Abstract
Plague, a life-threatening flea-borne zoonosis caused by Yersinia pestis, has most commonly been reported from eastern Africa and Madagascar in recent decades. In these regions and elsewhere, prevention and control efforts are typically targeted at fine spatial scales, yet risk maps for the disease are often presented at coarse spatial resolutions that are of limited value in allocating scarce prevention and control resources. In our study, we sought to identify sub-village level remotely sensed correlates of elevated risk of human exposure to plague bacteria and to project the model across the plague-endemic West Nile region of Uganda and into neighboring regions of the Democratic Republic of Congo. Our model yielded an overall accuracy of 81%, with sensitivities and specificities of 89% and 71%, respectively. Risk was higher above 1,300 meters than below, and the remotely sensed covariates that were included in the model implied that localities that are wetter, with less vegetative growth and more bare soil during the dry month of January (when agricultural plots are typically fallow) pose an increased risk of plague case occurrence. Our results suggest that environmental and landscape features play a large part in classifying an area as ecologically conducive to plague activity. However, it is clear that future studies aimed at identifying behavioral and fine-scale ecological risk factors in the West Nile region are required to fully assess the risk of human exposure to Y. pestis.
Introduction
Plague, caused by the bacterium Yersinia pestis, is a severe, primarily flea-borne, rodent-associated zoonosis. In humans, Y. pestis infection most commonly presents in one of three clinical forms.1 Bubonic plague, the most common of the three, is typically associated with the bite of an infectious flea or with direct contact between the infectious agent and open skin lesions; it is characterized by sudden onset of fever, chills, headache, malaise and regional lymphadenopathy.1–4 Occasionally, cutaneous exposure to Y. pestis may result in septicemic plague, which is characterized by fever and sepsis without the characteristic regional lymphadenopathy.1–4 Pneumonic plague is rare, but also is the most severe form; it is caused either by hematogenous spread of plague bacteria to the lungs or primary exposure through inhalation of the infectious agent.1,4,5 Although person-to-person transmission requires close contact between infectious and susceptible persons,6 primary pneumonic plague is a feared complication and focal outbreaks still occur.7–10 Fatality rates for untreated infections range from 50% to 60% for bubonic plague to nearly 100% for pneumonic infections,11,12 but outcome of infection is significantly improved by early diagnosis followed by appropriate antibiotic therapy.13
Because of the severity of the disease and its potential for epidemic spread, plague was one of only three internationally quarantinable diseases under previous International Health Regulations.14 Under current International Health Regulations15 (http://www.who.int/ihr/en/), pneumonic plague remains an internationally notifiable disease that is noted as a public health emergency of international concern, and other forms are reportable only from non-endemic localities. Thus, compliance with these new regulations requires a clear understanding of the locations of plague foci. Historical data on plague case occurrence, which were typically reported by country,16 provide an understanding of the coarse geographic distribution of the disease organism. However, risk is often spatially heterogeneous within plague-endemic countries. Geographic Information Systems (GIS) and Remote Sensing (RS) technology coupled with statistical modeling have been useful in generating risk maps that display this spatial heterogeneity and such outputs may aid in identifying previously uncharacterized plague foci.17–22
In recent decades, most human plague infections have been reported from eastern Africa and Madagascar.16,23 The GIS-based models have identified widespread areas of risk on the African continent, and they have determined that ecological predictors of risk are variable among these foci.21 Although informative for raising awareness in potentially endemic areas, the coarse scale of these risk maps limits their use in targeting scarce prevention and control resources. The objectives of the present study were to 1) identify remotely sensed correlates of plague risk in the previously identified endemic West Nile region of Uganda,22,24,25 2) extrapolate the model to a sub-village scale and to neighboring regions of the Democratic Republic of Congo, and 3) evaluate model performance at multiple spatial scales. We believe that the sub-village resolution of the model may aid in identifying villages at highest risk of plague case occurrence, and may be useful for guiding further efforts to more accurately identify environmental and behavioral risk factors for the disease.
Materials and Methods
Study area.
The study was conducted primarily in Vurra and Okoro Counties, located within Arua and Nebbi Districts, respectively, in the West Nile region of Uganda (Figure 1). The Rift Valley escarpment bisects the counties, dividing them into two distinct sections. The lower elevation section east of the escarpment is characterized by sandy soil and low rainfall, and the western, higher elevation area is comprised of lush vegetation, fertile soil, and numerous rivers and tributaries. Although villages are distributed on both sides of the escarpment, most human plague cases are reported from western portions of the counties located above 1,300 meters.22 More detailed descriptions of the ecological characteristics of the area have been reported.22,25,26
Figure 1.

Map of parishes in the West Nile region in northwestern Uganda (inset). Locations of clinics from which confirmed plague cases were reported during 2008–2009 are shown as crosses. Villages of residence of cases and controls are shown as solid or hatched polygons, respectively. Vurra County in Arua District and Okoro County in Nebbi District are shown in dark and light gray, respectively. This figure appears in color at www.ajtmh.org.
The West Nile region is divided into several administrative boundaries. From largest to smallest, these include districts, counties, sub-counties, parishes, and villages. With the exception of villages, boundaries of each have been geo-coded (International Livestock Research Institute, Nairobi, Kenya, 2006). Our study focuses on the village and sub-village level, and boundaries of villages and residences of interest were mapped for this study as described below.
Description of cases and controls.
