Abstract
Rats are known to be the most important reservoirs and transmission sources of leptospirosis. However, the status of leptospirosis in the Philippines regarding reservoirs and transmission remains unknown. A survey was conducted in Metro Manila and Laguna that analyzed samples obtained from 106 rats. Using the microscopic agglutination test, we found that 92% of rat serum samples were positive for anti-Leptospira antibodies; the most common infecting serovars were Manilae, Hebdomadis, and Losbanos. On the basis of pulsed-field gel electrophoresis and gyrase B gene sequence analyses, four groups of rat kidney isolates were found: L. interrogans serovar Manilae, serovar Losbanos, and serogroup Grippotyphosa, and L. borgpetersenii serogroup Javanica. Most isolates were lethal after experimental infection of golden Syrian hamsters. Results showed that these four Leptospira serovars and serogroups are circulating among rats, and that these animals may be one of the possible transmission sources of leptospirosis in the Philippines.
Introduction
Leptospirosis is a worldwide zoonosis that is common in countries with humid tropical and sub-tropical climates. It is also known to be a well-represented zoonotic infection occurring in humans and animals such as rats, dogs, water buffaloes, horses, and pigs.1 It is caused by spirochetes belonging to the pathogenic species of Leptospira. Humans and susceptible animals acquire this infection through direct contact with urine or body fluids of infected animals, especially rodents, or indirect contact with contaminated environments.2,3 Rodents, especially rats, are considered to be the most important reservoirs or maintenance hosts of Leptospira.3,4 Pathogenic leptospires that infect these maintenance hosts persist in their kidneys causing either slight or no harm. However, they cause mild to severe illness, and even death to susceptible hosts such as humans and other animals (also called accidental or incidental hosts).2
As with other bacterial diagnosis, isolation of leptospires is considered to be the gold standard in diagnosing leptospirosis. However, because isolation of leptospires is usually difficult, the microscopic agglutination test (MAT) is widely used as the reference test for leptospirosis diagnosis and is considered to be a useful tool in epidemiologic studies or surveillance on leptospirosis.2,5 Identification of serovars of Leptospira isolates is carried out by cross-agglutinin absorption test (CAAT). However, this method and the MAT are laborious, time-consuming, and require extensive collection of reference strains and their corresponding rabbit antisera.6,7 It is for these reasons that different molecular techniques were and are currently being established to classify isolates rapidly and with less labor. These molecular techniques and the MAT and CAAT have been used in the characterization of Leptospira isolates and in leptospirosis surveillance.1,8–31
Some of the DNA-based or molecular classification methods that have been developed and used for these purposes and for diagnosing leptospirosis are ribotyping,6,32 pulsed-field gel electrophoresis (PFGE),6,32 multilocus sequence typing,6,33–35 amplification and sequencing of different genes such as 16S ribosomal RNA,6,32 23S ribosomal RNA,6,32 rpoB,6 ligB,6 flaB,27,36 gyrB,6,26 and others.6,32 Pulsed-field gel electrophoresis has been widely used in epidemiologic studies of bacterial infections.37 It was also used to differentiate closely related serovars in genus Leptospira. DNA gyrase (topoisomerase type II) is one of the housekeeping enzymes found among bacteria, and analysis using its B subunit gene (gyrB) was found to differentiate closely related bacterial strains.38 Therefore, in our current study, we used PFGE and gyrB sequence analysis in the genotyping of rat isolates. Genotypic classification has grouped leptospires into 20 genomospecies based on DNA relatedness.6,39–41
The Philippines is known to be endemic for leptospirosis, with the Department of Health of the Philippines listing it as one of the priority re-emerging diseases that needs to be addressed.42 Most studies conducted on leptospirosis in the Philippines used the MAT to identify the prevailing Leptospira serovars among humans and animals.8–12,19,20 Antibodies against Leptospira were detected in serum samples of humans1,10–14,19,20 and animals.1,8,9,15,16,43 Some of these studies also reported isolation of strains. There are currently three well-characterized Leptospira isolates from the Philippines.44 These are strains LT 398 (L. interrogans serovar Manilae), LT 101-69 (L. interrogans serovar Losbanos), and C3 (L. interrogans serovar Carlos). The first two isolates were isolated from rats in Manila and Laguna (i.e., Los Baños), respectively, and the third strain was isolated from a toad. Although a number of studies have been conducted on leptospirosis in the Philippines, the status of leptospirosis in the Philippines in terms of prevalence, incidence, reservoirs, transmission, and pathogenicity is unclear. There is also little knowledge about the rodent–Leptospira relationship in the Philippines. Therefore, the purpose of our study was to identify and characterize prevailing Leptospira serovars among rats in the Philippines.
Materials and Methods
Description of study areas.
