we cannot expect the predicted power and false positive rate to be equal when 1 – s = 3.7/n (Appendix 5.2). However, the simulations can confirm that the two are approximately equal when μ1 =μf p and that value of 1 – s which results in equality is proportional to 1/n. When the power and false positive rate cross in Figure 4 for nM = 100, we find μ1 = 2.40, μf p = 2.41, and 1 – s = 0.088. Similarly for nM = 1000, we find μ1 = 2.65, μf p = 2.64, and 1 – s = 0.0089. For simulations under the second set of pRM j, we found the false positive rates to be 0.11, 0.30, and 0.61 when nM = 100 and 0.035, 0.15, and 0.51 when nM = 1000.
Figure 4:
The type I error rate for tests based on TL1 can exceed power. Type I error rate increases as the proportion, s, of ancestry independent SNPs decreases. Other parameters in the simulation were N=5000 (50,0000), , and nM = nR = 100(1000).
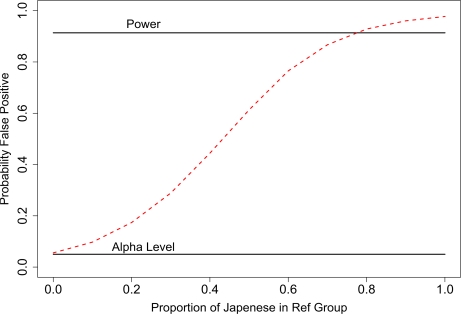
The false positive rate for the HapMap samples were calculated when the mixture and reference groups each had 10 subjects. Both the individual of interest and the subjects in the mixture were from the CEU population. As the ratio of Japenese:CEU individuals increased in the reference group, the false positive rate increased from the α-level, 0.05, to near 1. When the ratio exceeded 8:2, the false positive rate exceeded the estimated power.
4. Discussion
As the overall popularity of SNP microarray technology increases and the cost of the technology decreases, there will likely be a shift from STRs to genome-wide sets of SNPs as the preferred method for DNA identification. Therefore, databases designed to store genetic identifiers for individuals will likely store genotypes for sets of SNPs in the future. Coupled with our earlier discussion, there will be three main advantages of using high-density SNPs to determine whether an individual contributes DNA to a mixture: accessibility, higher resolving power and the ability to work with low quality, degraded, DNA.
Here, we have demonstrated that tests based on TL1 can suffer from inflated type I and type II errors if the mixture contains individuals with unknown ancestries. Therefore, we introduced a new test statistic, and an accompanying test that only require matching the ancestry of the reference group to that of the individual of interest. Even if the individual of interest has a mixture of ancestries, there should still be some subjects, with a similar mixture of ancestries, that can be used for comparison. This test is also robust to platform and sample size. We showed that both switching from the L1 to the L2 measure and switching to the new type of statistic increased the power to detect the individual of interest. Therefore, is not only more robust than TL1, it tends to have increased power.
Supplementary Material
additional power comparison
Figure 5:
The type I error rate for tests based on TL1 can exceed power in simulations based on HapMap samples. The individual of interest and individuals in the mixture group were from the CEU population. The individuals in the reference group were a mixture from the CEU and Japanese populations. The type I error rate increased with the percentage of individuals in the reference group that were from the Japanese population. Other parameters in the simulation were N=1000, nM = nR = 10, and
Acknowledgments
We would like to thank Dr. David Craig for providing us with data from his original article and to the anonymous reviewers for their helpful comments. This work was supported by NIH GM59507.
5. Appendix
5.1. Distribution of DL1 (j)
To estimate the distribution of DL1 (j), we make the following assumptions, where g ∈ {R, M}:
Model (4) is true.
βR j = βM j = 0
-
n ≡ nM = nR
And restrict the SNPs examined to those satisfying
-
|pg j – 0.5 | >δ
for some small δ such that
P(|p̂g j – pg j | > δ /2) ≈ 0.
P(|εg j | > δ /2) ≈ 0.
