Summary
The putative modular polyketide synthase (PKS) that prescribes biosynthesis of the bryostatin natural products from the uncultured bacterial symbiont of the marine bryozoan Bugula neritina possesses a discrete ORF (bryP) that encodes a protein containing tandem acyltransferase (AT) domains upstream of the PKS ORFs. BryP is hypothesized to catalyze in trans acylation of the PKS modules for polyketide chain elongation. To verify conservation of function, bryP was introduced into AT-deletion mutant strains of a heterologous host containing a PKS cluster with similar architecture, and polyketide production was partially rescued. Biochemical characterization demonstrated that BryP catalyzes selective malonyl-CoA acylation of native and heterologous acyl carrier proteins and complete PKS modules in vitro. The results support the hypothesis that BryP loads malonyl-CoA onto Bry PKS modules, and provide the first biochemical evidence of the functionality of the bry cluster.
Introduction
Evidence is mounting that many bioactive natural products isolated from marine invertebrates (e.g., sponges, ascidians, bryozoans) are produced by microbial symbionts [reviewed in (Konig et al., 2006; Piel, 2004; Salomon et al., 2004)]. Particularly compelling evidence indicates that the bryostatins, ecologically relevant bioactive compounds isolated from the temperate marine bryozoan, Bugula neritina, are produced by the uncultured microbial symbiont “Candidatus Endobugula sertula” that resides in B. neritina (Davidson et al., 2001; Lopanik et al., 2004; Lopanik et al., 2006). To date, different populations of B. neritina-“Ca. Endobugula sertula” have yielded 20 different bryostatins (Lopanik et al., 2004; Pettit, 1996). Each of these molecules bears an identical polyketide core, but is distinguished by its pendant acyl groups at two positions in the bryolactone ring system. The pharmacological activity of bryostatin 1 has been extensively studied; it has been shown to activate protein kinase C [reviewed in (Mutter and Wills, 2000)] and exhibits anticancer activity, as well as promise against neurodegenerative disorders, including Alzheimer’s disease (Etcheberrigaray et al., 2004; Sun and Alkon, 2005). To date, the National Cancer Institute lists 38 completed, on-going or planned clinical trials using bryostatin 1 as a single or combination therapeutic, and its importance as a molecular probe to understand specific neurological disorders is also increasing.
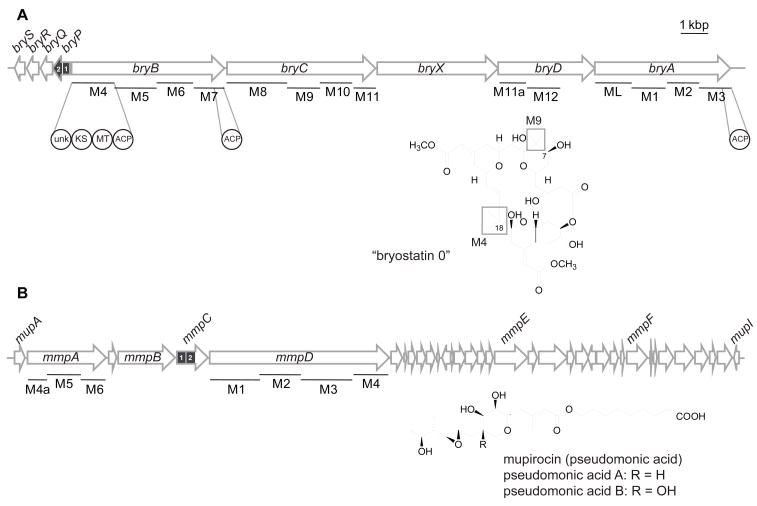
Recently, the 77 kb biosynthetic gene cluster (bry) that is putatively responsible for assembly and tailoring of the bryostatins was cloned and sequenced from two geographically distinct sibling species of “Ca. Endobugula sertula”/B. neritina (Fig. 1A) (Davidson and Haygood, 1999; Hildebrand et al., 2004; Sudek et al., 2007). The bry genes from the deep-water California (CA) sibling species are positioned on at least two loci of the chromosome, while the gene cluster from the shallow-water North Carolina (NC) sibling species appears to reside on a contiguous fragment of DNA. All evidence suggests that bry encodes the enzymes necessary for bryostatin biosynthesis, but as “Ca. Endobugula sertula” has been refractory to cultivation efforts, gene disruption and complementation have not been possible. Portions of bry have been shown by PCR to be absent in antibiotic-cured B. neritina (Davidson et al., 2001; Lopanik et al., 2006), and co-localization of 16S rRNA of “Ca. Endobugula sertula” and a fragment of the PKS genes using in situ hybridizations on B. neritina larvae (Davidson et al., 2001) provide evidence for their bacterial origin. Moreover, analysis of PKS fragments amplified from B. neritina-derived “Ca. Endobugula sertula” metagenome samples revealed that bry is the only biosynthetic locus large enough to specify assembly of bryostatin metabolites (Davidson et al., 2001).
Figure 1. Organization and orientation of biosynthetic gene clusters and discrete AT didomains.
(A) Presumed bryostatin gene cluster from shallow-water “Candidatus Endobugula sertula”-Bugula neritina populations (Sudek et al., 2007). Enzymatic domains overexpressed in this study are zoomed. Modules putatively responsible for the portions of the bryostatin precursor, “bryostatin 0”, are noted. (B) Mupirocin (pseudomonic acid) gene cluster from Pseudomonas fluorescens NCIMB 10586 (El-Sayed et al., 2003). Scale bar = 1 kbp.
Bacterial type I PKSs are large multifunctional enzymes that produce complex polyketide molecules in an assembly-line manner [reviewed in (Fischbach and Walsh, 2006)]. The group of enzymatic domains responsible for one chain elongation step is termed a module. Three enzymatic domains comprise a minimal module in PKS biosynthesis: the acyltransferase (AT), the acyl carrier protein (ACP), and the ketosynthase (KS). The AT catalyzes covalent linkage of the CoA-activated extender unit onto the ACP, and the KS domain catalyzes condensation of each extender unit, resulting in elongation of the polyketide chain by two carbons. Often, there are additional domains within a module, termed β-keto processing domains that act to modify the β–carbonyl: the ketoreductase (KR), the dehydratase (DH), and the enoyl reductase (ER). After full extension and reduction of the polyketide chain, it is released in either a cyclized or linear form, typically catalyzed by a thioesterase (TE) domain operating at the end of the pathway. Following release from the PKS, the natural product can be further modified by tailoring enzymes, such as oxygenases, methyltransferases, and glycosyltransferases [reviewed in (Rix et al., 2002)].
One significant feature of the putative bryostatin PKS is the absence of AT domains within the polypeptides. Instead, a single ORF (bryP) encoding two AT domains is present directly upstream of the PKS genes in the NC-derived gene cluster (Fig. 1A) (Sudek et al., 2007). There are several other PKS and hybrid PKS/nonribosomal peptide synthetase (NRPS) clusters that have discrete AT domains (termed “trans-AT”), including leinamycin (Cheng et al., 2003), lankacidin (Mochizuki et al., 2003), mupirocin (Fig. 1B) (El-Sayed et al., 2003), pederin (Piel, 2002), onnamide (Piel et al., 2004), rhizoxin (Partida-Martinez and Hertweck, 2007), mycosubtilin (Duitman et al., 1999), myxovirescin (Simunovic et al., 2006), virginamycin M (Pulsawat et al., 2007), difficidin (Chen et al., 2006), macrolactin (Chen et al., 2006; Schneider et al., 2007), and bacillaene (Butcher et al., 2007; Moldenhauer et al., 2007). In these AT-less modules, there are well-defined remnants of AT domains that are hypothesized to aid in docking of the trans-AT protein, although this has yet to be confirmed experimentally (Nguyen et al., 2008; Tang et al., 2004). The first in vitro experiment demonstrating that a trans-AT is able to load malonyl-CoA onto excised ACPs was performed with the discrete AT (LmnG) and ACPs from the leinamycin gene cluster (Cheng et al., 2003). Furthermore, LmnG was shown to load malonyl-CoA onto a tridomain portion of LmnJ (DH-ACP-KR). Subsequent studies have shown that discrete ATs are able to load malonyl-CoA onto cognate discrete and excised ACPs (Calderone et al., 2007; Calderone et al., 2006). It is proposed (Nguyen et al., 2008) that discrete AT domains act in trans, loading the extender unit onto the ACP of the AT-less module, but this has yet to be demonstrated in vitro with a complete native PKS module.
