Abstract
Hepatitis E is a worldwide public health problem, especially in areas with poor sanitation. This study examines the potential hepatitis E virus (HEV) animal reservoirs and the current status of HEV infection among animals and humans in an endemic area of Xinjiang, China. One thousand five hundred twenty-one serum samples from 12 different animal species and 296 sera from humans were detected for anti-HEV with an in-house enzyme immunoassay, and partial HEV RNA was amplified with a reverse transcription–nested polymerase chain reaction (RT-nPCR). All these distinct animal species, except jerboa and hoptoad, were positive for anti-HEV. However, HEV RNA was only amplified from pigs and a sporadic hepatitis E case in humans. The human HEV strain (CHN-XJ-HE29) shared 100% nucleotide identity with the swine HEV strain (CHN-XJ-SW50), both of which were collected from the same district; this indicates the possibility of HEV transmission from swine to humans in an endemic area.
Introduction
Hepatitis E virus (HEV), the causative agent of epidemic and sporadic hepatitis E, is no longer confined to developing countries but has also become a concern of developed countries.1 The virus is transmitted primarily by fecal–oral route. Water-borne epidemic is one of the characteristics of hepatitis E in developing countries where sanitation conditions are poor.2
HEV, as the sole member of the Hepevirus genus in Hepeviridae family, is a non-enveloped virus with a positive sense, single-stranded RNA genome approximately 7.2 kb in length. HEV has at least four distinct genotypes with one serotype; HEV genotypes 1 and 2 exclusively infect humans, and they are often associated with outbreaks or large epidemics in developing countries. However, HEV genotypes 3 and 4 infect both humans and animals, and they are often associated with sporadic hepatitis E.3 On mainland China, there are three HEV genotypes (1, 3, and 4) prevailing in humans and/or animals. Among them, HEV genotype 1 had once caused a large-scale epidemic in southern Xinjiang (1986–1988), and its causative strains were successively isolated from most parts of China from 1989 to 2003.4–7 HEV genotype 3, considered as an imported genotype, has been found in eastern China since 2006.8 HEV genotype 4, which circulates both in humans and animals, has become the dominant genotype instead of genotype 1 since 2004.7
Since the first swine HEV strain was isolated in 1997 by Meng,9 it has been documented by more and more studies that swine is the largest viral reservoir of HEV and that hepatitis E is a zoonosis.10–12 However, in India (an HEV-endemic area), human HEV belonged to genotype 1, whereas swine HEV was restricted to genotype 4. Additionally, the local human HEV isolate (AKL-90; genotype 1a) failed to infect specific pathogen-free pigs.13,14 Furthermore, hepatitis E outbreaks or large-scale epidemics mainly occurred in Southeast Asia, Central Asia, Middle East, and North Africa, where most local citizens are Muslims, Buddhists, and Hindu who seldom eat pork or hardly have contact with pigs. Therefore, swine may not play the only role in transmission of HEV in these endemic areas. In addition, anti-HEV was found to be prevalent among various animals such as horses, cows, rodents, cats, dogs, goats, and so on.1,15,16 Moreover, partial or complete genomes of HEV were detected in deer, wild boar, mongoose.17–19 Therefore, there should be some HEV animal reservoirs other than swine that cause zoonotic diseases in HEV-endemic areas.
Xinjiang Uighur Autonomous Region adjoins to Central Asia and is mostly occupied by Muslim populations. The largest hepatitis E outbreak occurred in the region from 1986 to 1988 with about 120,000 persons infected, and its causative agent was identified as genotype 1 HEV. The outbreak lasted for over 2 years and was associated with continuous drinking-water contamination.20 In recent years, swine HEV isolates (genotype 4) were also identified in this region.21,22 However, the significant genetic diversity between the swine and human HEV isolates suggested that the swine HEV was unlikely to be the major source of the hepatitis E outbreak in Xinjiang.
In this study, 12 different animal species and 296 persons from Xinjiang were examined to search for potential HEV animal reservoirs besides swine and to learn the current status of HEV infection among animals and humans in Xinjiang, China.
Materials and Methods
Sample collection.
One thousand five hundred twenty-one serum samples from 12 different animal species in southern Xinjiang were collected in this study. These 12 species were simply classified into four major categories according to their distinct living habits: slaughter swine, domestic herbivorous animals, rodents, and aquatic animals. The detailed classification and illustration for the animal serum samples are shown in Table 1.
Table 1.