In Uganda, the standard criteria for diagnosis of plague include sudden onset of fever, chills, malaise, headache or prostration accompanied by either painful regional lymphadenopathy (bubonic), hematemesis or hematochezia (septicemic), or cough with hemoptysis (pneumonic). As described,22 we compiled a database for 1999–2008 that included the health clinic where the patient was seen, village of residence, date of onset of plague signs and symptoms, and a classification of suspect or probable plague case. Suspect cases included all cases where the clinic log records stated that the patient was diagnosed and treated for plague. In contrast, a probable case was a patient seen at a referral hospital in the region, was diagnosed and treated for plague, and additional environmental data suggestive of an on-going plague epizootic (e.g., report of a rat die-off in the area where the patient was believed to have been exposed to Y. pestis) were provided.
During the 2008–2009 plague season (approximately August–March), laboratory confirmation was conducted for patients with clinically suspected plague. Case confirmation was provided by demonstration of Y. pestis-specific phage lysis of a culture isolated from primary specimens (i.e., lymph node aspirates, blood, or sputa) and/or demonstration of seroconversion.27 After laboratory confirmation of a case, the location of residence was recorded using a handheld global positioning system (GPS) receiver (Magellan Explorist 500; MiTAC Digital Corp., Santa Clara, CA; or Garmin GPSMap 60CSx; Garmin, Olathe, KS); these points comprise the case data included in the methods given below. All procedures involving human subjects were reviewed and approved by the Centers for Disease Control and Prevention Institutional Review Board (Protocol nos. 4696 and 5301), the Uganda Virus Research Institute Scientific Ethics Committee (GC/127/08/12/22), and the Uganda National Council for Science and Technology (HS 488).
To control for access to care and to maximize the likelihood that control locations had a low probability of plague occurrence, in our selection of control locations we visited each of the seven clinics that reported a laboratory-confirmed plague case in 2008–2009 (Figure 1). From the clinic log books, we extracted the name of the first village to appear before or after the confirmed plague case that was not represented on the list of villages that reported a suspect or probable plague case in 1999–2008. We aimed to identify two control villages for each case village.
The boundaries of each case and control village were mapped using a handheld GPS receiver while walking the perimeter of each village with a village leader; longitude and latitude data were used to create a shapefile delineating boundaries of case and control villages (Figure 1). Locations of human habitations, typically round or square earthen structures with thatch roofing, were digitized by overlaying the case–control village shapefiles on orthorectified Quickbird imagery collected from 2002–2009 or WorldView imagery collected on February 16–24, 2008. All layers were projected to universal transverse Mercator zone 36N WGS 1984. From each control village, a single hut location was randomly selected using Hawth's Random Selection Sampling Tool (Hawth's Analysis Tools version 3.27). The resulting shapefile comprised the control points used in the methods described below.
Predictive variables.
The GIS and RS data used in this analysis have been described in detail.22 Briefly, these data include 1) administrative boundaries within Uganda depicting district, county, and parish (International Livestock Research Institute, 2006), 2) a 90-meter digital elevation model (Shuttle Radar Topography Mission Elevation Data Set, 2008, accessed August 2008 at http://seamless.usgs.gov/), 3) Landsat 7 Enhanced Thematic Mapper Plus (ETM+) images captured January 1, 2007 (Row/Path: 58/172) and January 10, 2007 (Row/Path: 58/173). Landsat ETM+ images were acquired through a cooperative agreement with the National Geospatial Intelligence Agency and were captured during clear atmospheric conditions; radiometric and geometric distortions of the imagery were corrected. Landsat images from January 1 and 10, 2007 were mosaiced together as described by Winters and others.22
In our exploration of the relationship between plague risk and elevation, we treated elevation as continuous or dichotomous variables. According to the model of Winters and others,22 we created a raster layer defining areas as either above or below 1,300 meters. In addition to each individual Landsat ETM+ band value, we included 1) surface temperature, 2) the normalized difference vegetation index, 3) brightness, 4) greenness, and 5) wetness. Descriptions of the derivations of each variable were reported by Winters and others.22
Model construction.
The objective of our model was to classify the West Nile region of Uganda into areas of elevated or low risk of human exposure to plague bacteria. In accordance with reported methods,17 logistic regression models were constructed to evaluate the association between the probability of a grid cell being classified as high risk and its environmental attributes (described above). The first candidate model included variables that were previously identified as predictive of risk at a parish-level scale (i.e., elevation, brightness, greenness, wetness).22 Other candidate models were constructed after predictive variables were screened by forward stepwise logistic regression modeling to identify those with the greatest association with elevated risk. Spearman correlation tests were used to eliminate variables that were significantly associated with risk (P < 0.05) but highly correlated with other covariates (ρ = 0.85). Each of the candidate models (Table 1) may be described by the equation
Table 1.
Candidate models for associations of remotely sensed variables and likelihood of a pixel being classified as elevated risk of exposure to Yersinia pestis in the West Nile region of Uganda*
| Model ID | Negative log-likelihood | K | AIC | ΔAIC | AUC | Sensitivity | Specificity | PPV | NPV | Independent model variables |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 56.91 | 5 | 66.91 | 3.21 | 80 | 81 | 69 | 57 | 88 | 1,300 meters, wetness, greenness, brightness |
| 2 | 55.73 | 6 | 67.73 | 4.03 | 80 | 86 | 74 | 62 | 91 | 1,300 meters, band 3, band 7, ST, brightness |
| 3 | 55.60 | 5 | 65.60 | 1.90 | 81 | 89 | 71 | 60 | 93 | 1,300 meters, band 3, band 6, brightness |
| 4 | 55.60 | 4 | 63.70 | 0 | 81 | 86 | 71 | 60 | 91 | 1,300 meters, band 3, band 6 |
K = number of estimated parameters included in the model; AIC = Akaike information criterion; AUC = area under the receiving operator characteristic curve; PPV = positive predictive value; NPV = negative predictive value; ST = surface temperature.