In addition to bry, several other PKS and PKS/NRPS gene clusters encode multiple discrete AT domains. The pederin system from the microbial symbiont of the beetle Paederus fucipes has two AT domains on two different ORFs (Piel, 2002), while the bacillaene gene cluster from two Bacillus spp. has three AT domains on three different ORFs (Butcher et al., 2007; Chen et al., 2006; Moldenhauer et al., 2007). The bryostatin gene cluster has two AT domains encoded on a single ORF (BryP), and several other gene clusters also have trans-AT didomain ORFs, including the mupirocin gene cluster from Pseudomonas fluorescens (mmpC) (El-Sayed et al., 2003), the myxovirescin gene cluster from Myxococcus xanthus (taV) (Simunovic et al., 2006), and the rhizoxin gene cluster from Burkholderia rhizoxina (rhiG) (Partida-Martinez and Hertweck, 2007). Gene disruption studies of the complete didomain were performed for both taV (Simunovic et al., 2006) and rhiG (Partida-Martinez and Hertweck, 2007), and in both cases, secondary metabolite production was completely abolished. Similarly, when the second AT domain of MmpC (AT2) was deleted from the genome of P. fluorescens, no mupirocin was detected in the supernatant of the mutant strain (El-Sayed et al., 2003). Upon complementation with a plasmid bearing mmpC, mupirocin production was restored to wild type levels, demonstrating the necessity of MmpC AT2 for mupirocin biosynthesis. Although it is clear that the trans-AT domains are required for biosynthesis, the purpose of two AT domains on a single ORF remains enigmatic.
In this paper, we explore the substrate specificity of BryP in vivo and in vitro. Mono- and didomain constructs of BryP were introduced into P. fluorescens mmpC AT1 and AT2 deletion mutants, and restoration of mupirocin production for one AT resulted. We investigated the substrate specificity in vitro by assessing the ability of the bryP-encoded AT mono- and didomains to transfer malonyl- or methylmalonyl-CoA to a variety of carrier proteins from the bryostatin PKS and other heterologous PKS and NRPS systems. We demonstrate that BryP is able to transfer malonyl-CoA onto the ACP domain of a full native module from the bryostatin PKS. Furthermore, this work represents the first example of biochemical studies on enzymes from a microbial symbiont-derived natural product biosynthetic pathway.
Results & Discussion
Sequence and phylogenetic analysis of BryP AT1 and AT2
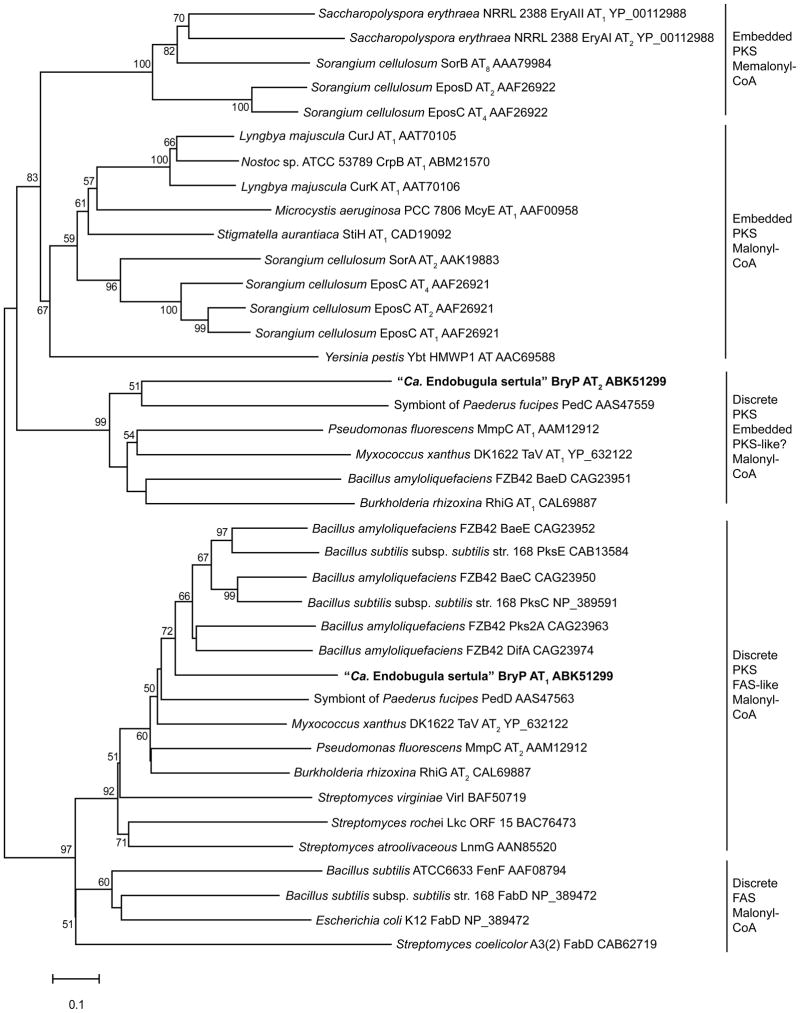
The DNA sequences of the shallow-water NC and deep-water CA AT didomain bryP are 98.5% identical, and the corresponding amino acid sequences are 96.8% identical (98.4% similar). BryP AT1 and AT2 from shallow-water NC populations share only 25.9% identity with each other at the amino acid level, but both domains contain the necessary signature sequences found in functional acyltransferases from fatty acid and polyketide synthases (Supp. Fig. 1). Notably, the N-terminal (P/S/T)GQGSQ loop that makes up one side of the active site binding cleft is present in both BryP AT1 (residues 8-13) and BryP AT2 (322-327). Additionally, the catalytic dyad (Ser-His) is present in both domains. Considerable investigation during the past few years (Petkovic et al., 2008; Reeves et al., 2001) has enabled prediction of the substrate preference for each AT domain, and an ability to correlate selectivity to key amino acid motifs. Based on multiple sequence alignments, BryP AT1 shares all characteristics of a malonyl-CoA specific acyltransferase (Supp. Fig. 1). BLAST analysis revealed that BryP AT1 is most similar to the PksC acyltransferase domain from the bacillaene gene cluster in Bacillus subtilis subsp. subtilis str. 168 (NP_389591; e-value: 8 × 10−77, 55% identity), and to BaeC from B. amyloliquefaciens FZB42 (YP_001421285; e-value: 2 × 10−76, 54% identity). BryP AT2 is most similar to an AT domain identified in the genome sequence of Clostridium cellulolyticum H10 (ZP_01574356.1; e-value: 8 × 10−56, 38% identity), to BaeD, a discrete AT from the bacillaene cluster of B. amyloliquefaciens FZB42 [CAG23951.1; e-value: 4 × 10−45, 33% identity (Chen et al., 2006)], and to PedC, a discrete AT encoded in the pederin gene cluster [AAS47559.1; e-value: 4 × 10−40, 34% identity; (Piel, 2002)]. However, due to lack of a structural model for BryP AT2 and its closest homologs, the role that specific residues lining the active site pocket play in substrate specificity is more difficult to predict.
Phylogenetic analysis of the amino acid sequences suggest that BryP AT1 is closely related to AT domains from bacterial FASs (Fig. 2). These discrete enzymes utilize malonyl-CoA as a substrate for fatty acid biosynthesis (Hopwood and Sherman, 1990). Other PKS trans-AT domains (MmpC AT2, PedD, RhiG AT2, LmnG, DifA) form a clade together with BryP AT1. Interestingly, BryP AT2 diverges from these FAS-related AT domains, and forms a clade with another group of trans-AT domains, including MmpC AT1 (El-Sayed et al., 2003), RhiG AT1 (Partida-Martinez and Hertweck, 2007), PedC (Piel, 2002), and BaeD (Chen et al., 2006) that appears to be more closely related to embedded PKS AT domains (Fig. 2). While all trans-AT PKSs that have been sequenced to date have a discrete AT domain from the former category (e.g., related to FAS AT), only a few have a discrete AT from the latter category (e.g., PKS embedded AT). Of those with two or three AT domains [e.g., bryostatin (Sudek et al., 2007), mupirocin (El-Sayed et al., 2003), bacillaene (Cheng et al., 2003), myxovirescin A1 (Simunovic et al., 2006), rhizoxin (Partida-Martinez and Hertweck, 2007), and pederin (Piel, 2002)], two AT domains are encompassed on a single ORF for most of these clusters. However, it is unclear why some PKS gene clusters contain more than one trans-AT domain. As “Ca. Endobugula sertula” remains refractory to laboratory culture, it is not possible to assess function of the two AT domains comprising bryP by traditional gene disruption and complementation assays. Instead, a related surrogate system was employed to test functional rescue of AT activity.
Figure 2. Phylogeny of discrete PKS ATs in relation to FAS and embedded PKS ATs.
Minimum evolution tree of amino acid sequences of AT domains from B. neritina-“Ca. Endobugula sertula” and related PKS and FAS gene clusters. Bootstrap analysis was performed 10,000 times, and nodes with percentages greater than 50% are labeled. Scale bar = 0.1 amino acid substitutions per site. GenBank accession numbers are listed after each sequence, and the sequences utilized in this study are in bold.