The prevalence of anti-HEV among 12 different animal species sera
| Classification | Animal species | Total samples | Positive samples | Positive rate of anti-HEV | Overall positive rate |
|---|---|---|---|---|---|
| Domestic animals | Slaughter swine | 78 | 66 | 84.61% | 84.61% |
| Domestic herbivorous animals | Donkey | 276 | 38 | 13.77% | 9.92% |
| Horse | 100 | 11 | 11.00% | ||
| Goat | 200 | 15 | 7.50% | ||
| Cow | 200 | 13 | 6.50% | ||
| Rodents | Mus musculus | 148 | 52 | 35.14% | 14.50% |
| Sorex caecutiens | 25 | 4 | 16.00% | ||
| Marmota monax | 190 | 9 | 4.74% | ||
| Gerbil mouse | 77 | 3 | 3.90% | ||
| Jerboa mouse | 29 | 0 | 0.00% | ||
| Aquatic animals | Fish | 88 | 1 | 1.14% | 0.51% |
| Hoptoad | 110 | 0 | 0.00% | ||
| Total | – | 1,521 | 212 | – | 13.93% |
Two hundred ninety-six serum specimens of humans were obtained from normal people in the check-up department of local hospitals in southern Xinjiang (56) and northern Xinjiang (240).
One hundred twenty-five swine fecal samples were collected from < 3-month-old swine in southern Xinjiang (100) and northern Xinjiang (25), and 350 fecal specimens from < 3-year-old horses (100) and donkeys (250) were collected from southern Xinjiang.
All specimens surveyed in this study were collected from November 2007 to January 2008 and kept in −80°C until use.
Detection of anti-HEV in serum specimens.
All serum specimens were detected for anti-HEV by using a commercial enzyme-linked immunosorbent assay (ELISA; Wantai Biological Pharmacy Co., Beijing, China). The ELISA kit, a sandwich-enzyme immunoassay, was used with a recombinant peptide corresponding to amino acid (aa) 394–606 of the major structure protein specified by open-reading frame 2 (ORF2) of the genotype 1 HEV genome.15
Serum biochemistry detection.
Two hundred ninety-six serum samples from humans were tested for alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TB), and conjugated bilirubin (CB) by using standard methods on a SmartSpec 3000 spectrophotometer (BioRad, CA). Those samples with ALT elevation were also detected for HEV RNA.
RNA extraction and reverse transcription–nested PCR.
Total RNA was extracted from 100 μL of serum samples or 10% fecal supernatants by using the Total RNA Isolation System (Promega, Madison, WI). The viral RNA was finally dissolved in 20 μL ribonuclease (RNase)-free water and subjected to complementary DNA (cDNA) synthesis with one of the specific external antisense primers using avian myeloblastosis virus (AMV) reverse transcriptase (Promega) at 42°C for 60 minutes. For detecting all four known HEV genotypes, two sets of universal nested primers were designed to amplify the partial fragments in the ORF1 (129–373 nt) and the ORF2 (5983–6349 nt) of the HEV genome (the sites based on AY594199), respectively. Set one (primer P) for amplification of ORF1 (129–373 nt) was external P1 (forward, 56–7 6nt; 5′-AGG CTC CTG GCR TYA CTA CTG 3′) and P4 (reverse, 567–550 nt; 5′-CAT NGC YTC NGC RAC ATC 3′) and internal P2 (forward 113–129 nt; 5′-YC YTK GCG AAT GCT GTG 3′) and P3 (reverse 392–373 nt; 5′-GTR TAC CAV CGC TGA ACR TC 3′). Set two (primer S) for amplification of ORF2 (5983–6349 nt) was external S1 (forward 5738–575 nt; 7 5′ TTA TGC CCA GTA RCG YGT TG3′) and S4 (reverse 6466–6447 nt; 5′-CCC TTR TCR TGC TGA GCG TT 3′) and internal S2 (forward 5965–5983 nt; 5′-GNT GGC GYT CNG TYG AGA C 3′) and S3 (reverse 6369–6349 nt; 5′-CDG CCG ACG AAA TYA ATT CTG). Amplification conditions for both sets of primers and both rounds of PCR were the same: 30 cycles of 94°C for 30 seconds, 50°C for 30 seconds, and 72°C for 40 seconds.
Sequencing and phylogenetic analysis.
The target second-round PCR products were purified with the HQ&Q gel extraction kit (U-gene Co., Anhui, China). Then, purified PCR products were double-ends sequenced by the automatic DNA sequencer (ABI model 3730 sequencer) at Sinogenomax Company (Beijing, China).
Nucleotide sequence alignments and percent identities were generated by Bioedit v7.0.9 software (Carlsbad, CA). Phylogenetic trees were constructed by the neighbor-joining method, and the interior-branch test method was used with the aid of the MEGA 4.0 software package (version 4.0); 1,000 tests of the data were used to calculate percentages of the branches obtained.