Rats were trapped in selected areas in Metro Manila (Manila, Pasay, Quezon, Makati, and Las Piñas) and in Laguna (Bay and Victoria), the Philippines. These two areas are where two previously serotyped strains were isolated (LT 398 and LT 101-69). Metro Manila refers to the metropolitan area, which is composed of the capital city (Manila) plus 16 surrounding cities and 1 municipality. It is the most populous area in the Philippines and is considered to be the commercial, industrial, and financial center of the country. Laguna is a province located southeast of Metro Manila. Part of this province is industrialized and towns in this province are still engaged in agricultural or cottage industries.
Trapping of rats and sample collection.
Trapping of rats was done during August 2006–May 2007. Rats were trapped in markets, commercial establishments (e.g., eateries), household areas, ricefields, and haystack storage rooms. Wire-mesh traps with baits were left overnight in the aforementioned trapping sites. The rats were anesthetized by using diethyl ether. Serum samples were collected for MAT and kidneys were obtained for Leptospira isolation.
Microscopic agglutination test.
The MAT was conducted as described2 by using a panel of 17 live leptospires representing 13 serovars and 5 species (Table 1). Three of the strains used in this study were local (Philippine) isolates such as LT 398 (serovar Manilae),44 LT 101-69 (serovar Losbanos),44 and UP-BL-FR13 (serovar Ratnapura, Yanagihara Y and others, unpublished data). The endpoint titer is the final serum-antigen dilution giving a > 50% agglutination at a titer ≥ 1:20. The cut-off titer was set at this level because in our previous experiment with specific pathogen–free rats, no antibodies against the panel of antigens were detected at this titer. Serum samples that did not show any reactivity were considered MAT-negative. Because our study involves isolation of leptospires from rats, we could only collect single, not paired, samples. Therefore, in this study, the serovar with the highest titer was considered to be the presumptive infecting serovar45,46 and serum with this result was classified as being infected with single Leptospira serovar. However, if two or more serovars induced the highest titer, then the infection was considered to be probably caused by multiple serovars.
Table 1.
Leptospira strains used in this study, the Philippines
| Species | Serogroup | Serovar | Strain |
|---|---|---|---|
| L. interrogans | Pyrogenes | Manilae | LT 398* |
| Pyrogenes | Pyrogenes | Salinem | |
| Canicola | Canicola | Hond Utrecht IV | |
| Autumnalis | Autumnalis | Akiyami A | |
| Bataviae | Losbanos | LT 101-69* | |
| Hebdomadis | Hebdomadis | Akiyami B | |
| Australis | Australis | Akiyami C | |
| Icterohaemorrhagiae | Copenhageni | M20 | |
| Icterohaemorrhagiae | Icterohaemorrhagiae | Ictero No. 1 | |
| Pomona | Pomona | Pomona | |
| Sejroe | Hardjo | Hardjoprajitno | |
| L. borgpetersenii | Tarassovi | Tarassovi | Perepelitsin |
| Javanica | Poi | Poi | |
| L. kirschneri | Grippotyphosa | Ratnapura | UP-BL-FR13* |
| Grippotyphosa | Grippotyphosa | Moskva V | |
| L. meyeri | Semaranga | Semaranga | Veldrat Semaranga 173 |
| L. biflexa | Semaranga | Patoc | Patoc I |
Isolates from the Philippines (local isolates).
Isolation of leptospires.
Rat kidneys were aseptically removed, macerated with a 2.5-mL syringe, and cultured in 5-fluorouracil–containing modified Korthof's medium supplemented with 10% rabbit serum and 5-fluorouracil–containing Ellinghausen, McCullough, Johnson, Harris (EMJH) medium. These cultures were incubated overnight at 30°C. The next day, 500 μL of the culture supernatant was sub-cultured into fresh medium and were kept at 30°C and examined for growth of leptospires weekly for three months.
Typing of rat isolates.
Pulsed-field gel electrophoresis analysis of Leptospira isolates.
Pulsed-field gel electrophoresis was carried out based on the method previously described.28 Briefly, DNA from 1 mL of Leptospira isolates was digested with Not I and subjected to electrophoresis in CHEF DR III (Bio-Rad, Hercules, CA) for 20 hr at 14°C with recirculating 0.5× Tris-borate-EDTA buffer. Electrophoresis conditions included switch times of 10 seconds and 60 seconds for L. interrogans or 10 seconds and 40 seconds for L. borgpetersenii, an angle of 120°, and a gradient of 6 V/cm. The PFGE band patterns of different rat isolates and reference Leptospira strains were then compared.
Gyrase B gene sequence analysis of rat isolates.