Complementation of Pseudomonas fluorescens NCIMB 10586 mmpC AT deletion mutant by BryP AT1 and BryP AT1AT2
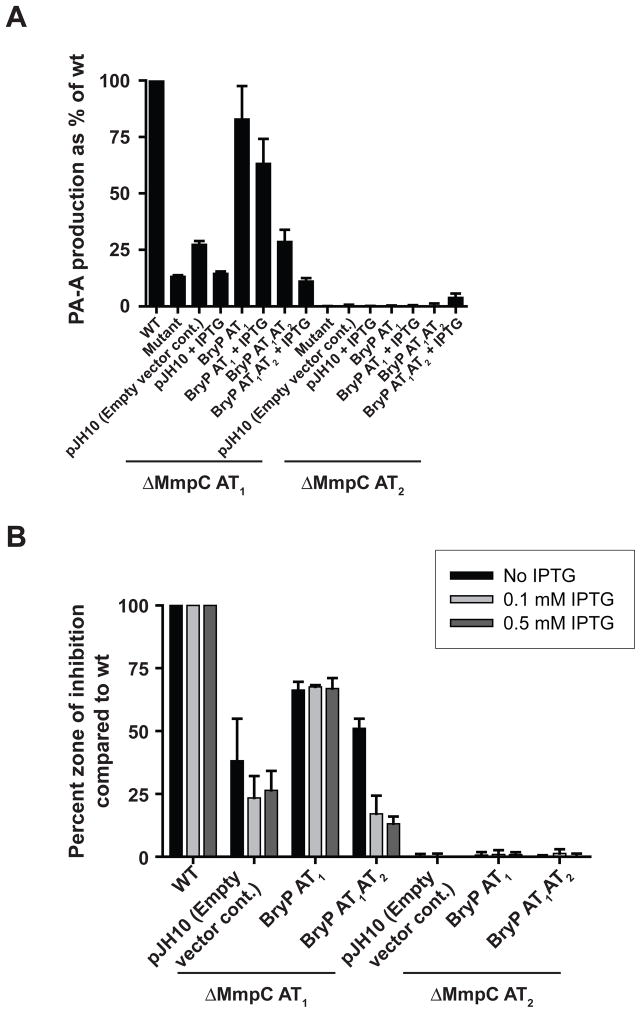
To test the hypothesis that the two bryP-encoded AT domains retain and display related functions, we assessed mutant complementation in the mupirocin biosynthetic system from Pseudomonas fluorescens [e.g., production of pseudomonic acid A (PA-A), Fig. 1B]. Of the two AT domains that comprise MmpC, MmpC AT2 is most similar to BryP AT1 phylogenetically (44.4% vs. 24.1% to BryP AT2 amino acid identity), while MmpC AT1 has a higher similarity to BryP AT2 (Fig. 2, 28.2% vs. 23.5% to BryP AT1 amino acid identity). Bioinformatic analysis has also revealed that MmpC likely contains a third C-terminal domain of unknown function. In frame deletion of mmpC-encoded AT1 results in the reduction of pseudomonic acid A (PA-A) to 13.5% of wt P. fluorescens, while no PA-A is detected in the mmpC AT2 deletion mutant (Fig. 3A). The disk-diffusion bioassays showed that antibiotic activity remaining in the MmpC ΔAT1 mutant was significant (Fig. 3B). Complementation of the ΔmmpC AT1 mutant by BryP AT1 resulted in restoration of PA-A to approximately 83% of wt levels in the HPLC assays (Fig. 3A). Similarly, in the disk-diffusion bioassays, complementation by bryP AT1 restored bioactivity to approximately 70% wt levels (controls were roughly 30% wt levels) and the addition of two different concentrations of IPTG (0.1 and 0.5 mM) did not affect the bioactivity of the extract (Fig. 3B). In contrast, expression of BryP AT1AT2 in the ΔMmpC AT1 mutant resulted in much lower production of PA-A. The addition of IPTG resulted in decreased levels of secondary metabolite production, suggesting that higher levels of BryP AT1AT2 can inhibit biosynthesis, possibly due to the formation of a non-functional protein-protein complex. However, similar results occurred when plasmid-borne mmpC AT2 was used to complement this mutant (El-Sayed et al., 2003), suggesting that intracellular levels of trans-AT may be important during biosynthesis. Interestingly, neither BryP AT1 nor BryP AT1AT2 was able to complement the ΔmmpC AT2 mutant, an in frame deletion removing only AT2 [leaving both AT1 and the third (unknown) domain of MmpC intact]. This result was surprising since BryP AT1 and MmpC AT2 cluster together in the FAS-like PKS trans-AT group. Since such AT domains clearly can retain the ability to cross-complement (but that this does not correlate with the specific phylotype), it appears that there may be no fundamental difference between the activity of the two acyltransferase domains except in the ad hoc way that they may have adapted to their genetic and biochemical context.
Figure 3. Complementation of Pseudomonas fluorescens MmpC mutants by BryP AT1 and BryP AT1AT2.
(A) HPLC detection of pseudomonic acid A (PA-A) production. Data are the mean of two experiments where PA-A in supernatants of WT and mutant bacterial cultures were compared, taking WT as 100% and expressing mutant levels relative to this. The error bars indicate differences between duplicate runs. Variations in absolute values between experiments were on average about 15%. (B) Disk diffusion assays of P. fluorescens antibiotic activity comparing mutant extracts against B. subtilis expressed as a percentage of wt activity which was taken as 100% as in (A).
In vitro substrate preference of BryP
Sequence analysis of the discrete AT domains suggests that BryP AT1 should utilize malonyl-CoA as a substrate as it has several amino acids that are thought to be involved in specificity for this precursor substrate [Met126 and Phe200 S. coelicolor FabD numbering, (Keatinge-Clay et al., 2003), Supp. Fig. 1]. BryP AT2 also has Phe200 (similar to ATs that mediate malonyl-CoA transfer), but has Leu126 and Ile56 that are typically found in AT domains selective for methylmalonyl-CoA (Keatinge-Clay et al., 2003), suggesting that it may use an alternative substrate. We therefore decided to investigate the substrate specificity of the two AT domains comprising BryP, as well as determine the ability of the AT domains to transacylate carrier proteins and complete PKS modules. AT-mediated transfer is a two-step reaction (Keatinge-Clay et al., 2003). In the first step, the extender-unit substrate (usually either malonyl-CoA or methylmalonyl-CoA) is covalently attached to the active site serine of the AT (termed “loaded”) and CoASH is released; in the second step, the extender unit undergoes transesterification from the AT active site serine to the phosphopantetheine prosthetic group of the ACP. In order to investigate the activity of BryP with PKS ACPs and modules, a series of gene constructs were cloned into Escherichia coli overexpression vectors, and the purified polypeptides (BryP, as mono- and didomain fusion proteins) employed for in vitro assays (see Supporting Information for details).
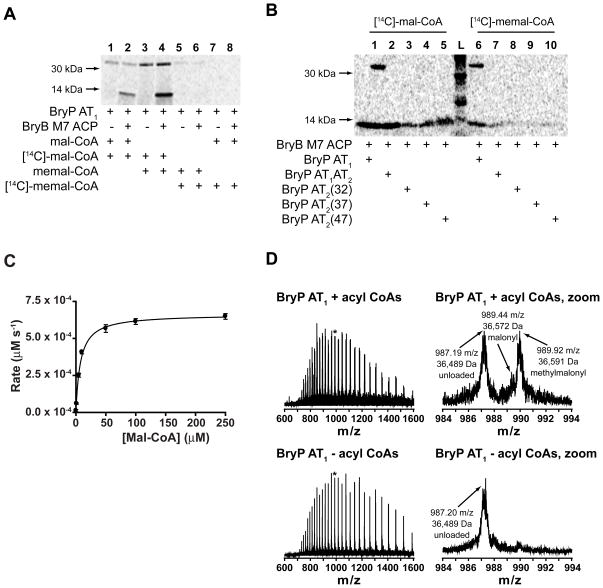
The acyl-CoA substrate loading preference of the BryP variants and their ability to transfer the substrates to an ACP from the bryostatin gene cluster (BryB M7 ACP) was assessed. BryP AT1 was able to transfer [14C]-malonyl-CoA onto holo BryB M7 ACP, but not onto the apo protein (Supp. Fig. 2). In addition no radioactivity was detected from the holo BryB M7 ACP that was incubated in the presence of [14C]-malonyl-CoA or [14C]-methylmalonyl-CoA but lacking BryP AT1, demonstrating that BryB M7 ACP is not able to self-acylate with either substrate. Substrate competition experiments were performed by incubating BryP AT1 alone or with BryB M7 ACP with a mixture of [14C]-labeled and unlabeled malonyl-CoA and methylmalonyl-CoA. The relative amount of labeling of each protein after SDS-PAGE autoradiography suggests that BryP AT1 prefers to load and transfer to BryB M7 ACP malonyl-CoA over methylmalonyl-CoA (Fig. 4A). The other BryP constructs [BryP AT1AT2 and three BryP AT2 constructs (32, 37, & 47; see Supporting Information)] were also able to transfer [14C]-malonyl- and [14C]-methylmalonyl-CoA to BryB M7 ACP, although it is evident that more malonyl-CoA is transferred than methylmalonyl-CoA in the same amount of time (20 min) (Fig. 4B). In a time course experiment, there was no apparent difference in the amount of labeling of both BryP AT1 and BryB M7 ACP, suggesting that malonate is not released (e.g., hydrolyzed) from the ACP phosphopantetheine arm after 60 min (data not shown). There was no significant difference in the activity of BryP AT1 in the range of pH tested (data not shown). In addition, there was no difference detected in the activity of BryP AT1 at pH 7.4 with and without EDTA and DTE. The kinetic parameters of BryP AT1 transferring varying concentrations of malonyl-CoA to BryB M7 ACP were determined. The Km was calculated to be 7 μM (± 0.6 SE) and kcat/Km was 0.1 μM−1 s−1 (Fig. 4C). Because of the decreased solubility of BryB M7 ACP at high concentrations, kinetic experiments that varied its concentration were not performed. The Km value of BryP AT1 with malonyl-CoA as the substrate (7 μM) is a little more than 5X lower than the value reported for FenF with MycA ACP2, but the kcat value is ~24X less than FenF (Aron et al., 2007). Both the Km and kcat values are much lower than those calculated for the S. coelicolor and E. coli FAS MAT and ACP (Szafranska et al., 2002), suggesting that although BryP AT1 has a low turnover rate, it has a high affinity for malonyl-CoA.