Assumptions 5 and 6 promise that n is large enough and the magnitude of εg j is small enough so that the signs inside the absolute values are determined by Y0 j – pg j. For much of the exposition, we will still leave the βR j and βM j terms in the equations, although we assume they are small enough to not affect the sign inside the absolute values, to demonstrate their effects on bias and variance had they not been assumed to equal 0. To start, divide DL1 (j) into three components
| (27) |
where
| (28) |
By definition pM j < 0.5. We next calculate the expected values of each component under the two assumptions, H1 and Hf p. We know that , where g indicates group. Therefore, assuming scenario H1, where without loss of generality, we let the individual of interest be subject 1 of the mixture,
| (29) |
| (30) |
| (31) |
We immediately see that
| (32) |
Let us assume that βM j = βR j, then we get
| (33) |
Next we calculate the expected value under H f p,
| (34) |
We immediately see that
| (35) |
For simplification, let us assume that βM j = βR j, then we get
| (36) |
Now, we turn our attention to the variance of DL1 (j) and recall that
| (37) |
We calculate the var(DL1 (j) | Y0 j), assuming all individuals within a group are unrelated,
| (38) |
Under H0, where E[DL1 (j) | Y0 j] = 0 and pR j = pM j, we see that
| (39) |
For simplification, we again assume that βM j = βR j and therefore
| (40) |
Under H1, we find that
| (41) |
and for large n, we see that E[var(DL1 (j) | Yj0, H1)] ≈ E[var(DL1 (j) | Yj0, H0)]. Similarly, because E[DL1 (j) | Y0 j, H1] = E[DL1 (j) |Y0 j, H0] + O( ), then . Therefore, we can assume that var(DL1 (j) | H1) ≈ var(DL1 (j) | H0).
Next, we turn to scenario Hf p. Clearly,
| (42) |
To calculate the var(E[DL1 (j) | Y0 j, Hf p]), we use the notation pΔj = pM j – pR j and βΔj =βM j – βR j, then under Hf p, if we let pR j < 0.5,
| (43) |
Now, if we assume that the minor allele in the mixture is the same as the minor allele in the reference group, or equivalently, that we have at least identified a reference population of moderately similar composition, then we can be satisfied that equation (43) is a satisfactory approximation of var(E[DL1 (j) | Y0 j, Hf p]). Again, with the simplification that βM j = βR j, we find
| (44) |
5.2. Estimating when μ1 = μ̄f p
Here, we provide a rough approximation of the percentage of SNPs that would need to be ancestry dependent for E[μ1 (j)] = (1 – s) E[μf p (j)]. We base this approximation on the assumption that pM j and pR j are uniform random variables over the intervals [0,0.5] and [0,1] respectively. Start by calculating E[DL1 (j) | pR j < 0.5] from equation 35, letting βM j = βR j where the expectation is over pR j and pM j.
| (45) |
Had we assumed pR j > 0.5, E[DL1 (j) | pR j > 0.5, Hf p] = 0.32. Combining the two equalities, we find E[DL1 (j) | Hf p] = 0.19. Obviously, from an evolutionary perspective, pR and pM should often be similar in value, but we ignore that here for simplicity. Furthermore, assume that only some proportion, 1 – s, of those N SNPs differ among ethnicities. Then E[μ f p (j)] = 0.19 and μ̄ f p ≡ (1 – s) E[μf p (j)] = 0.19(1 – s).
Next, from equation 33, we know nE [DL1 (j)] = 2pM j (1 – pM j)2, and again assuming pM j is a uniform random variable over the interval [0,0.5], we find E[DL1 (j) | H1] = .22/n. Therefore, across all SNPs, assuming no linkage, μ1 ≡ E[μ1 (j)] = .22/n.
Therefore, in order for μ1 = μ̄ f p, we would need .22/n = 0.19 (1 – s), which shows that false positives can be expected so long as the percentage of ancestry dependent SNPs, 1 – s, exceeds 1.16/n. Remember, that the difference between pR and pM is likely to be overstated by the above simplifications, and therefore this 1.16/n will be low.