Logit (P) = β0 + β1x1 + β2x2 + … + βkxk [expression 1]
where P is the probability that a grid cell will be classified as elevated risk and β0 is the intercept. The values β1 … βk represent coefficients assigned to each independent variable, x1, …, xk included in the regression model. The probability of each cell being classified as high or low risk was derived from expression 1 using the equation
P = e Logit (P)/(1 + e Logit (P)) [expression 2]
Whole model tests were used to assess statistical significance of each model. To determine
whether the covariates included in the model adequately described the distribution in the data, goodness of fit tests compared the pure error negative log-likelihood with the fitted model log-likelihood. When the χ2 test result was not significant (P > 0.05), sufficient explanatory variables were considered to be included in the model.
The overall discrimination ability of each model was assessed using area under the curve (AUC) estimates derived from receiver operating characteristic curves (ROCs). The AUC provides a threshold-independent measure of the overall accuracy of the model; values range from 0.5 to 1, where a value of 1 indicates that all points were correctly classified. We used ROCs to determine the optimal probability cut-off value for characterizing each grid cell as high or low risk. Based on the selected probability cut-off value that simultaneously maximized sensitivity and specificity,28 we compared sensitivity, specificity, and positive and negative predictive values (described below) for each of the candidate models (Table 1).
Akaike's information criterion (AIC) was used to compare candidate models and to select the most parsimonious model with the highest predictive power.29 For each candidate model an AIC value was calculated. The model with the lowest AIC value was deemed the best. However, models within 2 AIC units were considered to be competing models with substantial support.30 To select the best among competing models, we selected the model with highest sensitivity that had the best balance in sensitivity, specificity, and positive and negative predictive values. For the selected model, semivariograms of model residuals were constructed and spatial dependence was evaluated using Moran's I statistic. A leave-one-out method28 was used to evaluate how sensitive the model was to any particular case or control point. In summary, the best model was sequentially refitted by removing a single site, recording the AUC value, replacing the site and removing the next site in the data set. The average and range in AUC values are reported. Finally, the best model was extrapolated to all pixels contained within the two Landsat scenes by applying expression 2 using the raster calculator function of ArcGIS version 9.3 (Environmental Systems Research Institute, Redlands, CA).
Model evaluation.
To evaluate performance of the best model, sensitivity, specificity, and positive and negative predictive values were calculated based on the case and control points used to construct the model. To do so, expression 2 was applied to all pixels and risk probabilities were extracted for each case or control point. A site was assigned a value of elevated or low risk, if the probability value was above or below the threshold value identified by the ROC, respectively. Sensitivity is a measure of the percentage of case sites that were accurately classified by the model as elevated risk. Specificity summarizes the percentage of control sites that were accurately classified by the model as low risk. Positive and negative predictive values describe the percentages of sites that the model predicted to be elevated or low risk that were actual case or control locations, respectively.
In addition to the point-based evaluation, we calculated the percentage of each village that was classified as elevated risk. This secondary evaluation was conducted in part because exact exposure sites could not be determined. Given the amount of time inhabitants spend within their village of residence, it is expected that if the included variables are predictive of elevated risk, significant differences should be observed between case and control villages. Using Wilcoxon rank sums tests, we compared the median area of elevated risk between case and control villages. Because risk areas may be located within village boundaries, but not coincide with the locations of human habitations, we assessed the proportion of huts located within risk pixels. This assessment was accomplished by extracting the risk classification (i.e., elevated or low risk) for each hut that was digitized within the village boundaries. We then compared the median proportion of huts located within risk areas between case and control villages using a Wilcoxon rank sums test. We assumed that a significantly higher proportion of huts located within risk areas in case villages was supportive of our hypotheses that 1) the selected variables were indicative of plague risk and 2) most exposures occur in the peridomestic setting. To expand upon the village-level model evaluations, we calculated the percentage of each parish classified as elevated risk and used linear regression to explore the association between percent of the parish covered by areas of elevated risk and parish level incidence of human plague (based on suspect and probable plague cases reported in 1999–2007).
Results
Summary of epidemiologic data.
During 1999–2007, a median of 266 (range = 76–466) suspect or probable plague cases were reported per year. During August 2008–March 2009, a total of 163 cases were reported, of which 55 were confirmed by using standard laboratory diagnostic techniques. Thirteen of the 55 laboratory-confirmed cases were excluded because the patient was not seen in a health care clinic and therefore an appropriate control that would account for access to care could not be selected. Although we considered selecting controls from the nearest clinic to the patient's village, further investigation showed that patients do not consistently seek care in the nearest clinic, and thus these cases were ultimately excluded from our analysis. Significant errors in geocoding of the locations of residence of six laboratory-confirmed cases were identified; thus, these cases were excluded. In total, 36 case and 72 control points selected from 32 case villages and 61 control villages were included in our model development (Figures 1 and 2). The median coverage areas and the density of physical structures (number of digitized huts divided by the area encompassed by the village boundaries) were similar for case and control villages (χ2 ≤ 0.96, degrees of freedom [df] = 1, P ≥ 0.33, by Wilcoxon rank sums tests).