Figure 4. Substrate preference of BryP constructs.
(A) Autoradiography of SDS-PAGE of BryP AT1 substrate preference experiment. Lanes 1 & 2 = malonyl-CoA/[14C]-malonyl-CoA, lanes 3 & 4 = methylmalonyl-CoA/[14C]-malonyl-CoA, lanes 5 & 6 = methylmalonyl-CoA/[14C]-methylmalonyl-CoA, and lanes 7 & 8 = malonyl-CoA/[14C]-methylmalonyl-CoA. Lanes 1, 3, 5, 7 contain BryP AT1 only, and lanes 2, 4, 6, 8 contain BryP AT1 and BryB M7 ACP. (B) Autoradiography of SDS-PAGE of BryP constructs loading onto BryB M7 ACP. Lanes 1 & 6 = BryP AT1 + BryB M7 ACP, lanes 2 & 7 = BryP AT1AT2 + BryB M7 ACP, lanes 3 & 8 = BryP AT2(32) + BryB M7 ACP, lanes 4 & 7 = BryP AT2(37) + BryB M7 ACP, lanes 5 & 10 = BryP AT2(47) + BryB M7 ACP, and lane L = ladder. Lanes 1 – 5 = [14C]-malonyl-CoA and lanes 6–10 = [14C]-methylmalonyl-CoA. (C) Kinetics of BryP AT1 loading [14C]-malonyl-CoA onto loading onto BryB M7 ACP. Reactions and no enzyme controls were run in triplicate. (D) FT-ICR MS analysis (+37 charge state) of full BryP AT1 incubated with mixture of acyl-CoAs (top panels) or no substrate (bottom panels). Asterisk indicates peak that is magnified (right panels).
Fourier Transform Ion Cyclotron Mass Spectrometry (FT-ICR MS) was employed to qualitatively confirm the types of substrates loaded from a pool of acyl CoAs and to verify the predicted active-site serine of BryP AT1. Briefly, BryP AT1 was incubated with an equimolar mixture of malonyl-, methylmalonyl-, acetyl-, and propionyl-CoA, and analyzed intact by FT-ICR MS. Three peaks were detected in the repeating isotope pattern of the loaded BryP AT1 (Fig. 4D, top panel). For clarity, the mass shift in the +37 charge state is used in the following discussion, although similar shifts in mass were apparent for all charge states observed. Unloaded BryP AT1 at the expected mass was observed in the sample, as well as BryP AT1 with a shift in m/z of 2.25 or 83.2 Da likely due to the addition of malonyl-CoA. Additionally, a shift in m/z of 2.73 in the +37 charge state was observed, correlating to the addition of 101 Daltons, and the loading of methylmalonyl-CoA. The control reaction resulted in only unloaded BryP AT1 (Fig. 4D bottom panel). In a subsequent experiment, BryP AT1 was subjected to proteolytic digestion prior to FT-ICR MS analysis to ensure that loading occurs on the predicted active site serine residue. A Glu-C derived peptide, SHKPSYVAGHSLGE, was identified by MS/MS in the malonyl- and methylmalonyl-loaded forms. A partial tryptic peptide, TQFTQPALYIINALSFLDKIELESHKPSYVAGHSLGEYNALFAAGAFDFLTGLK, was also identified by MS/MS in the malonyl- and methylmalonyl-loaded forms. The predicted active site serine (bold) was determined to be the modified residue in both cases.
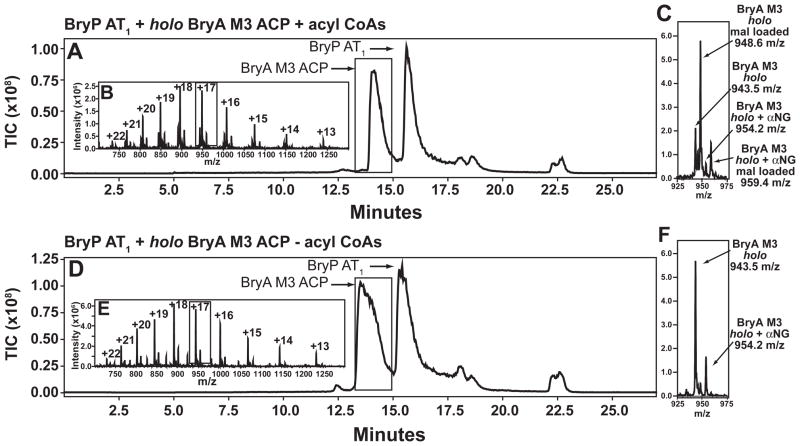
LC/MS was used to investigate acyl group transfer from BryP AT1 to another Bry ACP, BryA M3 ACP (Fig. 1A). Both proteins were incubated together in the presence or absence (control) of equimolar amounts of acetyl-, malonyl-, methylmalonyl-, and propionyl-CoA. Experiments in the absence of BryP AT1 showed that the holo ACP did not self-load to an appreciable degree (data not shown). In the presence of BryP AT1 and the acyl-CoAs, BryA M3 ACP peaks increased in mass by 5.1 m/z in the +17 charge state, an 86.7 Da mass increase consistent with a preference for malonyl-CoA (Fig. 5). An additional peak with a mass shift of 179 Da is observed in the LC-MS data; this is likely α-N-6-phosphogluconoylation, a common post-translational modification observed on fusion proteins with a 6X His affinity tag (Geoghegan et al., 1999). This additional peak exhibits the same mass shift and preference as the holo ACP.
Figure 5. LC/MS analysis of BryP AT1 transfer of acyl-CoAs to holo BryA M3 ACP.
Reactions with (A–C) and without (D–F) acyl-CoAs shown. (A, D) Reversed-phase HPLC TIC chromatogram. (B, E) Zoom of full spectrum. (C, F) Zoom on single charge state with assignments. mal = malonyl, αNG = α-N-6-phosphogluconoylation.
Taken together, these data suggest that BryP AT1 can load both malonyl- and methylmalonyl-CoA (Fig. 4), but that malonate is preferentially transferred to the Bry ACPs. The preference for malonyl-CoA over methylmalonyl-CoA has been demonstrated for other trans-AT gene clusters. In vitro assays with the myxovirescin gene cluster from Myxococcus xanthus DK1622 trans-AT didomain (TaV) (Simunovic et al., 2006) have shown that TaV AT2 [which is a FAS-like trans-AT (Fig. 2)], prefers malonyl-CoA over methylmalonyl-CoA, acetyl-CoA, and propionyl-CoA (Calderone et al., 2007). In addition, TaV AT2 was able to acylate both discrete and embedded ACP domains from the gene cluster (Calderone et al., 2007). No in vitro assays were performed on TaV AT1 (Calderone et al., 2007), which is more similar to BryP AT2 (Fig. 2). The trans-AT associated with the bacillaene cluster in B. subtilis (PksC) (Butcher et al., 2007), which forms a clade with BryP AT1 (Fig. 2), was also shown to preferentially load malonyl-CoA over acetyl-CoA and methylmalonyl-CoA, and transfer malonyl-CoA onto a cognate discrete ACP (AcpK) (Calderone et al., 2006).