For partial-genome phylogenetic analysis, the criteria of identity for different genotypes and subgenotypes classification were 80% and 90%, respectively.
Forty standard HEV strains used as references for analysis in this study were retrieved from GenBank as follows:
Genotype 1: Bur86 (D10330); Abb-2B (AF185822); Uigh179 (D11093); FHF (X98292); Morocco (AY230202); T3-Chad (AY204877).
Genotype 2: Mexican (M74506).
Genotype 3: US1 and US2, (AF060668-69); JKN-Sap (AB074918); HE-JA10 (AB089824); JBOAR-1Hyo04 (AB189070); Osh-205 (AF455784); swArkell (AY115488).
Genotype 4: subtype 4a: swGX32 (EU366959); ChH-S-1 (EF077630); JYI-ChiSai01C (AB197674); JKO-ChiSai98C (AB197673); Sh-hu-et1 (FJ373295); XJ3-2 (EF488818-488819); subtype 4b: swDQ (DQ279091); swGX40(EU676172); Ch181 (AJ344188); LZ-105 (AF103940); subtype 4c: HE-JK4 (AB099347); JSN-Sap-FH02C, (AB200239); swJ13-1 (AB097811); subtype 4d: swCH25 (AY594199); T1-China (AJ272108); Ch108 (AJ344181); Ch202 (AJ344184); subtype 4e: IND-SW1, IND-SW2, and IND-SW3 (AF324501-3); subtype 4f: HE-JA2 (AB082558); subtype 4g: CCC220 (AB108537); subtype 4n: Ch-shw1 (EU034707); estw2 (EU375320); HC10-44 (EU620651); HC1-88 (EU620636); HK104-2004 (FJ438418).
Nucleotide sequence accession number.
The nucleotide sequences of HEV isolates reported in this study have been deposited in GenBank under these accession numbers: FJ775165–FJ775171, GQ305997–GQ306000 (the region of partial ORF1), FJ775172–FJ775178, and GQ306001–GQ306004 (the region of partial ORF2).
Results
Prevalence of anti-HEV in animal sera.
The prevalence of anti-HEV among 12 different animal species in Xinjiang is shown in Table 1, and slaughter swine were found to have the highest anti-HEV prevalence (up to 84.61%). Domestic herbivorous animals and rodents were found to have lower anti-HEV prevalence (6.50–13.77% and 0–35.14%, respectively). Among aquatic animals, the prevalence of anti-HEV in hoptoad and fishes was 0% and 1.14%, respectively.
Detection of anti-HEV and HEV RNA among Xinjiang populations.
The prevalence of anti-HEV among southern and northern Xinjiang populations was 46.42% (26/56) and 18.33% (44/240), respectively, which showed a significant difference (P < 0.05). The prevalence of anti-HEV among Muslim and non-Muslim populations was 21.01% (29/138) and 25.95% (41/158), respectively, which showed no significant difference (P > 0.05). Of 56 serum samples from southern Xinjiang, one serum sample of a 41-year-old male Chinese (Han nationality) was HEV RNA positive both in partial ORF1 and partial ORF2 fragments. This human HEV strain was designated as CHN-XJ-HE29. The clinical laboratory information of the sample is as follows: ALT = 790 IU/L, AST = 191 IU/L, TB = 233.73 μmol/L, and CB = 92.3 μmol/L. Furthermore, the sample was positive for anti-HEV but negative for immunoglobulin M antibody against hepatitis A virus (anti-HAV IgM), immunoglobulin M antibody against hepatitis B core antigen (anti-HBc IgM), antibody against hepatitis C virus (anti-HCV), and HCV RNA. Thus, it can be clinically identified as an acute sporadic hepatitis E.
Detection of HEV RNA among animal samples.
Sixty-two of one hundred swine fecal samples (62%) were positive for HEV RNA in southern Xinjiang, whereas 8 of 25 swine fecal samples (32%) were positive for HEV RNA in northern Xinjiang. All the remaining 1,771 fecal and serum samples from the other 11 distinct animal species were negative for HEV RNA.
Thirty-eight swine HEV strains (30 from southern Xinjiang and 8 from northern Xinjiang) were sequenced both in partial ORF1 and partial ORF2 fragments. Only 10 of 38 HEV strains showed distinct sequences either in partial ORF1 (244 nt) or in partial ORF2 (365 nt), and they were designated as follows: CHN-XJ-SW5, CHN-XJ-SW6, CHN-XJ-SW10, CHN-XJ-SW12, CHN-XJ-SW13, CHN-XJ-SW16, CHN-XJ-SW36, CHN-XJ-SW50, CHN-XJ-SW61, and CHN-XJ-SW66.
Phylogenetic analysis.