Figure 2.
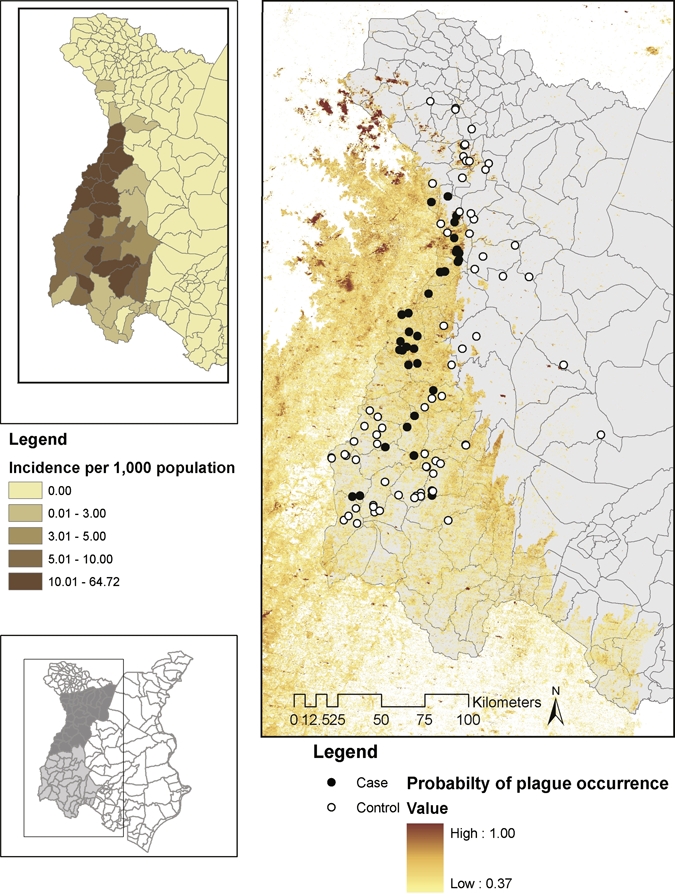
Predicted distribution of areas at elevated risk for exposure to Yersinia pestis within the area of interest (AOI) in the West Nile region of Uganda and in neighboring regions of the Democratic Republic of Congo. Shaded areas represent pixels classified as elevated risk; the color gradient indicates the probability of case occurrence within areas of elevated risk based on dichotimization at a probability value of 0.37. Input variables included in the predictive model include positive associations with elevation above 1,300 meters, brightness, and Landsat Enhanced Thematic Mapper Plus (ETM+) band 3, and a negative association with Landsat ETM+ band 6. Insets show the AOI and parish level incidence for Ugandan parishes within the AOI. This figure appears in color at www.ajtmh.org.
Summary of plague risk model.
Among the candidate models, two were identified as competing (models 3 and 4, Table 1). Model 3 was selected as the best because sensitivity and negative predictive value estimates were slightly higher for that model. The model included positive associations with the 1,300-meter elevation cut-off, brightness, and band 3, and a negative association with band 6 (Table 2). Similar to the other models tested, model 3 indicated that plague risk was higher above 1,300 meters than below. In addition, plague risk increased with increasing brightness values, which is often used as a measure of bare soil. However, it is important to note that brightness had only a small effect on model performance, as seen by comparing models 3 and 4 (Table 1). Band 3 was strongly negatively correlated with greenness (ρ = −0.834, P < 0.0001) and thus, indicates that plague risk is lower in areas with a lower green vegetation index during the month of January when agricultural lands are typically fallow and rainfall is scarce. Finally, band 6 had a strong negative correlation with wetness (ρ = −0.951, P < 0.0001), indicating that plague risk is higher in wetter areas. The model was extrapolated across the coverage area (Figure 2) to identify the distribution in risk. Model residuals were randomly distributed (Moran's I = 0.45, P = 0.20) and the leave-one-out evaluation showed that the model was not particularly sensitive to any individual point used to construct the model (average AUC = 0.81, range = 0.80–0.83).
Table 2.
Parameter estimates for the selected multivariate logistic regression model (model 3, Table 1) predicting areas of elevated risk for exposure to Yersinia pestis in the West Nile region of Uganda*
| Model covariates | Parameter estimate | Likelihood ratio test | |||
|---|---|---|---|---|---|
| Estimate | SE | χ2 | df | P | |
| Intercept | −5.00 | 2.31 | 5.18 | 1 | 0.02 |
| 1,300 meter elevation cut-off | 2.06 | 0.65 | 20.79 | 1 | < 0.0001 |
| Landsat ETM+ band 3 | 0.16 | 0.06 | 6.61 | 1 | 0.01 |
| Landsat ETM+ band 6 | −0.13 | 0.05 | 6.60 | 1 | 0.01 |
| Brightness | 0.01 | 0.03 | 0.14 | 1 | 0.71 |
df = degrees of freedom. ETM+ = Enhanced Thematic Mapper Plus. Whole model test χ2 = 25.53, df = 4, P < 0.0001; goodness of fit χ2 = 111.15, P = 0.15.
Model 3 had an overall accuracy of 81%. In total, 32 of 36 case locations were correctly classified by the model as elevated risk, yielding a sensitivity of 89%. Specificity was 71%, with 51 of 72 control locations classified as low risk. The model predicted that 53 of the hut locations were expected to fall within areas of elevated risk and 32 of these corresponded with case locations, thus yielding a positive predictive value of 60%. Given the low frequency of plague cases in this region and the single year of data used to evaluate the model, we believe this is an acceptable value. The negative predictive value was 93% with 51 of the 55 points expected to be situated within low risk areas identified as controls.