BryP exhibits flexibility for transferring malonate onto ACPs
The mono- and didomain constructs of BryP were assayed for their ability to transfer substrates onto an ACP from another PKS [pikromycin PikAIII M5 ACP (Xue et al., 1998)], and a peptidyl carrier protein (PCP) from an aminocoumarin (clorobiocin) biosynthetic gene cluster (Pojer et al., 2002). BryP AT1 transferred [14C]-malonyl-CoA onto a variety of carrier proteins, including the BryB M7, PikAIII M5 ACPs, as well as the holo (but not the apo) forms of CloN5 (Supp. Fig. 2). The BryP didomain (BryP AT1AT2) and the didomain single mutants BryP AT1°AT2 and BryP AT1AT2° were able to catalyze loading of each of the carrier proteins presented. The double mutant BryP AT1°AT2°, as expected, did not catalyze acyl transfer to any of the carrier proteins. One BryP AT2 construct (37) was also able to catalyze transfer of malonyl-CoA to each of the carrier proteins (data not shown). The flexibility of BryP in transferring malonate to ACPs from two PKS gene clusters (bryostatin and pikromycin) was similar to that of FenF, a discrete FAS-like AT in the mycosubtilin gene cluster (Duitman et al., 1999). FenF did not exhibit significant preference in vitro for the substrate (malonyl-CoA) ACP compared to the loading (palmitoyl-CoA) ACP in MycA (Aron et al., 2007). Conversely, while BryP was also able to transfer malonate onto a PCP from the clorobiocin biosynthetic gene (Supp. Fig. 3), FenF was eight times less efficient loading malonyl-CoA onto one of the peptidyl carrier protein (PCP) domains in MycA (Aron et al., 2007). Moreover, the discrete AT (LnmG) in the leinamycin gene cluster was not able to load malonyl-CoA onto an excised PCP from that PKS/NRPS gene cluster (Cheng et al., 2003), suggesting that either the PCP domain is unable to bind to malonyl-CoA, or that the discrete ATs are unable to recognize or interact with the PCP. As both the hybrid PKS/NRPS leinamycin and mycosubtilin gene clusters contain ACP and PCP domains, it would be advantageous for the discrete ATs to discriminate between the two types of carrier proteins to avoid biosynthetic derailment by acylating the incorrect thiolation domain. However, because the bryostatin gene cluster is only composed of PKS modules BryP may not have evolved to selectively interact with ACP versus PCP domains.
BryP-catalyzed acyl transfer onto PKS modules from the pikromycin, erythromycin, and bryostatin gene clusters
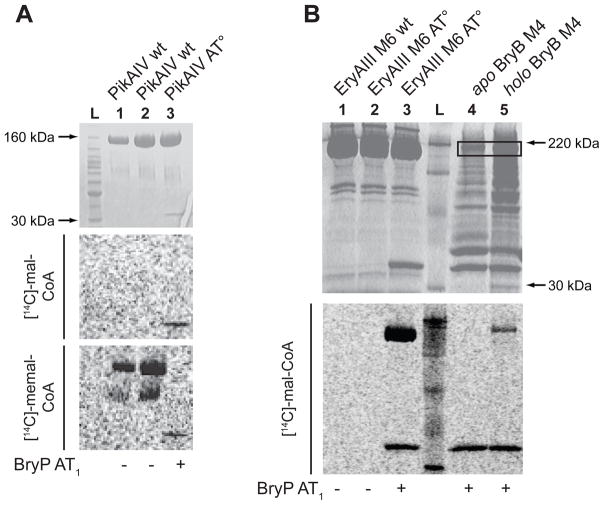
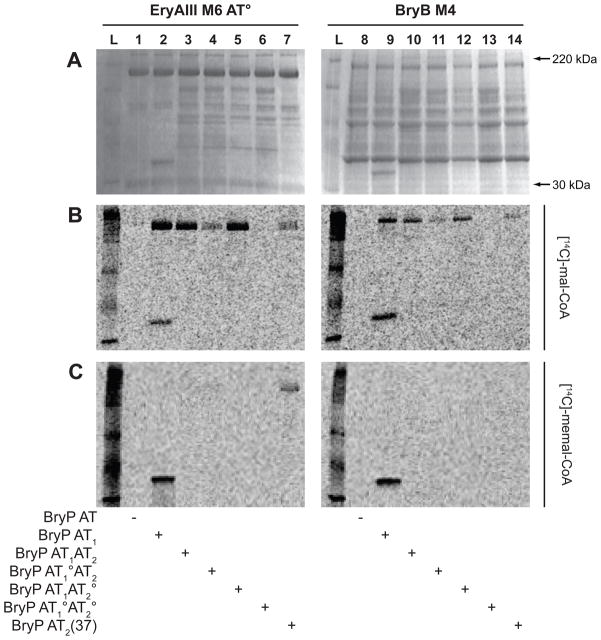
A previous study demonstrated the ability of the S. coelicolor FAS MAT to catalyze transfer of malonyl-CoA onto the ACP of an EryAIII module 6 AT° mutant (Kumar et al., 2003). We decided to extend this type of trans-AT substrate delivery analysis using BryP toward a series of PKS modules, including PikAIV (Pik module 6), EryAIII M6 and BryB M4. BryP AT1 was not able to load malonyl- or methylmalonyl-CoA onto the PikAIV M6 AT° module (Fig. 6A) whose native substrate is methylmalonyl-CoA (Xue et al., 1998). The wt PikAIV M6 was able to self-load and transfer methylmalonyl-CoA, but not malonyl-CoA. Similarly, wt EryAIII M6 was able to self-load methylmalonyl-CoA [its native substrate (Donadio et al., 1991)] (data not shown), but not malonyl-CoA (Fig. 6B). In contrast to the PikAIV M6 AT°, BryP AT1 was able to effectively transfer malonyl-CoA onto EryAIII M6 AT° (Fig. 6). Most significantly, BryP AT1 was able to transfer [14C]-malonyl-CoA onto the holo form of the native module BryB M4, but was unable to do so onto the apo preparation of BryB M4 (Fig. 6B). The BryP AT didomain was able to transfer malonyl-CoA onto both EryAIII M6 AT° and the native module BryB M4 (Fig. 7A, B). Both single mutant BryP AT didomain proteins were able to load malonyl-CoA onto these individual modules, although based on relative signal strengths of the BryP AT1AT2 point mutants, it appears that AT1 is more active than AT2 (Fig. 7B, lanes 4, 5, 11, 12). The BryP AT double mutant was not able to transfer malonyl-CoA onto the module. Neither BryP AT1 nor the didomain was able to transfer methylmalonyl-CoA onto either EryAIII M6 AT° or the native BryB M4 module (Fig. 7C). Interestingly, BryP AT2(37) was able to transfer methylmalonyl-CoA onto the EryAIII M6 AT° module, but was unreactive toward BryB M4 (Fig. 7C, lane 7). In vivo, these trans-ATs are hypothesized to acylate ACP domains that are part of a multidomain module. While the mono- and didomain constructs of BryP were able to transfer malonyl-CoA onto modules BryB M4 and EryAIII M6 AT° (Fig. 6B, 7B), BryP AT1 was unable to transfer malonyl-CoA onto PikAIV AT° (Fig. 6A). These data suggest that there may be some specificity regarding the interaction between the trans-AT and the module.
Figure 6. Autoradiography of SDS-PAGE of PKS module loading by BryP AT1.
(A) Loading of PikAIV M6 wt and AT°. Lanes 1 & 2 = PikAIV M6 wt + BryP AT1, lane 3 = PikAIV M6 AT° + BryP AT1, and lane L = ladder. Substrate for middle panel is [14C]-malonyl-CoA, and substrate for lower panel is [14C]-methylmalonyl-CoA. (B) Loading of EryAIII M6 wt and AT°, and BryB M4 with [14C]-malonyl-CoA by BryP AT1. Lane 1 = EryAIII M6 wt + no BryP AT1, lane 2 = EryAIII M6 AT° + no BryP AT1, lane 3 = EryAIII M6 AT° + BryP AT1, lane L = ladder, lane 4 = apo BryB M4 + BryP AT1, and lane 5 = BryB M4 + BryP AT1.
Figure 7. Loading of substrates onto PKS modules by BryP.
(A) Coomassie brilliant blue stained gels, and autoradiography with (B) [14C]-malonyl-CoA and (C) [14C]-methylmalonyl-CoA substrates. Lane L = ladder, lanes 1–7 = EryAIII M6 AT°, lanes 8–14 = BryB M4. Lanes 1 & 8 = no BryP, lanes 2 & 9 = BryP AT1, lanes 3 & 10 = BryP AT1AT2, lanes 4 & 11 = BryP AT1°AT2, lanes 5 & 12 = BryP AT1AT2°, lanes 6 & 13 = BryP AT1°AT2°, and lanes 7 & 14 = BryP AT2(37).