Polymerase chain reaction amplification of the DNA gyrase B subunit gene (gyrB) was performed by using primer sets UP1TL and UP2rTL as described in Table 2 under the following conditions: 40 cycles at 95°C (denaturing) for 30 seconds, 48°C (annealing) for 30 seconds, and 72°C (extension) for 60 seconds. Direct cycle sequencing was performed by using a BigDye termination version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). For the sequencing of gyrB, primers shown in Table 2 were used. DNA sequencing analysis was performed by using the 3130 genetic analyzer (Applied Biosystems). Alignment of sequences and Neighbor-joining (NJ) phylogenetic tree construction and bootstrap analysis were carried out by using the Lasergene version 7.1 software package (DNASTAR Madison, WI). The NJ tree constructed was rearranged by using the software NJ plot (www.ddbj.nig.ac.jp). The gyrB sequences of Leptospira strains and isolates in the Philippines have been deposited and assigned DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank accession numbers AB45524–AB455527.
Table 2.
Polymerase chain reaction and sequence primers of gyrB gene of Leptospira strains, the Philippines
| Primer | Target gene* | Sequence (5′→3′)† | Position (accession no.) | Reference |
|---|---|---|---|---|
| UP1TL‡§ | gyrB | CAYGCNGGNGGNAARTTYGA | 301–320 (AF434658) | 50 |
| UP2rTL‡§ | gyrB | TCNACRTCNGCRTCNGTCAT | 1520–1501 (AF434658) | 50 |
| LgyrF§ | gyrB | GGTCTTTCCGGAGAAGATG | 940–958 (AF434658) | 29 |
| LgyrR§ | gyrB | GAATTGAATTGAGGTTGAGG | 1016–997 (AF434658) | 29 |
gyrB = gyrase B.
D = A, G, or T; M = A or C; N = A, G, T, or C; R = A or G; V = A, C, or G; W = A or T; Y = C or T.
This primer was used for amplification of DNA fragment on the target gene.
This primer was used only for sequencing of amplified DNA fragments.
Identification of serogroups of rat isolates.
Serogroups of the isolates were identified by the MAT by using a panel of anti-Leptospira rabbit sera for 23 serovars.
Pathogenicity of isolates in golden Syrian hamsters.
Pathogenicity of isolates from rat kidney cultures was tested in four-week old male golden Syrian hamsters (Japan SLC, Inc., Shizuoka, Japan). Bacterial concentrations of 105 or 107/mL of each strain were injected intraperitoneally and the animals were observed for 21 days.47–49 Hamsters inoculated with either Korthof's or EMJH media only were used as negative controls. Kidney samples from dead or killed hamsters were cultured as mentioned above.
Animal ethics.
All animal experiments were reviewed and approved by the Ethics Committee on Animal Experiment at the Faculty of Medical Sciences, Kyushu University and the Ethics Committee of the University of the Philippines in Manila. The experiments were carried out under the conditions stipulated in the Guideline for Animal Experiments of Kyushu University and The Law (No. 105) and Notification (No. 6) of the Government of Japan.
Statistical analysis.
The chi-square test of homogeneity was used to test whether there was a difference in the MAT-positive rate for serum samples and the virulence of isolates from rats obtained in Metro Manila and Laguna. P values < 0.05 were considered significant. Survival analysis was conducted by using the Kaplan-Meier and log rank tests.
Results
Microscopic agglutination test.
A total of 106 rats were caught in Metro Manila (56 rats) and Laguna (50 rats). Antibodies were detected in 98 (92%) of 106 rat serum samples in Metro Manila and Laguna, and reciprocal agglutination titers ranged from 20 to 5,120 (Table 3). Seventy-six of these samples reacted with 1 Leptospira serovar and 22 samples reacted with multiple serovars. Most of the rats had antibodies against serovar Manilae (15), followed by serovars Hebdomadis (11), and Losbanos (9). Eight of the 106 rats were MAT-negative (i.e., non-reactive with any of the panel of antigens used).
Table 3.