Nucleotide sequence alignments and percent identities were generated based on partial ORF1 and partial ORF2 fragments, respectively. In the regions of ORF1 (244 bp) and ORF2 (365 bp), 11 HEV isolates (10 swine HEV and 1 human HEV isolates) shared 83.5–100% of nucleotide identities with each other, and they shared nucleotide similarities as 71.7–79.6%, 76.9–79.4%, 72.8–80%, and 80.5–96.0% with standard HEV isolates representing genotype 1, 2, 3, and 4, respectively. Therefore, all 11 HEV isolates belonged to genotype 4.
Phylogenetic trees were constructed based on partial ORF2 (281 bp) and partial ORF1 (195 bp) fragments, respectively (Figures 1A and 1B). The phylogenetic tree (Figure 1A) showed that these 11 HEV isolates were classified into two main clades. One clade included seven HEV isolates (six swine HEV and one human HEV isolates) from southern Xinjiang, sharing identity as 91.8–97.8% with the standard HEV isolates of subtype 4a. Therefore, these seven HEV isolates belonged to subtype 4a. The other clade included four swine HEV isolates from northern Xinjiang that had formed a new subgroup with standard swine HEV isolates (HC10-44, HC1-88, estw2, ch-shsw1, and SWXJ03). The similarities within this subgroup and with other subgroups (4a–4g) in genotype 4 were 92.4–99.6% and 80.7–88.2%, respectively. Therefore, this new subgroup was considered to be a novel subtype. The phylogenetic tree based on ORF1 (195 bp) (Figure 1B) also showed that seven HEV isolates from southern Xinjiang formed one branch with standard HEV isolates of subtype 4a, and the other four swine HEV isolates from northern Xinjiang formed a new subgroup with a human HEV strain (HK104-2004) isolated from Hong Kong that shared 95.3–95.8% nucleotide identities within the subgroup.
Figure 1.
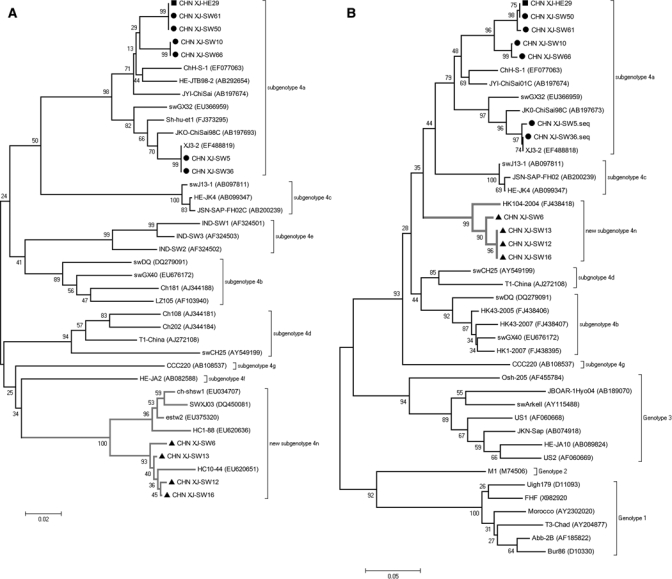
A, Phylogenetic trees depicting genotypic status of 51 HEV isolates based on partial ORF2 sequences (281 bp). B, Phylogenetic trees depicting genotypic status of 51 HEV isolates based on partial ORF1 sequences (195 bp). Human HEV and swine HEV strains from Southern Xinjiang were signed with ▪ and ▲, respectively; swine HEV strains from Northern Xinjiang were signed with •. The internal node numbers indicate the bootstrap values as a percentage of trees obtained from 1000 replicates.
Human HEV strain CHN-XJ-HE29 and swine HEV strain CHN-XJ-SW50 shared identical nucleotide sequences both in partial ORF1 and partial ORF2 fragments. In addition, the human HEV strain showed high similarities with another swine HEV isolate CHN-XJ-SW61 with 100% in ORF2 and 99.6% in ORF1, respectively.
Discussion
Twelve different animal species from Xinjiang were examined for anti-HEV in this study. Except for jerboa and hoptoad, the other 10 animal species were all positive for anti-HEV. Among them, swine showed the highest positive rate of anti-HEV (84.61%), and domestic herbivorous animals (including horses, donkeys, goats, and cows) showed a relatively low positive rate (9.92%). It has been reported that anti-HEV prevalence among horses in Egypt and eastern China is 13.0% and 16.6%, respectively, and partial HEV genomes were also identified in horses from these two areas.23,24 However, no more studies could independently confirm it, and our study failed to amplify HEV RNA from donkeys and horses in Xinjiang, although similar anti-HEV prevalence rates were found in donkeys (13.77%) and horses (11.00%). Consequently, the occasional reports of isolation HEV RNA from horses seem to represent laboratory contamination rather than genuine infection. The prevalence of anti-HEV among goats was 7.50% in this study, lower than the 24% in eastern China reported by Zhang and others.23 Experimental infection on lambs with human HEV strain (Osh-225 and Osh-228) was successfully performed by Usmanov and others,25 but so far, HEV RNA has not been detected in domestic sheep with HEV infection. Cows showed the lowest positive rate (6.50%) for anti-HEV among domestic herbivorous animals with a rate similar to the 6.00% reported in eastern China.23 This suggests that cows might be less susceptible to HEV than horses, donkeys, and goats.