For each village, we calculated the percentage of the total area that was classified by the model as elevated risk. For case villages, the median percentage of risk area was 76.5% (range = 0.01–100%), whereas the median percentage of risk areas within control villages was 41.6% (range = 0.002–100%; χ2 = 25.63, df =1, P < 0.0001). Risk pixels were contained within all villages, regardless of its designation as a case or control. To determine if the proportion of huts falling within risk areas differed between case and control villages, we calculated the percentage of huts within each village that were located within an area identified as elevated risk. On average, 79% (median value with range = 0–90%) of huts in case villages were located within areas identified by the model as elevated risk, whereas only 38% (range = 0–91%) of huts in control villages were located within pixels classified as elevated risk (χ2 = 25.97, df = 1, P < 0.0001). Thus, although all villages evaluated in our study contained areas of elevated risk, the distribution of risk areas with respect to locations of housing appears to have differed between case and control villages.
Finally, to determine whether risk coverage was significantly associated with plague incidence during 1999–2007, we constructed a linear regression of parish-level incidence on the proportion of each parish classified as elevated risk. This construction yielded a significant positive association in which the proportion of the parish at risk explained 27% of the variation in incidence (incidence = –0.378 + 17.96 × proportion of parish classified as elevated risk; F = 74.26, df = 1, 205, P < 0.0001). Similar to the model of Winters and others,22 Ayavu parish was identified as an outlier, with the reported incidence being much higher than expected by the model. When this parish was excluded from the analysis, 42% of the variation in incidence was explained by the proportion of the parish classified as elevated risk (incidence = –0.778 + 18.55 × proportion of parish classified as elevated risk; F = 145.23, df = 1, 204, P < 0.0001).
Discussion
Plague prevention and control activities typically include conducting surveillance of host and vector populations to identify evidence of epizootic activities and educating the public to seek care for signs and symptoms indicative of Y. pestis infection and to eliminate or reduce food and harborage for rodents in and around homes and workspaces.31 In Uganda, the implementation of these activities is most often conducted at the village or even sub-village spatial scale, and typically as a reactive activity to human cases. Therefore, fine-scale spatial representation of areas posing an elevated risk of plague activity is valuable for prioritizing such efforts. Because of privacy concerns and reporting conventions, it has been common practice to present epidemiologic data based on administrative boundaries such as countries, counties, or sub-county boundaries.16,22,25,32,33 Such aggregating of data obscures significant spatial patterns and falsely portrays risk as homogeneous across the coverage area.
In contrast to previous GIS-based spatial risk models of plague in Africa that were created at continental21 or regional spatial scales,22 our model identified remotely sensed correlates of elevated risk at the sub-village scale. This fine resolution model may be used to more precisely identify risk factors for human exposure to Y. pestis and potentially to focus limited surveillance, prevention, and control resources and, ideally, will enable increased proactive measures and an enhanced index of suspicion for plague symptoms in patients from areas described as being at risk. Similar to a previous plague risk model created at the parish level within the plague-endemic West Nile region of Uganda,22 our model showed that at the sub-village scale, plague risk was higher at elevations above 1,300 meters compared with those below this elevation. Furthermore, remotely sensed covariates that were included in the model implied that localities that are wetter, with less vegetative growth and more bare soil during the dry month of January (when agricultural plots are typically fallow) pose an elevated risk of plague case occurrence. The associations with elevated plague risk in higher and wetter areas are consistent with qualitative observations made by Hopkins in 1949.34
The reasons why these variables are indicative of elevated plague risk have not been evaluated explicitly. However, we speculate that the spectral signatures used in our model may be detecting areas with more intensive agriculture. To confirm this possibility, future studies are needed to classify fine-scale land use patterns and their association with plague risk. Nonetheless, we believe such an association with agricultural intensity and plague case occurrence is biologically plausible. According to a trophic cascade hypothesis,35–37 areas with increased primary production of food crops may increase the carrying capacity of rodents. Several quantitative models have demonstrated that the probability of plague epizootics, which represent periods when humans are at greatest risk for exposure to infectious fleas,38 is dependent on reaching critical thresholds of key rodent hosts.39,40 If the crops produced in these plots are later stored within housing structures, as is common practice in this region,25 infected rodents and their fleas may be drawn into human habitations, thus increasing the likelihood of contact between humans and infectious fleas. In addition, the positive association with wetness, as expressed by the inclusion of band 6, may be indicative of conditions that are conducive to flea survival. In general, fleas are susceptible to desiccation-induced mortality,41 particularly when temperatures are fairly high as they would be during the month of January when the Landsat images were captured. Vectorial capacity models suggest that as flea survivorship increases, the probability of flea-borne transmission also increases.42,43
Overall, the model performed well at the point, village, and even parish evaluation levels. However, it is important to note that the point-level model was evaluated by using data included in the build set that may optimistically bias the model evaluation. As surveillance activities continue over the years, model performance should be re-evaluated, and the model should be fine-tuned, if necessary. At the point scale, the model was 89% accurate in classifying areas from which plague cases were reported as elevated risk. However, 11% of case homes were situated in areas classified as low risk. We assumed that, similar to other plague foci,17,18,44–47 most plague cases occur in the peridomestic setting. This finding was supported by the observation that a higher proportion of case homes, relative to controls, were located within pixels classified as elevated risk. However, we did not interview patients or conduct environmental investigations to evaluate whether every case included in our study was most likely exposed in or near his or her home. Additionally, this error could be attributed to the fact that not all risk factors could be identified by available input variables. By the corollary, the high model sensitivity could arise when the elevated risk area is over-estimated. For example, if 100% of the coverage area is classified as elevated risk, all cases should occur within areas of elevated risk. This occurrence does not appear to be the case in our model, which significantly reduced the area of elevated risk (Figure 2) and reported a specificity of 71%.