MAT results of 106 rat serum samples, the Philippines*
| Serovar | Area | No. serum samples with specific agglutination titers | No. positive | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | 40 | 80 | 160 | 320 | 640 | 1, 280 | 2, 560 | 5, 120 | |||
| Single serovar | |||||||||||
| Manilae | MM | 1 | 1 | 1 | 4 | 2 | 4 | 1 | 1 | 15 | |
| L | 0 | ||||||||||
| Hebdomadis | MM | 2 | 2 | 1 | 1 | 1 | 1 | 8 | |||
| L | 1 | 1 | 1 | 3 | |||||||
| Losbanos | MM | 3 | 2 | 2 | 2 | 9 | |||||
| L | 0 | ||||||||||
| Ratnapura | MM | 0 | |||||||||
| L | 2 | 3 | 1 | 1 | 7 | ||||||
| Poi | MM | 1 | 1 | 2 | |||||||
| L | 1 | 3 | 1 | 5 | |||||||
| Pomona | MM | 1 | 1 | 1 | 3 | ||||||
| L | 1 | 1 | 2 | ||||||||
| Hardjo | MM | 1 | 1 | 2 | |||||||
| L | 2 | 1 | 3 | ||||||||
| Semaranga | MM | 0 | |||||||||
| L | 1 | 1 | 1 | 1 | 4 | ||||||
| Tarassovi | MM | 1 | 1 | 2 | |||||||
| L | 1 | 1 | |||||||||
| Icterohaemorrhagiae | MM | 1 | 1 | ||||||||
| L | 1 | 1 | 2 | ||||||||
| Patoc | MM | 1 | 1 | ||||||||
| L | 1 | 1 | |||||||||
| Australis | MM | 1 | 1 | ||||||||
| L | 0 | ||||||||||
| Grippotyphosa | MM | 1 | 1 | ||||||||
| L | 0 | ||||||||||
| Canicola | MM | 0 | |||||||||
| L | 1 | 1 | |||||||||
| Autumnalis | MM | 0 | |||||||||
| L | 1 | 1 | |||||||||
| Copenhageni | MM | 1 | 1 | ||||||||
| L | 0 | ||||||||||
| Pyrogenes | MM | 0 | |||||||||
| L | 0 | ||||||||||
| Multiple serovars | MM | 3 | 1 | 2 | 1 | 1 | 8 | ||||
| L | 13 | 1 | 14 | ||||||||
| Sub-total | MM | 5 | 4 | 7 | 10 | 10 | 6 | 7 | 4 | 1 | 54 |
| L | 14 | 6 | 6 | 8 | 4 | 3 | 3 | 44 | |||
| Total | 19 | 4 | 13 | 16 | 18 | 10 | 10 | 7 | 1 | 98 | |
There were 8 MAT-negative serum samples. MAT = microscopic agglutination test; MM = Metro Manila; L = Laguna.
Antibodies were more frequently detected in rats from Metro Manila (54 of 56, 96%) than in those from Laguna (44 of 50, 88%). However, the difference in MAT-positive rates in these two areas was not statistically significant (χ2 = 2.6894, P = 0.101). Some of the reactive serovars among rats from Metro Manila were different from those in Laguna. Antibodies against serovars Manilae and Losbanos were detected among serum samples of rats in Metro Manila, but no antibodies against these serovars were found among rats caught in Laguna. Conversely, antibodies against serovars Ratnapura and Semaranga were present among rats from Laguna but not among rats from Metro Manila (Table 3).
Isolation of Leptospira from rat kidneys.
Results of leptospiral isolation from the kidneys of 106 rats are shown in Table 4. Leptospires were isolated from the kidneys of 46 rats (43%). Twenty-nine (63%) of the isolates were from rats in Metro Manila and 17 (37%) were from Laguna. As mentioned in the Materials and Methods, rat kidneys were cultured in both liquid modified Korthof's and EMJH media. One rat from Metro Manila and three from Laguna had mixed infections because the isolates that grew in Korthof's medium were serovar Manilae and those that grew in EMJH medium were serogroup Javanica.
Table 4.
Leptospira isolates classified by gyrase B gene sequencing and pulsed-field gel electrophoresis*
| Location | No. (%) of rats† | No. of isolates identified | |||
|---|---|---|---|---|---|
| Group A L. interrogans (sv Manilae) | Group B L. interrogans (sv Losbanos) | Group C L. interrogans (sg Grippotyphosa) | Group D L. borgpetersenii (sg Javanica) | ||
| Metro Manila | 29/56 (52) | 16 | 10 | 0 | 4 |
| Laguna | 17/50 (34) | 0 | 0 | 7 | 13 |
| Total | 46/106‡ (43) | 16 | 10 | 7 | 17 |
sv = serovar; sg = serogroup.
No. of rats with positive kidney culture/total no. of captured rats.
Forty-six rat kidneys were culture-positive. However, four rats had isolates that were of different serovars when cultured in Korthof's medium and in Ellinghausen, McCullough, Johnson, Harris medium. Therefore, there were 46 culture-positive rat kidneys but 50 isolates.
Typing of isolates.
PFGE analysis of Leptospira isolates.
PFGE analysis of different rat isolates showed that the band patterns were categorized into four groups (groups A–D). The Not I digestion patterns of groups A, B, and D were identical or similar to L. interrogans serovars Manilae and Losbanos and some strains of L. borgpetersenii serogroup Javanica, respectively (Figure 1 and Table 4). The smaller DNA fragments of the isolates from group C were similar to those of serovar Manilae (Figure 1 and Table 4), whereas their larger DNA fragments were different.
Figure 1.