Five distinct species of rodents were examined in this study, and the positive rates for anti-HEV ranged from 0.00% to 35.14%; these rates were similar to those reported of wild rats in Japan (0.00–31.5%).26 Among them, Mus musculus accounted for the highest positive rate of anti-HEV (35.14%), whereas jerboa mouse and gerbil mouse showed much lower positive rates (0% and 3.9%, respectively). It is interesting to find a wide range of anti-HEV prevalence in different species of rodents. One possible explanation for this might be different living habits in rodents. For example, Mus musculus live in towns or villages; therefore, they have a greater chance of contact with humans than gerbil mouse and jerboa mouse, which mainly live in remote grasslands or deserts. Trials to experimentally infect laboratory rats or mice with human HEV led to contradictory results, because some studies reported successful infection and others did not.27,28 Recently, an HEV-related agent was detected from wild rats, and sequence comparison to human and avian HEV strains revealed only 59.9% and 49.9% nucleotide identity, respectively.29 The low nucleotide sequence identities between rat HEV and human HEV may indicate an independent evolution of both viruses with no transmission between both hosts. However, the presence of regions with highly conserved aa sequences within the capsid protein between rat HEV and human HEV may explain the previously observed serological reactions of rodent samples to human HEV strains because of the presence of cross-reacting antibodies.29
As mentioned above, domestic herbivorous animals and some rodents showed positive for anti-HEV, but HEV RNA was not detected from these animal species in this study. This may be attributed to the low HEV RNA titer in sera and feces, the short duration of viremia and viral excretion, and the impaired RT-nPCR detection caused by HEV genetic diversity. Therefore, it is necessary to develop a highly effective broad-spectrum PCR primer to amplify the possible HEV-like agents in these animals. Moreover, by analogy with HEV-like virus in rats, it is also probable that the presence of HEV-related viruses in domestic herbivorous animals producing cross-reacting antibodies with human HEV accounts for the positivity of anti-HEV.
Hepatitis E outbreaks were considered to be associated with contaminated water. However, the overall positive rate of anti-HEV among aquatic animals (including fishes and hoptoad) was only 0.51% (1/198). This indicated that aquatic animals may not be susceptible to HEV or that HEV-like agents in aquatic animals have a long evolutionary distance from human HEV. In this way, the ELISA kits used in this study were not suitable to detect the anti-HEV antibody among aquatic animals.
Many studies have showed that swine is the largest viral reservoir of HEV. In this study, the positive rates of HEV RNA among swine in southern Xinjiang and northern Xinjiang were 62% (62/100) and 32% (8/25), respectively. From these tests, 10 swine HEV isolates with distinct nucleotide sequences were isolated. Based on sequence alignments and phylogenetic analysis, all 10 swine HEV isolates belonged to genotype 4. Among them, six swine HEV isolates from southern Xinjiang belonged to subtype 4a because of sharing high nucleotide identities of 91.8–97.8% with standard isolates from the same group. The other four strains from northern Xinjiang were clustered into a novel group (Figure 1A) along with swine HEV isolates (HC10-44, HC1-88, estw2, ch-shsw1, and SWXJ03) from Hunan Province and Shanghai. The novel group did not belong to any subtypes (4a–4g) reported by Lu and others30 but was consistent with subtype 4n reported by Li and others (unpublished data). In addition, 13 swine HEV isolates of subtype 4d were previously identified in northern Xinjiang by Xun and Cheng-ping.22 Therefore, there are at least three distinct HEV subtypes (4a, 4d, and 4n) prevailing among swine in Xinjiang.