It is noteworthy that 29% of controls were located within areas of elevated risk. This finding may be attributed in part to the fact that our study focused on only one year in which human plague cases were confirmed. It is possible that these localities did not experience epizootic activity in the year of interest, but could pose a threat over the long-term. Previous studies in this region have shown that the distribution of zoonotic hosts and their vectors are not evenly distributed across the landscape or over time.26,48 Thus, some areas that contain the environmental characteristics that may support epizootic activity may be quiescent at any given point in time. Additionally, this finding emphasizes that case occurrence is not dictated entirely by environmental or landscape factors identified by our model. Preventative behaviors such as avoidance of sick or dead animals, reducing rodent food and harborage in and around homes, and avoiding contact with fleas31,45,46 may have reduced the likelihood of human infection in some areas. Our study did not evaluate prevention and control measures that may have been implemented and environmental surveillance activities were not conducted as part of this study to determine if any of these areas were experiencing plague epizootics. However, our model results may be used to control for known environmental and landscape risk factors within future case–control studies aimed at understanding either human behavioral risk factors, or fine-scale environmental factors that cannot be detected using remote-sensing technology (e.g., spatial heterogeneity in the abundance and diversity of rodent hosts and their vector fleas26). Targeted investigations of areas classified as high risk that continue not to report plague cases may provide insight into subtle differences in housing, crops or behavior that prove protective.
It is important to note that the positive predictive value for the model was only 60%. That is, in 60% of point locations where the model classified an area to be elevated risk, a case occurred in that location. This finding may be explained by the reasons given above, namely that plague epizootics do not occur every year in localities that are conducive to such activities and human behavior can enhance or reduce the probability of human case occurrence. Not surprisingly for a rare disease, the negative predictive value was quite high, indicating that 93% of locations that were classified as low risk did not yield a case. Although this information may be informative in allocating prevention and control resources, it is worth emphasizing that areas of elevated risk occur throughout the West Nile region, and health care providers and the public should remain vigilant in their attempts to recognize epizootic activity regardless of whether a particular area is categorized as low or elevated risk. This point is underscored by the observation that, although case villages contained a higher proportion of homes located within elevated risk pixels, areas of elevated risk were detected in all villages, regardless of whether a plague case had been reported.
Finally, extrapolation of our model into neighboring regions in the Democratic Republic of Congo agrees with previous reports of continued epizootic activity across the international border in Orientale Province.23,49 Thus, successful efforts to control plague in the West Nile region require regional implementation of prevention and control measures. However, it is important to note that because of a lack of available fine-resolution data on where laboratory-confirmed plague cases are occurring in the Democratic Republic of Congo, the model has not been evaluated in that country and our model extrapolation serves simply as a hypothetical risk surface for that area. Our study follows World Health Organization recommendations49 to integrate GIS technology and statistical modeling to better define the areas of elevated risk in this region, which may reduce the costs of plague surveillance by targeting high-risk areas. Future studies are required to evaluate the accuracy of model predictions in the Democratic Republic of Congo and to identify risk factors for human exposure to Y. pestis that were not included in our model. We believe that the model presented here demonstrates that environmental and landscape features play a large part in classifying an area as ecologically conducive to plague activity. However, it is clear that future studies aimed at identifying behavioral and fine-scale ecological risk factors in the West Nile region are required to fully assess the risk of human exposure to Y. pestis.
Acknowledgments
We thank Joseph Tendo and Linda Atiku for assistance in the field.
Footnotes
Authors' addresses: Rebecca J. Eisen, Kevin S. Griffith, Jeff N. Borchert, Katherine MacMillan, Emily Zielinski-Gutierrez, Anna M. Winters, Russell E. Enscore, Martin E. Schriefer, Charles B. Beard, Kenneth L. Gage, and Paul S. Mead, Division of Vector-Borne Infectious Diseases, Centers for Disease Control and Prevention, Fort Collins, CO. Titus Apangu, Nicholas Owor, Sara Acayo, and Rogers Acidri, Uganda Virus Research Institute, Entebbe, Uganda.