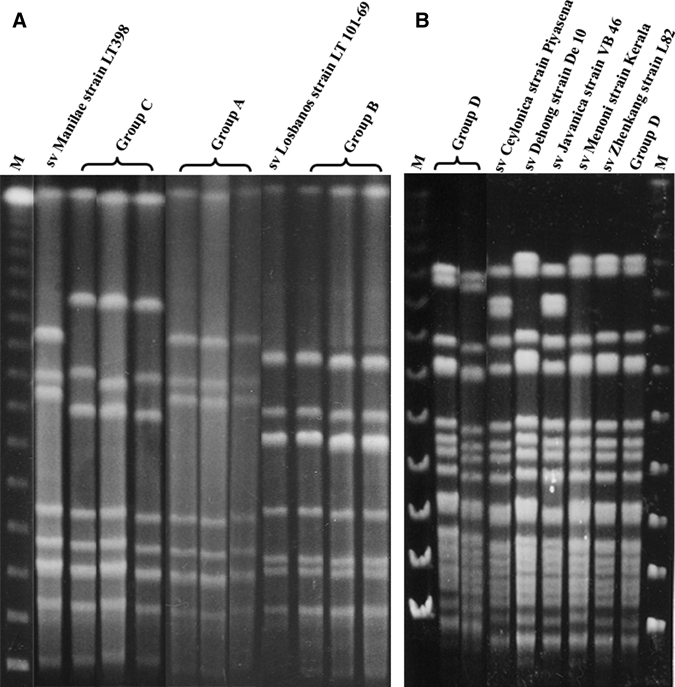
Representative pulse-filed gel electrophoresis band patterns of the rat kidney isolates and reference strains, the Philippines. Group A = rat kidney isolates (represented by K59, K60, and K64) having identical band patterns as Leptospira interrogans serovar Manilae strain LT 398. Group B = rat kidney isolates (represented by K37, E38, and E41) having the same band patterns as L. interrogans serovar Losbanos strain LT 101-69. Group C = rat kidney isolates (represented by K5, E7, and K16) with band patterns of smaller DNA similar to serovar Manilae; isolates were classified as L. interrogans serogroup Grippotyphosa. Group D = rat kidney isolates (represented by K6, K35, and E63) with band patterns identical to several strains belonging to L. borgpetersenii serogroup Javanica. M = lambda marker; sv = serovar; VB 46 = Veldrat Batavia 46.
Gyrase B sequence analysis of rat isolates.
Leptospira isolates in the Philippines were phylogenetically characterized by the DNA gyrB gene. These isolates were classified into four groups, three of which were L. interrogans and one was L. borgpetersenii (Figure 2 and Table 4). Strains belonging to groups A (represented by isolate K59) and B (represented by isolate E37) were classified into L. interrogans and had sequences identical to those of serovar Manilae strain LT 398 and serovar Losbanos strain LT 101-69, respectively. The strains belonging to group C (represented by isolate E5) had a sequence of a one-base substitution identical to that of serovar Manilae. Strains identified as L. borgpetersenii (group D, represented by isolate E10) showed identical sequences to that of L. borgpetersenii serovars Ceylonica strain Piyasena, Dehong strain De10, Javanica strain Veldrat Batavia 46, Menoni strain Kerala, and Zhenkang strain L82.
Figure 2.
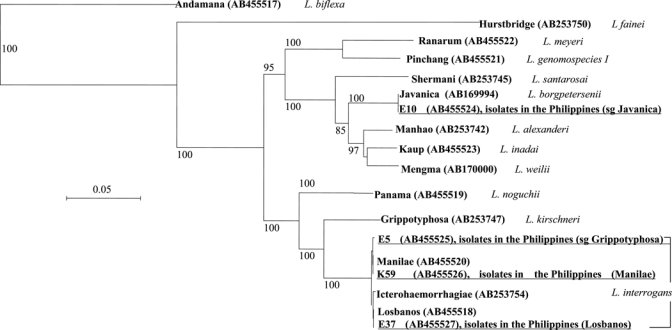
Phylogenetic tree based on gyrase B gene sequences. Sequence accession numbers are indicated in parenthesis. Underlined strains represent the isolates obtained in this study. Bar = sequence divergence of 5%. sg = serogroup.
Identification of serogroups of rat isolates.
Serogroups of the isolates were identified by MAT by using a panel of anti-Leptospira rabbit sera for 23 serovars. Strains from group A reacted with antisera for serovars Manilae and Pyrogenes (both serovars belong to serogroup Pyrogenes), and the PFGE patterns of these isolates were identical to that of the reference strain of serovar Manilae (Figure 1). Thus, strains belonging to group A were identified as L. interrogans serovar Manilae. Strains from group B reacted with antisera for serovars Losbanos and Bataviae (both serovars belong to serogroup Bataviae). The PFGE pattern of group B isolates was identical to that of the Losbanos reference strain (Figure 1) and serovar Bataviae.7 Therefore, we conducted an absorption test. Antisera that absorbed with group B isolates reacted with the serovar Bataviae reference strain but not with the Losbanos reference strain, indicating that the isolates were L. interrogans serovar Losbanos. Group C strains reacted with antiserum for serovar Grippotyphosa, but the PFGE pattern of the isolates was different from that of serovar Grippotyphosa reference strain (data not shown). Therefore, group C strains were identified as L. interrogans serogroup Grippotyphosa. Isolates from group D were reacted with antisera for serogroup Javanica (serovars Javanica and Poi). The PFGE pattern of group D strains was identical to some reference strains belonging to serogroup Javanica. Because we did not carry out CAAT, group D strains were identified as L. borgpetersenii serogroup Javanica. The number of isolates belonging to groups A–D was 16, 10, 7, and 17, respectively (Table 4).