China was generally considered as an HEV-endemic area. However, hepatitis E outbreaks were only once recorded in southern Xinjiang as opposed to other parts of the country, and HEV epidemic strains (genotype 1) have been seldom identified from any parts of the country since 2004. Therefore, with the improvement of sanitation in big cities in China, it is not appropriate to consider all of China as an HEV-endemic area. The issue was also confirmed in present study. The positive rate of anti-HEV in the general population of southern Xinjiang (46.42%) was significantly higher than that of northern Xinjiang (18.33%). This might be attributed to the hepatitis E outbreak from 1986 and 1988 in southern Xinjiang, which induced widespread subclinical HEV infection among the population in the region. On the contrary, northern Xinjiang, without a history of hepatitis E outbreaks or epidemics, showed similar positive rates to that of eastern China (17.2%).31
One human HEV strain, CHN-XJ-HE29, isolated from an individual of Han nationality with acute sporadic hepatitis E shared 100% nucleotide identity with swine HEV strain CHN-XJ-SW50. Furthermore, both the HEV isolates were identified from the same district in this study. Swine are raised and consumed by non-Muslim populations in Xinjiang, and its excreta are frequently used to fertilize crops without special treatment. Considering that consumption of undercooked meat or contact with pigs are the high risks for HEV infection,32–34 the acute sporadic hepatitis E case might correlate with the local swine. Unfortunately, the leftover food and general sanitation environment could not be examined. Yet, as an individual of Han nationality, the patient might have a great chance of contact with pigs or consumption of HEV-contaminated pork. Therefore, swine-to-human transmission of sporadic hepatitis E in an endemic area of southern Xinjiang was suggested. In addition, swine HEV CHN-XJ-SW5 and CHN-XJ-SW36 from southern Xinjiang showed high nucleotide identities (96.9–97.8%) comparable with Japanese human HEV isolates (JKO-ChiSai98C) imported from China. Additionally, four swine HEV strains isolated from northern Xinjiang shared 95.3–95.8% nucleotide identities with human HEV strain HK104-204 isolated from Hong Kong. This finding also provided molecular evidence for zoonotic transmission of hepatitis E.
In this study, the anti-HEV prevalence among Muslims seemed to be lower than among non-Muslim populations, but there was no significant difference. However, Zhang and others23 reported that there was a significantly higher prevalence of anti-HEV among Han nationality populations (73%; 147/200) than among age- and geography-matched Muslim populations (15.77%; 59/374).35 This indicates that differences in ethnic culture background and diet habits, especially consumption of pork, greatly influence the prevalence of HEV infection. However, more samples from Muslim and non-Muslim populations are needed for the further confirmation of this point of view. Although Muslims seldom have exposure to swine or its excreta, the HEV seroprevalence among this population was rather high. HEV isolates identified from Muslim populations belonged to genotype 1, but the isolates from animals were restricted to genotypes 3 and 4. The high degree of genetic diversity between them indicated that HEV infections among Muslim populations were rarely associated with animals. We now consequently suggest that individuals with unapparent infections may be a continuously circulating pool of HEV infection among Muslims, and it is the poor sanitation in Xinjiang that makes the existence of the HEV circulation possible.
In summary, HEV genotype 4 has been the dominant strain among swine and individuals of non-Muslin populations in Xinjiang. This study suggested the possibility of swine-to-human transmission of sporadic hepatitis E in an endemic area.
Acknowledgment
We are grateful to Mr. Melvin Fong Wai Hon for proofreading the manuscript.
Footnotes
Financial support: This work was funded by the National Natural Science Foundation of China (Grant 30570063) and the 863 National High Technology Research and Development Program of China (Grant 2006A A02Z453).
Authors' addresses: Hongwei Fu, Lingjun Li, Yonghong Zhu, Ling Wang, Jiabao Geng, Yibin Chang, and Hui Zhuang, Department of Microbiology, Peking University Health Science Center, Beijing, China, E-mails: frankson@bjmu.edu.cn, lingjunli@nus.edu.sg, zhuyh@bjmu.edu.cn, lingwang@bjmu.edu.cn, Gengjiabao666@126.com, Ningbing265@163.com, and zhuangbmu@126.com. Cheng Xue, Clinical Laboratory, People's Hospital of Qiemo County, Qiemo, Xinjiang Uighur Autonomous Region, China, E-mail: xuecheng@163.com. Gang Du and Yaning Li, Clinical Laboratory, Central Hospital of Kelamayi, Xinjiang Uighur Autonomous Region, Kelamayi, China, E-mails: dugang12345678@126.com and liyaning1220@163.com.