All of the bryostatins characterized to date have two gem-dimethyl groups (at C8 and C18, Fig. 1). The modules that are putatively responsible for the assembly of those regions contain methyltransferase domains (M4 and M9) (Sudek et al., 2007) that could add either one or two methyl groups to the α-carbon. We were motivated to investigate the substrate preference of the two AT domains of BryP as they could possibly load different substrates (malonyl-CoA or methylmalonyl-CoA) onto the ACPs from these modules, which could then be methylated either once or twice by the embedded MT enzymatic domains resulting in the geminal dimethylated carbon atoms. Two other natural products that have gem-C-dimethyl groups are the mixed PKS/NRPS compounds epothilone (Molnar et al., 2000) and yersiniabactin (Gehring et al., 1998), and both have embedded MT domains within the PKS module that is responsible for the elongation and modification of the corresponding portion of the molecule. The biosynthetic origins for the methyl groups have been investigated, and interestingly, two different mechanisms have been described. In yersiniabactin biosynthesis, data from in vivo feeding studies (Gehring et al., 1998), and in vitro assays with purified enzymes (Miller et al., 2002) suggest that the polyketide chain is elongated by malonyl-CoA and both methyl groups originate from S-adenosyl-methionine. However, in epothilone biosynthesis, several lines of evidence including feeding studies (Gerth et al., 2000) and bioinformatic analysis of the AT domain substrate preference (Molnar et al., 2000; Petkovic et al., 2008) support the hypothesis that methylmalonyl-CoA is incorporated into the polyketide chain by the embedded AT, followed by C-methylation on the α-carbon. One of the Bry MT-containing modules was targeted (BryB M4) (Sudek et al., 2007) in efforts to determine the likely mode of methylation. When methylmalonyl-CoA was tested as a substrate, the single BryP AT2 domain was able to label only EryAIII M6 AT°, whose natural substrate is methylmalonyl-CoA (Donadio et al., 1991). As neither the BryP didomain nor the BryP AT1°AT2 mutant was able to acylate EryAIII M6 AT° with methylmalonyl-CoA, having the additional domain (AT1) may interfere with the ability of AT2 to load the module with methylmalonyl-CoA. However, since BryP was not able to load methylmalonyl-CoA onto BryB M4, it seems more likely that the MT adds two methyl groups as in yersiniabactin biosynthesis. The di-methylation at the C-8 and C-18 positions on the growing bryostatin chain elongation intermediate is consistent with the general malonyl-CoA selectivity of all known trans-AT domains investigated from various Gram-positive, Gram-negative, and microbial symbiont natural product pathways (Nguyen et al., 2008). This is also consistent with the lack of a methylmalonyl-CoA precursor pool in all knownγ–proteobacteria that includes the uncultured bacterial symbiont “Candidatus Endobugula sertula” that is responsible for the biosynthesis of the bryostatin anticancer natural products.
Significance
In this study, we demonstrate that BryP exhibits flexibility when loading malonyl- or methylmalonyl-CoA, but prefers to transfer malonate to excised and modular ACPs. Furthermore, BryP is able to transfer the predicted extender unit, malonyl-CoA, onto a complete native module from the presumed bryostatin biosynthetic gene cluster. Results from this investigation suggest that the geminal dimethyl groups at C-8 and C-18 on the bryolactone ring system are derived from double C-methylation of a malonate extender unit during chain elongation. While several large PKS gene clusters from uncultivated symbiotic microbes have been cloned and sequenced (Partida-Martinez and Hertweck, 2007; Piel, 2002; Piel et al., 2004), this is the first instance that a large portion of a PKS gene cluster from an uncultivated symbiotic microbe has been overexpressed and demonstrated to function with cognate enzymatic domains. In vivo assays with P. fluorescens mupirocin ΔAT mutants showed that BryP is only able to complement one of the AT domain mutant strains. A number of questions remain to be explored about how these domains physically interact with PKS modules and full PKS polypeptides and megacomplexes. Finally, this is the first step towards demonstrating unequivocally that the assigned Bry metabolic system produces the bryostatins.
Experimental Procedures
Sequence and phylogenetic analysis of the AT domains
Shallow and deep bryP sequences (Sudek et al., 2007) were analyzed by BLAST (Altschul et al., 1990). The amino acid sequences were aligned with the sequences of other trans-AT domains as well as integrated AT domains from PKS gene clusters using ClustalX (Thompson et al., 1997). A minimum evolution phylogenetic tree was generated using MEGA version 4 (Tamura et al., 2007). Subjecting the alignment to alternative algorithms (Neighbor Joining and Maximum Parsimony) resulted in similar topology.
Generation of mmpC AT in-frame deletion mutants in the mupirocin producer, Pseudomonas fluorescens NCIMB10856
Construction of an in-frame deletion in MmpC AT2 has been previously described (El-Sayed et al., 2003). Deletion of MmpC AT1 was carried out with suicide plasmid pJHAT101 that contains an mmpC deletion of 907 bp (23214 – 24120 bp inclusive, database numbering). This creates a frame shift in the remaining MmpC AT1 sequence such that a 24 codon ORF will deliver ribosomes to the ribosome binding site and ATG start codon at the beginning of the AT2 domain of mmpC. Production of pseudomonic acids by this mutant was verified, suggesting that MmpC AT2 was still functional.
Construction of P. fluorescens expression plasmids
For in trans expression of bryostatin acyltransferases in P. fluorescens AT deletion strains, domains were cloned into the IncQ vector pJH10 under the control of the tac promoter (El-Sayed et al., 2003). BryP AT1 was restricted from pNL020 (Supp. Table 1) and ligated into pJH10 to give pJS261. The AT didomain (BryP AT1AT2) was PCR-amplified from a fosmid subclone (MM5_2_BO7) using primers BryP_AT1_F and BryP_AT2_R (Supp. Table 2), and cloned into pJH10 to give pJS262. Inserts were verified by sequencing. Complementation plasmids were introduced into the MmpC mutant strains of P. fluorescens by conjugation with plasmid-bearing E. coli S17-1 as described previously (Hothersall et al., 2007). Positive colonies were screened for metabolite production. Antibiotic disk-diffusion bioassays using B. subtilis cultures were used to screen for mupirocin, which is a mixture of pseudomonic acids (PA), dominated by PA-A (90%). Direct quantification of PA-A was performed by HPLC analysis of strain extracts. Both experiments were run in duplicate.
In vitro substrate preference of BryP
Radioactive assays of BryP activity
To determine the ability of BryP to acylate various carrier proteins and PKS modules, each was individually incubated with the BryP proteins and radiolabeled substrate (Cloning and overexpression details in Supplemental Information). In general, the reactions were run in 50 mM HEPES buffer, pH 7.4, 5–10 μM carrier protein or module protein, 1–8 μM BryP protein, and 0.4–0.5 mM substrate. The enzymes were equilibrated at RT for 5 min before the substrate was added. Reactions proceeded for 5–20 min at RT, and were quenched by the addition of 2X SDS-PAGE loading buffer with no reducing agent. The samples were mixed and loaded onto SDS-PAGE gels. After electrophoresis, the gels were stained with either Coomassie blue or SimplyBlue (Invitrogen) stains, and destained. The gels were dried (Bio-Rad), and placed into a phosphoimager cassette (Amersham Biosciences). After 5 days, the phosphoimager screen was scanned, and the images were analyzed.
To determine the substrate preference of BryP AT1, radiolabeled and unlabeled malonyl-and methylmalonyl-CoA were mixed in separate reactions. [14C]-malonyl- and [14C]-methylmalonyl-CoA (55 mCi/mmol, ARC, Inc.) were mixed with unlabeled malonyl- and methylmalonyl-CoA to result in 0.5 mM total concentration of the substrates. After a 5 min incubation at RT, the amount of labeling on BryP AT1 (2 μM) and BryB M7 ACP (10 μM) was visualized by SDS-PAGE autoradiography. The BryP AT didomain (BryP AT1AT2) and three BryP AT2 constructs [AT2(32), AT2(37), AT2(47)] were assayed for the ability to transfer [14C]-malonyl-CoA and [14C]-methylmalonyl-CoA (0.5 mM) to BryB M7 ACP (20 μM). As the BryP didomain and BryP AT2 constructs could not be purified to homogeneity, their concentration could not be determined accurately. Six μL of the eluted protein from the Ni-NTA column was used in each reaction; BryP AT1 was added to a final concentration of 4 μM. The reaction was quenched after 20 min. In a time course experiment, 7.5 μM of BryB M7 ACP was incubated with 1 μM BryP AT1 and 0.4 mM [14C]-malonyl-CoA, and the reactions were quenched after 5, 10, 15, 30, and 60 min.
A protein precipitation assay was used to quantify the activity of BryP AT1 in the presence of BryB M7 ACP. The activity of BryP AT1 was determined in HEPES buffer (50 mM) with different pH values ranging from 6.0 to 9.2. These assays were conducted by incubating BryP AT1 (1 nM) with [14C]-malonyl-CoA (0.1 mM) and BryB M7 ACP (50 μM), and measuring the amount of radiolabel bound to BryB M7 ACP after acid precipitation and scintillation counting based on the method previously described (Koppisch and Khosla, 2003). EDTA and DTE (2 mM each) was added to each reaction; one duplicate set of reactions at pH 7.4 was run without EDTA or DTE to determine if they affected BryP AT1 activity. The reactions and no enzyme controls (BSA 10 mg/mL) were run in duplicate. The reaction mixture was equilibrated at RT for 5 min before the enzyme (or BSA) was added, the reaction proceeded at RT for 5 min. The amount of radiolabel incorporation vs. background was compared among the duplicate reactions at the different pHs, and between the pH 7.4 samples run with and without DTE and EDTA.