Pathogenicity of isolates in golden Syrian hamsters.
The median survival time of hamsters experimentally infected with the 105 bacteria/mL dose of rat kidney isolates from Metro Manila was 12 days and the time for hamsters infected with isolates from Laguna was 14 days (Table 5). However, the difference was not statistically significant (χ2 = 0.0031, P = 0.956). For hamsters infected with the 107 bacteria/mL dose of isolates from Metro Manila, the median survival time was 9 days. For hamsters infected with isolates from Laguna, the time was 12 days. There was no statistically significant difference between these median survival times (χ2 = 3.183, P 0.074).
Table 5.
Pathogenicity of Leptospira isolates in golden Syrian hamsters, the Philippines
| Species | Serovar or serogroup | Challenge dose | Days to death post-infection* | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15–20 | ≥ 21† | |||
| Metro Manila | |||||||||||||||
| L. interrogans | Manilae | 105 | • | ••• | • | • | ••• | • | • | ••••○ | |||||
| 107 | •••••• | ••• | • | • | • | • | • | •• | |||||||
| Losbanos | 105 | •○ | • | •○ | • | ○ | ••• | ||||||||
| 107 | • | ••• | •• | •○ | •• | ||||||||||
| L. borgpetersenii | Javanica | 105 | • | •• | |||||||||||
| 107 | • | ○ | ○ | ||||||||||||
| Laguna | |||||||||||||||
| L. interrogans | Grippotyphosa | 105 | • | ••• | • | • | |||||||||
| 107 | ••• | •• | • | ||||||||||||
| L. borgpetersenii | Javanica | 105 | • | • | • | • | • | ••••○○ | |||||||
| 107 | •• | •• | •••••○○ | ||||||||||||
Individual hamsters from which Leptospira were recovered (•) or not recovered (○) from kidney cultures.
Survived ≥ 21 days post-infection.
Leptospires were recovered from the kidneys of most dead hamsters (Table 5). However, we isolated leptospires from the kidneys of some of the hamsters that survived.
On the basis of the log rank test on the survival of infected hamsters, the estimated median survival time of hamsters infected with the 107 bacteria/mL dose of isolates from Metro Manila was not different from that of isolates from Laguna (χ2 = 2.307, P = 0.129). There was also no difference observed in the survival of hamsters infected with the 105 bacteria/mL dose of isolates from both areas (χ2 = 0.000, P = 0.990).
Discussion
The Philippines is known to be one of the countries in Southeast Asia endemic for leptospirosis. However, little is known about the status of leptospirosis in this country in terms of its prevalence and incidence among humans and animals, circulating/prevailing Leptospira species and serovars, reservoirs, transmission, and pathogenicity. The purpose of this study was to obtain information on the status of leptospirosis in the Philippines in terms of its prevalence among rats. Evidence strongly suggests that rats are the most important reservoir of leptospires. Although these animals may harbor the organisms, they do not get sick or die of leptospirosis.2 However, they become chronically infected and continuously shed the organisms for more than seven months,47 thereby contaminating the environment and making it possible for the leptospires to come in contact with other animals or humans who are exposed to this environment.2,3
Our study provided information regarding the infecting Leptospira serovars and serogroups among rats in the Philippines on the basis of results of different tests. This study is the first published study on leptospirosis in the Philippines in which isolates were typed according to PFGE and gyrB gene sequence analysis. The pathogenicity test of rat isolates in hamsters also provided information on the virulence of isolates from the Philippines, which was only previously reported in a study by Yanagihara and others.12
In diagnosing leptospirosis by using MAT, testing of paired serum samples is ideal for discriminate current or past infection and cross-reactions. However, in animal studies, it is often impossible to collect and test paired sera. Thus, animal studies are sometimes limited to the collection and testing of single serum samples. In our study, Leptospira serovars that had the highest reactivity (i.e., titer) were considered to be the possible infecting serovar.45,46 However, we cannot determine if this finding is caused by past or current infections because we only had single serum samples. Furthermore, cross-reactions, especially between serogroups, are often observed in the MAT.6 However, the cross-reactivity is not observed within weeks or months after infection. Therefore, it is necessary that paired sera be tested to truly determine the infection status of subjects. The eight non-reactive rat serum samples may be negative for leptospirosis or may have antibodies against serovars that were not included in our panel.