References
- 1.Dalton HR, Bendall R, Ijaz S, Banks M. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis. 2008;8:698–709. doi: 10.1016/S1473-3099(08)70255-X. [DOI] [PubMed] [Google Scholar]
- 2.Mushahwar IK. Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J Med Virol. 2008;80:646–658. doi: 10.1002/jmv.21116. [DOI] [PubMed] [Google Scholar]
- 3.Meng XJ. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol. 2009;140:256–265. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aye TT, Uchida T, Ma XZ, Iida F, Shikata T, Zhuang H, Win KM. Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986–1988) of China. Nucleic Acids Res. 1992;20:3512. doi: 10.1093/nar/20.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang H, Li K, Zhu W. Partial nucleotide sequences of hepatitis E viruses isolated from patients in 14 cities of China. Zhonghua Yixue Zazhi. 2000;80:893–896. [PubMed] [Google Scholar]
- 6.Li RC, Ge SX, Li YP, Zheng YJ, Nong Y, Guo QS, Zhang J, Ng MH, Xia NS. Seroprevalence of hepatitis E virus infection, rural southern People's Republic of China. Emerg Infect Dis. 2006;12:1682–1688. doi: 10.3201/eid1211.060332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Y, Ge S, Zhang J, Guo Q, Ng MH, Wang F, Xia N, Jiang Q. Swine as a principal reservoir of hepatitis E virus that infects humans in eastern China. J Infect Dis. 2006;193:1643–1649. doi: 10.1086/504293. [DOI] [PubMed] [Google Scholar]
- 8.Ning H, Yu S, Zhu Y, Dong S, Yu R, Shen S, Niu Z, Li Z. Genotype 3 hepatitis E has been widespread in pig farms of Shanghai suburbs. Vet Microbiol. 2008;126:257–263. doi: 10.1016/j.vetmic.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, Haynes JS, Thacker BJ, Emerson SU. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishizawa T, Takahashi M, Mizuo H, Miyajima H, Gotanda Y, Okamoto H. Characterization of Japanese swine and human hepatitis E virus isolates of genotype IV with 99% identity over the entire genome. J Gen Virol. 2003;84:1245–1251. doi: 10.1099/vir.0.19052-0. [DOI] [PubMed] [Google Scholar]
- 11.Ji Y, Zhu Y, Liang J, Wei X, Yang X, Wang L, Li L, Chang Y, Tang R, Zhuang H. Swine hepatitis E virus in rural southern China: genetic characterization and experimental infection in rhesus monkeys (Macaca mulatta) J Gastroenterol. 2008;43:565–570. doi: 10.1007/s00535-008-2196-3. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Zhu Y, Fu H, Wei X, Wang L, Liang J, Ji Y, Tang R, Zhuang H. Full-genome nucleotide sequence and analysis of a Chinese swine hepatitis E virus isolate of genotype 4 identified in the Guangxi Zhuang autonomous region: evidence of zoonotic risk from swine to human in South China. Liver Int. 2009;29:1230–1240. doi: 10.1111/j.1478-3231.2009.02012.x. [DOI] [PubMed] [Google Scholar]
- 13.Shukla P, Chauhan UK, Naik S, Anderson D, Aggarwal R. Hepatitis E virus infection among animals in northern India: an unlikely source of human disease. J Viral Hepat. 2007;14:310–317. doi: 10.1111/j.1365-2893.2006.00815.x. [DOI] [PubMed] [Google Scholar]
- 14.Arankalle VA, Chobe LP, Joshi MV, Chadha MS, Kundu B, Walimbe AM. Human and swine hepatitis E viruses from western India belong to different genotypes. J Hepatol. 2002;36:417–425. doi: 10.1016/s0168-8278(01)00297-5. [DOI] [PubMed] [Google Scholar]
- 15.Wang YC, Zhang HY, Xia NS, Peng G, Lan HY, Zhuang H, Zhu YH, Li SW, Tian KG, Gu WJ, Lin JX, Wu X, Li HM, Harrison TJ. Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J Med Virol. 2002;67:516–521. doi: 10.1002/jmv.10131. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto H, Takahashi M, Nishizawa T, Usui R, Kobayashi E. Presence of antibodies to hepatitis E virus in Japanese pet cats. Infection. 2004;32:57–58. doi: 10.1007/s15010-004-3078-0. [DOI] [PubMed] [Google Scholar]