The reaction kinetics of BryP AT1 with malonyl-CoA and purified BryB M7 ACP were determined using the TCA precipitation method at pH 7.4. The final concentration of BryP AT1 was 1 nM, and the final concentration of BryB M7 ACP was 50 μM. The concentration of [14C]-malonyl-CoA varied from 0.2 to 250 μM. The loading reaction proceeded for 5 min on ice, after which the reaction was quenched. The reaction and controls (1 nM BSA) were performed in triplicate. Kinetic parameters were not obtained for varying concentrations of BryB M7 ACP as this protein would precipitate out of solution at high concentrations.
FT-ICR MS analysis of BryP AT1
BryP AT1 (5 μM) was reacted with a pool of 250 μM acetyl-, malonyl-, methylmalonyl-, and propionyl-CoA in 50 mM HEPES pH 7 buffer and 1 mM TCEP. After incubation for 30 min, samples were acidified with 1% formic acid. Intact protein samples were desalted with Handee Microspin columns (Pierce) packed with 20 μL of 300Å polymeric C4 resin (Vydac). Samples were loaded onto the columns and washed with 30 column volumes of 0.1% formic acid prior to elution with 10 column volumes of 50% acetonitrile + 0.1% formic acid. Intact protein samples were analyzed by an FT-ICR MS (APEX-Q with Apollo II ion source and actively shielded 7T magnet, Bruker Daltonics). Data was gathered from m/z 250–2000 utilizing direct infusion electrospray ionization in positive ion mode. Electrospray was conducted at 3,600 volts with 24–60 scans per spectra utilizing 0.5s external ion accumulation in the hexapole prior to analysis in the FT-ICR using a loop value of 15. All CID MS/MS was performed external to the FT-ICR cell with quadrapole mass selection. In order to confirm that the modification occurred on the active site serine, BryP AT1 was then subjected to proteolytic digestion after reaction with the acyl CoA pool and FT-ICR MS analysis. Glu-C (Worthington Biochemical) or trypsin (Promega) (1:100/w:w) was added to the samples, and digestion proceeded for 4 hrs at 37°C. Proteolytic peptides were desalted as above with the exception that C18 resin was utilized. FT-ICR MS analysis was performed as above except that the loop value was 4 and the ion accumulation time was 1s.
LC-MS Analysis of BryA M3 ACP
BryP AT1 (2 μM), and BryP AT1 (20 μM) were reacted with a pool of 250 μM acetyl-, malonyl-, methylmalonyl-, and propionyl-CoA. Reactions were incubated for 45 minutes in 50 mM HEPES pH 7 and 1 mM TCEP, and were quenched by diluting 2X in 6 M urea, 100 mM HEPES, pH 6. Ten μL of this mixture was analyzed by LC-MS with a Shimadzu LCMS-2010EV (Columbia, MD) after separation on a PLRP-S 2 × 50 mm 4000A 8 μm polymeric RP-HPLC column (Varian, Palo Alto, CA) heated to 50°C. Samples were desalted online for 5 min with 95% Buffer A (98.9% water, 1% acetonitrile, 0.1% formic acid) and 5% Buffer B (1% water, 98.9% acetonitrile, and 0.1% formic acid) followed by gradient elution over 20 min. Profile mode data was gathered from m/z 400–2000 utilizing electrospray ionization in positive ion mode.
BryP transfer to other ACPs
The ability of BryP AT1 to acylate a variety of carrier proteins was assayed by in vitro incubation of the proteins with radiolabeled substrates followed by SDS-PAGE autoradiography as described previously. Purified BryP AT1 (2 μM) was incubated with both apo and holo BryB M7 ACP (10 μM), 0.4 mM [14C]-malonyl-CoA and [14C]-methylmalonyl CoA for 5 min at RT. The ability of the other BryP constructs (BryP AT1AT2, BryP AT2(37), and site mutants BryP AT1°AT2, AT1AT2°, AT1°AT2°) to load [14C]-malonyl-CoA onto a variety of carrier proteins was assessed in a similar fashion. The carrier proteins used were BryA M3 ACP (apo and holo), PikAIII M5 ACP, and CloN5 (apo and holo). Each purified carrier protein preparation (20 μM) was incubated with the BryP protein and [14C]-malonyl-CoA (0.2 mM) for 10 min at RT. The BryP concentrations added were 4 μM BryP AT1, 0.9 mg/mL BryP didomains, and 0.6 mg/mL of BryP AT2(37). The reaction proceeded for 10 min at RT.
BryP loading onto modules from the pikromycin, erythromycin, and bryostatin gene clusters
Wt PikAIV M6 and the AT mutant (PikAIV M6 AT°) were incubated with BryP AT1 and [14C]-malonyl-CoA or [14C]-methylmalonyl-CoA. Wt PikAIV was tested at 5 and 10 μM, and PikAIV AT° was tested at 10 μM. BryP AT1 (2 μM) was added, and after equilibration, either radiolabeled substrate was added (0.4 mM). After 5 min at RT, the reaction was quenched. Next, BryP AT1 was assayed with malonyl-CoA and EryAIII M6 AT° or BryB M4. BryP AT1 (4μM) and [14C]-malonyl-CoA (0.5 mM) were incubated with EryAIII M6 AT° (10 μM) for 10 min at RT. The amylose resin elutions of BryB M4 were used without further purification; the pooled fractions were assayed at a final concentration of 2.2 mg/mL total (holo BryB M4) or 1.1 mg/mL (apo BryB M4). The other BryP constructs were tested for their ability to load both malonyl- and methylmalonyl-CoA onto the EryAIII M6 AT° and BryB M4 modules. The final concentration of EryAIII M6 AT° was 5 μM, BryP AT1 was 4 μM, and substrate was 0.5 mM. The final concentration of the BryB M4 preparation was 3.3 mg/ml, the BryP AT didomain preps were 0.9 mg/mL, and BryP AT2(37) was 0.6 mg/mL. The reactions were quenched after 15 min at RT.