Determining the cut-off titer (also referred to as significant titer) is another issue in the MAT.13 As previously mentioned, we were not able to detect any antibodies against Leptospira in specific pathogen–free rats even at a reciprocal titer of 20. Therefore, we set the significant titer at 20. Nineteen of the rat serum samples had titers at this level (Table 3). Of these 19 serum samples, 16 had antibodies against multiple Leptospira serovars. We believe that some of the antibodies with a titer of 1:20 are probably non-specific.2
In this study, leptospires isolated from rats in Metro Manila and Laguna were classified into four groups on the basis of molecular tests (Table 4, and Figures 1 and 2). This finding suggests that these four genotypes had a strong tendency of persistence in the renal tubules. Thus, they were isolated from the kidneys of rats. It is known that upon infection, leptospires appear in the blood and are spread to other tissues and organs.2,3 They are eventually cleared by the immune response of hosts. However, they may remain in the kidney tubules and may be shed in the urine of infected hosts for weeks to more than seven months.47 There may also be a bacterial strain difference in the ability to chronically infect the hosts. In a study by Thiermann,47 Norway rats that were experimentally infected with serovars Icterohemorrhagiae and Grippotyphosa had different lengths of chronicity. Chronic infection developed faster and organisms were shed longer (220 days) in rats infected with serovar Icterohemorrhagiae than in rats infected with serovar Grippotyphosa (40 days). Another hypothesis is that the four groups of isolates in our study may be easily cultured. Thus, only these serovars and serogroups were isolated from the 46 rats but other serovars were not isolated.
As previously mentioned, one rat from Metro Manila and three rats from Laguna had mixed infections because isolates obtained from these rats had sequences and band patterns similar to serovar Manilae (for isolates that grew in Korthof's medium) and serogroup Javanica (for isolates that grew in EMJH medium). Although we do not have an evidence to support our finding, we believe that that media can be strain-selective. On the basis of our experience, L. interrogans strain Ictero No. 1 grew well in Korthof's medium but not in EMJH medium.
The MAT results showed that rats had antibodies against the four Leptospira serovars and serogroups. The MAT results of rats from Metro Manila were consistent with the molecular typing results because most antibodies detected in these rats were against serovars Manilae and Losbanos (Table 6). However, most specificities of antibodies detected in rats from Laguna were not consistent with serovars and serogroups that the rats carried (Table 7). Thus, antibodies against serovars homologous and non-homologous to the isolates were detected in rats from Metro Manila and Laguna. A possible reason for this finding may be that rats used in this study may have been previously infected with these serovars (as shown by their positive MAT results). However, because these organisms may have been already cleared from the animals, leptospires belonging to these serovars could not be isolated but antibodies remained in the rats.
Table 6.
MAT results of rats culture-positive for Leptospira, Metro Manila, the Philippines*
| Strain no. of rat isolates | MAT result | |
|---|---|---|
| Serovar | Titer | |
| L. interrogans serovar Manilae | ||
| E47 | Manilae | 160 |
| E53 | Manilae and Autumnalis | 640 |
| K55 | Hebdomadis and Hardjo | 80 |
| E56 | Manilae | 1,280 |
| E57 | Manilae | 160 |
| E58 | Manilae | 320 |
| K59 | Poi | 160 |
| E60 | Manilae | 2,560 |
| K62 | Manilae | 320 |
| K64 | Manilae | 640 |
| K72 | Manilae | 1,280 |
| K74 | Tarassovi | 320 |
| E76 | Manilae | 320 |
| E78 | Manilae | 5,120 |
| K79 | Manilae | 640 |
| K83 | Manilae | 1,280 |
| L. interrogans serovar Losbanos | ||
| K37 | Losbanos and Hebdomadis | 160 |
| E38 | Losbanos | 640 |
| E40 | Losbanos | 640 |
| E41 | Losbanos | 2,560 |
| K42 | Hardjo | 80 |
| E43 | Pyrogenes, Losbanos, Patoc, and Semaranga | 20 |
| K44 | Losbanos | 1,280 |
| E45 | Losbanos | 1,280 |
| K46 | Manilae | 160 |
| K68 | Pomona | 160 |
| L. borgpetersenii serogroup Javanica | ||
| E63 | Poi | 320 |
| E75 | Tarassovi | 2,560 |
| E80 | Manilae | 1,280 |
MAT = microscopic agglutination test.
Table 7.