5.3. Inequality of Absolute Values (proof)
Theorem: Let , , g(X) = | 0.5–X |, X1 ⊥ X2, , and p < 0.5. Then
| (46) |
Let Z1 = F1 (X1) and Z2 = F2 (X2), where F1 and F2 are their respective cumulative distribution functions. Then
| (47) |
There are two possibilities. Case A) Assume . Then, by linearity of g(X),
| (48) |
Clearly, by similar logic,
| (49) |
Case B: . Then, clearly
| (50) |
Therefore, for case B we also see
| (51) |
Taking the expectation over both scenarios gives the desired result, E[g (X2)] – E[g (X1)] < 0.
5.4. Distribution of DL2 (j) and
The distribution for DL2 (j) and can be described by the variances and covariances of terms in the following vector:
| (52) |
by simple calculation, the var(V1) can be described by:
In this section, as we focus on only a single locus, we have dropped the subscript ‘j’ from Yi j, pj and . Next, by expanding the terms, and assuming n ≡ nM = nR, and βR = βM = 0, we get
| (53) |
and
| (54) |
By substituting the appropriate values from the variance matrix for V1, we get the variance given in section 2.3. Using the same concept of expanding the terms and straight forward calculation, we were able to find covar( ).
References
- Balding David J. Likelihood-based inference for genetic correlation coefficients. Theoretical Population Biology. 2003;63(3):221–230. doi: 10.1016/S0040-5809(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Couzin Jennifer. Genetic privacy: Whole genome data not anonymous, challenging assumptions. Science. 2008 Sep;321(5894):1268–1374. doi: 10.1126/science.321.5894.1278. [DOI] [PubMed] [Google Scholar]
- Foreman LA, Champod C, Evett JA, Lambert S Pope. Interpreting dna evidence: A review. International Statistical Review. 2003;71:473–495. [Google Scholar]
- Fung Wing K, Hu Yue-Qing. Evaluating mixed stains with contributors of different ethnic groups under the nrc-ii recommendation 4.1. Statistics in Medicine. 2002 Nov;21:3583–3593. doi: 10.1002/sim.1313. [DOI] [PubMed] [Google Scholar]
- Homer Nils, Szelinger Szabolcs, Redman Margot, Duggan David, Tembe Waibhav, Muehling Jill, Pearson John V, Stephan Dietrich A, Nelson Stanley F, Craig David W. Resolving individuals contributing trace amounts of dna to highly complex mixtures using high-density snp genotyping microarrays. PLoS Genet. 2008 Aug;4(8):e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling Mark A, Gill Peter. Encoded evidence: Dna in forensic analysis. Nat Rev Genet. 2004 Oct;5:739–751. doi: 10.1038/nrg1455. [DOI] [PubMed] [Google Scholar]
- Kidd Kenneth K, Pakstis Andrew J, Speed William C, Grigorenko Elena L, Kajuna Sylvester LB, Karoma Nganyirwa J, Kungulilo Selemani, Kim Jong-Jin, Lu Ru-Band, Odunsi Adekunle, Okonofua Friday, Parnas Josef, Schulz Leslie O, Zhukova Olga V, Kidd Judith R. Developing a snp panel for forensic identification of individuals. Forensic Science International. 2006;164(1):20–32. doi: 10.1016/j.forsciint.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Macgregor Stuart, Zhao Zhen Zhen, Henders Anjali, Nicholas Martin G, Montgomery Grant W, Visscher Peter M. Highly cost-efficient genome-wide association studies using dna pools and dense snp arrays. Nucleic Acids Res. 2008;36:e35. doi: 10.1093/nar/gkm1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JV, Huentelman MJ, Halperin RF, Tembe WD, Melquist S, et al. Identification of the genetic basis for complex disorders by use of poolingbased genomewide single-nucleotide-polymorphism association studies. American Journal of Human Genetics. 2007;80:126–139. doi: 10.1086/510686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M, Hedgecock D, Higuchi RG, Vigilant L, Erlich HA. Population variation of human mtdna control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. American Journal of Human Genetics. 1991;48(2):370–382. [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The international hapmap project. Nature. 2003 Dec;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
additional power comparison
additional power comparison