- 17.Sonoda H, Abe M, Sugimoto T, Sato Y, Bando M, Fukui E, Mizuo H, Takahashi M, Nishizawa T, Okamoto H. Prevalence of hepatitis E virus (HEV) Infection in wild boars and deer and genetic identification of a genotype 3 HEV from a boar in Japan. J Clin Microbiol. 2004;42:5371–5374. doi: 10.1128/JCM.42.11.5371-5374.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto H. Genetic variability and evolution of hepatitis E virus. Virus Res. 2007;127:216–228. doi: 10.1016/j.virusres.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura M, Takahashi K, Taira K, Taira M, Ohno A, Sakugawa H, Arai M, Mishiro S. Hepatitis E virus infection in wild mongooses of Okinawa, Japan: demonstration of anti-HEV antibodies and a full-genome nucleotide sequence. Hepatol Res. 2006;34:137–140. doi: 10.1016/j.hepres.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Huang RT, Li DR, Wei J, Huang XR, Yuan XT, Tian X. Isolation and identification of hepatitis E virus in Xinjiang, China. J Gen Virol. 1992;73:1143–1148. doi: 10.1099/0022-1317-73-5-1143. [DOI] [PubMed] [Google Scholar]
- 21.Liang JR, Wei X-F, Xue C, Ji Y-L, Zhu Y-H. Genotype and subgenotype of swine HEV. Zhonghua Weishengwuxue He Mianyixue Zazhi. 2007;27:747–750. [Google Scholar]
- 22.Xun MA, Cheng-ping LU. Analysis of the complete genome of a strain swCH25 of swine hepatitis E virus isolated from Xinjiang. Zhongguo Nongye Kexue. 2005;38:1669–1774. [Google Scholar]
- 23.Zhang W, Shen Q, Mou J, Gong G, Yang Z, Cui L, Zhu J, Ju G, Hua X. Hepatitis E virus infection among domestic animals in eastern China. Zoonoses Public Health. 2008;55:291–298. doi: 10.1111/j.1863-2378.2008.01136.x. [DOI] [PubMed] [Google Scholar]
- 24.Saad MD, Hussein HA, Bashandy MM, Kamel HH, Earhart KC, Fryauff DJ, Younan M, Mohamed AH. Hepatitis E virus infection in work horses in Egypt. Infect Genet Evol. 2007;7:368–373. doi: 10.1016/j.meegid.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Usmanov RK, Balaian MS, Dvoinikova OV, Alymbaeva DB, Zamiatina NA, Kazachkov IA, Belov VI. An experimental infection in lambs by the hepatitis E virus. Vopr Virusol. 1994;39:165–168. [PubMed] [Google Scholar]
- 26.Hirano M, Ding X, Li TC, Takeda N, Kawabata H, Koizumi N, Kadosaka T, Goto I, Masuzawa T, Nakamura M, Taira K, Kuroki T, Tanikawa T, Watanabe H, Abe K. Evidence for widespread infection of hepatitis E virus among wild rats in Japan. Hepatol Res. 2003;27:1–5. doi: 10.1016/s1386-6346(03)00192-x. [DOI] [PubMed] [Google Scholar]
- 27.Maneerat Y, Clayson ET, Myint KS, Young GD, Innis BL. Experimental infection of the laboratory rat with the hepatitis E virus. J Med Virol. 1996;48:121–128. doi: 10.1002/(SICI)1096-9071(199602)48:2<121::AID-JMV1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 28.Li TC, Suzaki Y, Ami Y, Tsunemitsu H, Miyamura T, Takeda N. Mice are not susceptible to hepatitis E virus infection. J Vet Med Sci. 2008;70:1359–1362. doi: 10.1292/jvms.70.1359. [DOI] [PubMed] [Google Scholar]
- 29.Johne R, Plenge-Bonig A, Hess M, Ulrich RG, Reetz J, Schielke A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol. 2010;91:750–758. doi: 10.1099/vir.0.016584-0. [DOI] [PubMed] [Google Scholar]
- 30.Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- 31.Dong C, Dai X, Shao JS, Hu K, Meng JH. Identification of genetic diversity of hepatitis E virus (HEV) and determination of the seroprevalence of HEV in eastern China. Arch Virol. 2007;152:739–746. doi: 10.1007/s00705-006-0882-0. [DOI] [PubMed] [Google Scholar]
- 32.Meng XJ, Wiseman B, Elvinger F, Guenette DK, Toth TE, Engle RE, Emerson SU, Purcell RH. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J Clin Microbiol. 2002;40:117–122. doi: 10.1128/JCM.40.1.117-122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsubayashi K, Kang JH, Sakata H, Takahashi K, Shindo M, Kato M, Sato S, Kato T, Nishimori H, Tsuji K, Maguchi H, Yoshida J, Maekubo H, Mishiro S, Ikeda H. A case of transfusion-transmitted hepatitis E caused by blood from a donor infected with hepatitis E virus via zoonotic food-borne route. Transfusion. 2008;48:1368–1375. doi: 10.1111/j.1537-2995.2008.01722.x. [DOI] [PubMed] [Google Scholar]
- 34.Kulkarni MA, Arankalle VA. The detection and characterization of hepatitis E virus in pig livers from retail markets of India. J Med Virol. 2008;80:1387–1390. doi: 10.1002/jmv.21220. [DOI] [PubMed] [Google Scholar]
- 35.Zhang CX, Li W, Xiang Y, Arsya Investigation on the distribution of HEV in different nationalities and animals from Urumqi. Difangbing Tongbao. 2003;18:33–35. [Google Scholar]