References
- 1.Poland JD, Dennis DT. In: Plague Manual: Epidemiology, Distribution, Surveillance and Control. Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Geneva: World Health Organization; 1999. pp. 43–54. (Diagnosis and clinical manifestations). [Google Scholar]
- 2.Campbell GL, Dennis DT. In: Harrison's Principles of Internal Medicine. Fauci AS, Braunwald E, Isselbacher KJ, Wilson JD, Martin JB, Kasper DL, Hauser SL, Longo DL, Harrison TR, editors. New York: McGraw-Hill; 1998. pp. 975–980. (Plague and other Yersinia infections). [Google Scholar]
- 3.Butler T. In: Current Topics in Infectious Disease. Greenough WB, Merigan TC, editors. New York: Plenum; 1983. pp. 71–92. (Plague and other Yersinia infections). [Google Scholar]
- 4.Dennis DT, Gage KL. In: Infectious Diseases. Cohen J, Powderly WG, editors. London: Mosby; 2003. pp. 1641–1648. (Plague). [Google Scholar]
- 5.Dennis DT, Meier FA. In: Pathology of Emerging Infections. Horsburgh CR, Nelson AM, editors. Washington, DC: American Society for Microbiology Press; 1997. pp. 21–48. (Plague). [Google Scholar]
- 6.Garner JS. Guideline for isolation precautions in hospitals. The Hospital Infection Control Practices Advisory Committee. Infect Control Hosp Epidemiol. 1996;17:53–80. doi: 10.1086/647190. [DOI] [PubMed] [Google Scholar]
- 7.Begier EM, Asiki G, Anywaine Z, Yockey B, Schriefer ME, Aleti P, Ogden-Odoi A, Staples JE, Sexton C, Bearden SW, Kool JL. Pneumonic plague cluster, Uganda, 2004. Emerg Infect Dis. 2006;12:460–467. doi: 10.3201/eid1203.051051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention Bubonic and pneumonic plague—Uganda. MMWR Morb Mortal Wkly Rep. 2009;58:778–781. [PubMed] [Google Scholar]
- 9.Ratsitorahina M, Chanteau S, Rahalison L, Ratsifasoamanana L, Boisier P. Epidemiological and diagnostic aspects of the outbreak of pneumonic plague in Madagascar. Lancet. 2000;355:111–113. doi: 10.1016/S0140-6736(99)05163-6. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization Weekly epidemiological record. Wkly Epidemiol Rec. 2006;81:397–408. [Google Scholar]
- 11.Levy CE, Gage KL. Plague in the United States, 1995–1997. Infect Med. 1999;16:54–64. [Google Scholar]
- 12.Poland JD, Barnes AM. In: CRC Handbook Series in Zoonoses, Section A: Bacterial, Rickettsial and Mycotic Diseases. Steele JH, editor. Boca Raton, FL: CRC Press; 1979. pp. 515–559. (Plague). [Google Scholar]
- 13.Crook LD, Tempest B. Plague. A clinical review of 27 cases. Arch Intern Med. 1992;152:1253–1256. doi: 10.1001/archinte.152.6.1253. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization . International Health Regulations. First edition. Geneva: World Health Organization; 1983. pp. 26–29. [Google Scholar]
- 15.World Health Organization . International Health Regulations. Second edition. Geneva: World Health Organization; 2005. pp. 43–46. [Google Scholar]
- 16.World Health Organization Human plague in 2002 and 2003Wkly Epidemiol Rec 79301–308.15369044 [Google Scholar]
- 17.Eisen RJ, Enscore RE, Biggerstaff BJ, Reynolds PJ, Ettestad P, Brown T, Pape J, Tanda D, Levy CE, Engelthaler DM, Cheek J, Bueno R, Jr, Targhetta J, Montenieri JA, Gage KL. Human plague in the southwestern United States, 1957–2004: spatial models of elevated risk of human exposure to Yersinia pestis. J Med Entomol. 2007;44:530–537. doi: 10.1603/0022-2585(2007)44[530:hpitsu]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 18.Eisen RJ, Reynolds PJ, Ettestad P, Brown T, Enscore RE, Biggerstaff BJ, Cheek J, Bueno R, Targhetta J, Montenieri JA, Gage KL. Residence-linked human plague in New Mexico: a habitat-suitability model. Am J Trop Med Hyg. 2007;77:121–125. [PubMed] [Google Scholar]
- 19.Holt AC, Salkeld DJ, Fritz CL, Tucker JR, Gong P. Spatial analysis of plague in California: niche modeling predictions of the current distribution and potential response to climate change. Int J Health Geogr. 2009;8:38. doi: 10.1186/1476-072X-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakazawa Y, Williams R, Peterson AT, Mead P, Staples E, Gage KL. Climate change effects on plague and tularemia in the United States. Vector Borne Zoonotic Dis. 2007;7:529–540. doi: 10.1089/vbz.2007.0125. [DOI] [PubMed] [Google Scholar]
- 21.Neerinckx SB, Peterson AT, Gulinck H, Deckers J, Leirs H. Geographic distribution and ecological niche of plague in sub-Saharan Africa. Int J Health Geogr. 2008;7:54. doi: 10.1186/1476-072X-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winters AM, Staples JE, Ogen-Odoi A, Mead PS, Griffith K, Owor N, Babi N, Enscore RE, Eisen L, Gage KL, Eisen RJ. Spatial risk models for human plague in the West Nile region of Uganda. Am J Trop Med Hyg. 2009;80:1014–1022. [PubMed] [Google Scholar]
- 23.Neerinckx S, Bertherat E, Leirs H. Human plague occurrences in Africa: an overview from 1877 to 2008. Trans R Soc Trop Med Hyg. 2010;104:197–103. doi: 10.1016/j.trstmh.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 24.Kilonzo BS. Plague epidemiology and control in eastern and southern Africa during the period 1978 to 1997. Cent Afr J Med. 1999;45:70–76. [PubMed] [Google Scholar]
- 25.Orochi-Orach S. Plague Outbreaks: the Gender and Age Perspective in Okoro County, Nebbi District, Uganda. Nebbe. Uganda: Agency for Accelerated Regional Development; 2002. [Google Scholar]