Supplementary Material
Acknowledgments
We would like to thank Niels Lindquist for providing sampling and laboratory support, and W. Clay Brown and Jim DelProposto at the LSI High Throughput Protein core facility for generating and screening the BryP didomain and AT2 overexpression constructs, and for supplying the pRARE-CDF coexpression plasmid. We would like to thank Jeff Kittendorf for cloning the EryAIII M6 construct, Brian Beck for cloning the PikAIV M6 wt and mutant constructs, and Sabine Grüschow for cloning the sfp coexpression vector, pSG701. Sylvie Garneau-Tsodikova generously provided purified apo and holo preparations of CloN5. N.B.L was supported by a National Institutes of Health NRSA fellowship (5F32CA110636). Research support was generously provided by NIH R01 GM076477 to D.H.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aron ZD, Fortin PD, Calderone CT, Walsh CT. FenF: Servicing the mycosubtilin synthetase assembly line in trans. ChemBioChem. 2007;8:613–616. doi: 10.1002/cbic.200600575. [DOI] [PubMed] [Google Scholar]
- Butcher RA, Schroeder FC, Fischbach MA, Straightt PD, Kolter R, Walsh CT, Clardy J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc Nat Acad Sci USA. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone CT, Iwig DF, Dorrestein PC, Kelleher NL, Walsh CT. Incorporation of nonmethyl branches by isoprenoid-like logic: Multiple beta-alkylation events in the biosynthesis of myxovirescin A1. Chem Biol. 2007;14:835–846. doi: 10.1016/j.chembiol.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Convergence of isoprene and polyketide biosynthetic machinery: Isoprenyl-S-carrier proteins in the pksX pathway of Bacillus subtilis. Proc Nat Acad Sci USA. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XH, Vater J, Piel J, Franke P, Scholz R, Schneider K, Koumoutsi A, Hitzeroth G, Grammel N, Strittmatter AW, et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol. 2006;188:4024–4036. doi: 10.1128/JB.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YQ, Tang GL, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Nat Acad Sci USA. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG. Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl Environ Microbiol. 2001;67:4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SK, Haygood MG. Identification of sibling species of the bryozoan Bugula neritina that produce different anticancer bryostatins and harbor distinct strains of the bacterial symbiont “Candidatus Endobugula sertula”. Biol Bull. 1999;196:273–280. doi: 10.2307/1542952. [DOI] [PubMed] [Google Scholar]
- Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular Organization of Genes Required for Complex Polyketide Biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- Duitman EH, Hamoen LW, Rembold M, Venema G, Seitz H, Saenger W, Bernhard F, Reinhardt R, Schmidt M, Ullrich C, et al. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: A multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc Nat Acad Sci USA. 1999;96:13294–13299. doi: 10.1073/pnas.96.23.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed AK, Hothersall J, Cooper SM, Stephens E, Simpson TJ, Thomas CM. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem Biol. 2003;10:419–430. doi: 10.1016/s1074-5521(03)00091-7. [DOI] [PubMed] [Google Scholar]
- Etcheberrigaray R, Tan M, Dewachter I, Kuiperi C, Van der Auwera I, Wera S, Qiao LX, Bank B, Nelson TJ, Kozikowski AP, et al. Therapeutic effects of PKC activators in Alzheimer’s disease transgenic mice. Proc Nat Acad Sci USA. 2004;101:11141–11146. doi: 10.1073/pnas.0403921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, Blattner FR, Walsh CT, Perry RD. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- Geoghegan KF, Dixon HBF, Rosner PJ, Hoth LR, Lanzetti AJ, Borzilleri KA, Marr ES, Pezzullo LH, Martin LB, LeMotte PK, et al. Spontaneous α-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: The cause of extra mass of 258 or 178 Da in fusion proteins. Anal Biochem. 1999;267:169–184. doi: 10.1006/abio.1998.2990. [DOI] [PubMed] [Google Scholar]
- Gerth K, Steinmetz H, Hofle G, Reichenbach H. Studies on the biosynthesis of epothilones: The biosynthetic origin of the carbon skeleton. Journal of Antibiotics. 2000;53:1373–1377. doi: 10.7164/antibiotics.53.1373. [DOI] [PubMed] [Google Scholar]
- Hildebrand M, Waggoner LE, Liu HB, Sudek S, Allen S, Anderson C, Sherman DH, Haygood M. bryA: An unusual modular polyketide synthase gene from the uncultivated bacterial symbiont of the marine bryozoan Bugula neritina. Chem Biol. 2004;11:1543–1552. doi: 10.1016/j.chembiol.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Sherman DH. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- Hothersall J, Wu J, Rahman AS, Shields JA, Haddock J, Johnson N, Cooper SM, Stephens ER, Cox RJ, Crosby J, et al. Mutational analysis reveals that all tailoring region genes are required for production of polyketide antibiotic mupirocin by Pseudomonas fluorescens - Pseudomonic acid B biosynthesis precedes pseudomonic acid A. J. Biol. Chem. 2007;282:15451–15461. doi: 10.1074/jbc.M701490200. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Shelat AA, Savage DF, Tsai SC, Miercke LJW, O’Connell JD, Khosla C, Stroud RM. Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure. 2003;11:147–154. doi: 10.1016/s0969-2126(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Konig GM, Kehraus S, Seibert SF, Abdel-Lateff A, Muller D. Natural products from marine organisms and their associated microbes. ChemBioChem. 2006;7:229–238. doi: 10.1002/cbic.200500087. [DOI] [PubMed] [Google Scholar]
- Koppisch AT, Khosla C. Structure-based mutagenesis of the malonyl-CoA:Acyl carrier protein transacylase from Streptomyces coelicolor. Biochemistry. 2003;42:11057–11064. doi: 10.1021/bi0349672. [DOI] [PubMed] [Google Scholar]
- Kumar P, Koppisch AT, Cane DE, Khosla C. Enhancing the modularity of the modular polyketide synthases: Transacylation in modular polyketide synthases catalyzed by malonyl-CoA:ACP transacylase. J Am Chem Soc. 2003;125:14307–14312. doi: 10.1021/ja037429l. [DOI] [PubMed] [Google Scholar]
- Lopanik N, Gustafson KR, Lindquist N. Structure of bryostatin 20: A symbiont-produced chemical defense for larvae of the host bryozoan, Bugula neritina. J Nat Prod. 2004;67:1412–1414. doi: 10.1021/np040007k. [DOI] [PubMed] [Google Scholar]
- Lopanik N, Lindquist N, Targett N. Potent cytotoxins produced by a microbial symbiont protect host larvae from predation. Oecologia. 2004;139:131–139. doi: 10.1007/s00442-004-1487-5. [DOI] [PubMed] [Google Scholar]
- Lopanik NB, Targett NM, Lindquist N. Isolation of two polyketide synthase gene fragments from the uncultured microbial symbiont of the marine bryozoan Bugula neritina. Appl Environ Microbiol. 2006;72:7941–7944. doi: 10.1128/AEM.01277-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopanik NB, Targett NM, Lindquist N. Ontogeny of a symbiont-produced chemical defense in Bugula neritina (Bryozoa) Mar Ecol Prog Ser. 2006;327:183–191. [Google Scholar]
- Miller DA, Luo LS, Hillson N, Keating TA, Walsh CT. Yersiniabactin synthetase: A four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- Mochizuki S, Hiratsu K, Suwa M, Ishii T, Sugino F, Yamada K, Kinashi H. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol Microbiol. 2003;48:1501–1510. doi: 10.1046/j.1365-2958.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- Moldenhauer J, Chen XH, Borriss R, Piel J. Biosynthesis of the antibiotic bacillaene, the product of a giant polyketide synthase complex of the trans-AT family. Angew Chem Internat Ed. 2007;46:8195–8197. doi: 10.1002/anie.200703386. [DOI] [PubMed] [Google Scholar]
- Molnar I, Schupp T, Ono M, Zirkle RE, Milnamow M, Nowak-Thompson B, Engel N, Toupet C, Stratmann A, Cyr DD, et al. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem Biol. 2000;7:97–109. doi: 10.1016/s1074-5521(00)00075-2. [DOI] [PubMed] [Google Scholar]
- Mutter R, Wills M. Chemistry and clinical biology of the bryostatins. Bioorg Med Chem. 2000;8:1841–1860. doi: 10.1016/s0968-0896(00)00150-4. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- Partida-Martinez LP, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 2007;8:41–45. doi: 10.1002/cbic.200600393. [DOI] [PubMed] [Google Scholar]
- Petkovic H, Sandmann A, Challis LR, Hecht HJ, Silakowski B, Low L, Beeston N, Kuscer E, Garcia-Bernardo J, Leadlay PF, et al. Substrate specificity of the acyl transferase domains of EpoC from the epothilone polyketide synthase. Org Biomol Chem. 2008;6:500–506. doi: 10.1039/b714804f. [DOI] [PubMed] [Google Scholar]
- Pettit GR. Progress in the discovery of biosynthetic anticancer drugs. J Nat Prod. 1996;59:812–821. doi: 10.1021/np9604386. [DOI] [PubMed] [Google Scholar]
- Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Nat Acad Sci USA. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J, Hui DQ, Wen GP, Butzke D, Platzer M, Fusetani N, Matsunaga S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Nat Acad Sci USA. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pojer F, Li SM, Heide L. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology. 2002;148:3901–3911. doi: 10.1099/00221287-148-12-3901. [DOI] [PubMed] [Google Scholar]
- Pulsawat N, Kitani S, Nihira T. Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae. Gene. 2007;393:31–42. doi: 10.1016/j.gene.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Reeves CD, Murli S, Ashley GW, Piagentini M, Hutchinson CR, McDaniel R. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry. 2001;40:15464–15470. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]
- Rix U, Fischer C, Remsing LL, Rohr J. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat Prod Rep. 2002;19:542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- Salomon CE, Magarvey NA, Sherman DH. Merging the potential of microbial genetics with biological and chemical diversity: an even brighter future for marine natural product drug discovery. Nat Prod Rep. 2004;21:105–121. doi: 10.1039/b301384g. [DOI] [PubMed] [Google Scholar]
- Schneider K, Chen XH, Vater J, Franke P, Nicholson G, Borriss R, Sussmuth RD. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J Nat Prod. 2007;70:1417–1423. doi: 10.1021/np070070k. [DOI] [PubMed] [Google Scholar]
- Simunovic V, Zapp J, Rachid S, Krug D, Meiser P, Muller R. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. ChemBioChem. 2006;7:1206–1220. doi: 10.1002/cbic.200600075. [DOI] [PubMed] [Google Scholar]
- Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu HB, Patel A, Sherman DH, Haygood MG. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- Sun MK, Alkon DL. Dual effects of bryostatin-1 on spatial memory and depression. Eur J Pharmacol. 2005;512:43–51. doi: 10.1016/j.ejphar.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Szafranska AE, Hitchman TS, Cox RJ, Crosby J, Simpson TJ. Kinetic and mechanistic analysis of the malonyl CoA: ACP transacylase from Streptomyces coelicolor indicates a single catalytically competent serine nucleophile at the active site. Biochemistry. 2002;41:1421–1427. doi: 10.1021/bi012001p. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tang GL, Cheng YQ, Shen B. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem Biol. 2004;11:33–45. doi: 10.1016/j.chembiol.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue YQ, Zhao LS, Liu HW, Sherman DH. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc Nat Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.