- 26.Amatre G, Babi N, Enscore RE, Ogen-Odoi A, Atiku LA, Akol A, Gage KL, Eisen RJ. Flea diversity and infestation prevalence on rodents in a plague-endemic region of Uganda. Am J Trop Med Hyg. 2009;81:718–724. doi: 10.4269/ajtmh.2009.09-0104. [DOI] [PubMed] [Google Scholar]
- 27.Chu MC. Laboratory Training Manual of Plague Diagnostics Tests. Atlanta: Centers for Disease Control and Prevention and Geneva: World Health Organization; 2000. [Google Scholar]
- 28.Fielding AH, Bell JF. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ Conserv. 1997;24:38–49. [Google Scholar]
- 29.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr AC- 1974;19:716–723. [Google Scholar]
- 30.Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. New York: Springer; 2002. [Google Scholar]
- 31.Gage KL. In: Plague Manual: Epidemiology, Distribution, Surveillance and Control. Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Geneva: World Health Organization; 1999. pp. 167–171. (National health services in prevention and control). [Google Scholar]
- 32.Eisen L, Eisen RJ. Need for improved methods to collect and present spatial epidemiologic data for vectorborne diseases. Emerg Infect Dis. 2007;13:1816–1820. doi: 10.3201/eid1312.070211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisen RJ, Eisen L. Spatial modeling of human risk of exposure to vector-borne pathogens based on epidemiological versus arthropod vector data. J Med Entomol. 2008;45:181–192. doi: 10.1603/0022-2585(2008)45[181:smohro]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 34.Hopkins GH. Report on Rats, Fleas and Plague in Uganda. Nairobi. Kenya: East African Standard, Ltd; 1949. [Google Scholar]
- 35.Collinge SK, Johnson WC, Ray C, Matchett R, Grensten J, Cully JF, Gage KL, Kosoy M, Loye JE, Martin A. Testing the generality of the trophic-cascade model for plague. EcoHealth. 2005;2:102–112. [Google Scholar]
- 36.Enscore RE, Biggerstaff BJ, Brown TL, Fulgham RE, Reynolds PJ, Engelthaler DM, Levy CE, Parmenter RR, Montenieri JA, Cheek JE, Grinnell RK, Ettestad PJ, Gage KL. Modeling relationships between climate and the frequency of human plague cases in the southwestern United States, 1960–1997. Am J Trop Med Hyg. 2002;66:186–196. doi: 10.4269/ajtmh.2002.66.186. [DOI] [PubMed] [Google Scholar]
- 37.Parmenter RR, Yadav EP, Parmenter CA, Ettestad P, Gage KL. Incidence of plague associated with increased winter-spring precipitation in New Mexico. Am J Trop Med Hyg. 1999;61:814–821. doi: 10.4269/ajtmh.1999.61.814. [DOI] [PubMed] [Google Scholar]
- 38.Gage KL, Kosoy MY. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol. 2005;50:505–528. doi: 10.1146/annurev.ento.50.071803.130337. [DOI] [PubMed] [Google Scholar]
- 39.Davis S, Begon M, De Bruyn L, Ageyev VS, Klassovskiy NL, Pole SB, Viljugrein H, Stenseth NC, Leirs H. Predictive thresholds for plague in Kazakhstan. Science. 2004;304:736–738. doi: 10.1126/science.1095854. [DOI] [PubMed] [Google Scholar]
- 40.Davis S, Trapman P, Leirs H, Begon M, Heesterbeek JA. The abundance threshold for plague as a critical percolation phenomenon. Nature. 2008;454:634–637. doi: 10.1038/nature07053. [DOI] [PubMed] [Google Scholar]
- 41.Cavanaugh DC, Marshall JD., Jr The influence of climate on the seasonal prevalence of plague in the Republic of Vietnam. J Wildl Dis. 1972;8:85–94. doi: 10.7589/0090-3558-8.1.85. [DOI] [PubMed] [Google Scholar]
- 42.Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci U S A. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisen RJ, Gage KL. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet Res. 2009;40:1. doi: 10.1051/vetres:2008039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akiev AK. Epidemiology and incidence of plague in the world, 1958–79. Bull World Health Organ. 1982;60:165–169. [PMC free article] [PubMed] [Google Scholar]
- 45.Gratz N. In: Plague Manual: Epidemiology, Distribution, Surveillance and Control. Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Geneva: World Health Organization; 1999. pp. 97–134. (Control of plague transmission). [Google Scholar]
- 46.Mann JM, Martone WJ, Boyce JM, Kaufmann AF, Barnes AM, Weber NS. Endemic human plague in New Mexico: risk factors associated with infection. J Infect Dis. 1979;140:397–401. doi: 10.1093/infdis/140.3.397. [DOI] [PubMed] [Google Scholar]
- 47.Tikhomirov E. In: Plague Manual: Epidemiology, Distribution, Surveillance and Control. Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Geneva: World Health Organization; 1999. pp. 11–37. (Epidemiology and distribution of plague). [Google Scholar]
- 48.Eisen RJ, Borchert JN, Holmes JL, Amatre G, Van Wyk K, Enscore RE, Babi N, Atiku LA, Wilder AP, Vetter SM, Bearden SW, Montenieri JA, Gage KL. Early-phase transmission of Yersinia pestis by cat fleas (Ctenocephalides felis) and their potential role as vectors in a plague-endemic region of Uganda. Am J Trop Med Hyg. 2008;78:949–956. [PubMed] [Google Scholar]
- 49.World Health Organization International meeting on preventing and controlling plague: the old calamity still has a future. Wkly Epidemiol Rec. 2006;28:279–284. [PubMed] [Google Scholar]