MAT results of rats culture-positive for Leptospira, Laguna, the Philippines*
| Strain no. of rat isolates | MAT result | |
|---|---|---|
| Serovar | Titer | |
| L. interrogans serogroup Grippotyphosa | ||
| K5 | Hardjo | 1,280 |
| E7 | Ratnapura | 1,280 |
| K18 | Poi | 80 |
| K25 | Hebdomadis | 2,560 |
| K32 | – | NA |
| K93 | Hebdomadis | 320 |
| L. borgpetersenii serogroup Javanica | ||
| K6 | Poi, Pyrogenes, Grippotyphosa, Autumnalis, Ratnapura, Australis, Icterohaemorrhagiae, Semaranga and Pomona | 20 |
| K10 | Poi | 160 |
| E11 | Poi | 160 |
| E16 | Hardjo | 320 |
| E20 | – | NA |
| E26 | Canicola | 1,280 |
| E28 | – | NA |
| E31 | Grippotyphosa and Pomona | 80 |
| K33 | – | NA |
| K35 | Poi | 160 |
| E94 | Ratnapura | 320 |
MAT = microscopic agglutination test; – = non-reactive to the panel of antigens used in MAT (MAT negative); NA = not applicable (non-reactive).
Phylogenetic analysis of gyrB has been applied to the taxonomic classification of Pseudomonas, Acetinobacter, and Mycobacterium.50–52 The gyrB gene was used to identify Leptospira isolates from the Amami islands in Japan.26 It was also used by Slack and others38 to develop a molecular technique for identifying pathogenic Leptospira species by conventional or real-time polymerase chain reaction amplification and sequencing of a partial fragment of the gyrB gene. The molecular evolution of this gene was faster than that of the rrs gene, as shown by its sequence and an average base substitution for rrs of 1% per 50 million years; that of gyrB at synonymous sites was 0.7–0.8% per 1 million years.51,52 This finding indicated that analysis of gyrB was able to differentiate closely related bacterial strains, although tested strains were identical in rrs gene sequencing. By using PFGE and gyrB sequence analysis, we determined that isolates in our study were segregated into four groups. This finding shows that these two methods were sensitive in discriminating leptospires. Furthermore, the PFGE results identified the high clonality of isolates within the same serovars as described.7,53,54
As mentioned earlier in this report, previous seroepidemiologic studies in the Philippines had detected antibodies against several Leptospira serovars such as Tarassovi, Sejroe, Poi, Javanica, Manilae, Losbanos, and Grippotyphosa in different human and animal serum samples.1,8–16,43 The antibodies detected in humans and animals in previous studies were also detected among the rats in our current study, which indicates that these serovars may have been present in the Philippines for several decades.
Although our sample size and sampling areas were limited, on the basis of the typing of isolates, we believe that rats in Metro Manila may be the sources of serovars Manilae, Losbanos, and Javanica, and rats in Laguna may be the sources of serogroups Grippotyphosa and Javanica. We also hypothesize that other Leptospira serovars such as Hebdomadis, Ratnapura, and Pomona, may also be prevalent in the Philippines, as shown by the presence of antibodies against these serovars in the MAT. Furthermore, isolation of serovars Manilae and Losbanos and serogroups Grippotyphosa and Javanica from rats in our study and detection of antibodies against these serovars and serogroups in humans and animals in previous studies indicate that rats may be the sources of leptospirosis transmission in the Philippines.
In terms of pathogenicity of rat isolates, hamsters that were infected with these isolates died within two weeks. The route used in this study was the intraperitoneal route, which is the most widely used infection route in most leptospiral studies reported. However, leptospires usually enter the body through cuts/wounds on the skin. It would therefore be interesting to know if the pathogenicity of the isolates would be the same using the subcutaneous route and using lower doses (i.e., less than 105). In general, injected organisms (leptospires) should be isolated from tissues or body fluids of infected animals, especially those that died from infection. However, in this study, we found that this is not always the case. As shown in Table 5, leptospires were also isolated from hamsters that were able to survive experimental leptospirosis. In contrast, leptospires were not recovered from hamsters that died after interaperitoneal injection with leptospires. This finding may show that the leptospiral doses used in our study may be sub-lethal because infected hamsters were able to survive and recover with no signs of infection. Survival of hamsters from experimental infection and the absence of recovery of injected leptospires may also indicate that isolates from the Philippines used may be non-pathogenic to the animals.
In this study, we did not definitively identify or type species of rats that we used. However, we observed that the rats obtained in Metro Manila were mostly Rattus norvegicus and those obtained in Laguna were mostly R. tanezumi. In previous studies, carriage of leptospirosis was found to be correlated to the species of rats and its age.3 However, we were not also able to determine the age and sex of the captured rats. It would therefore be useful if the rat species, age, and sex were identified in future investigations.
The CAAT, although time-consuming and laborious, is an indispensable method for typing Leptospira isolates.2,6 Because of lack of resources needed for CAAT (i.e., reference antisera against Leptospira), our laboratory was not able to perform this method. However, our use of PFGE and gyrB sequence analysis as alternative methods for typing isolates have been done in other studies of leptospirosis.6,7,22,26,28,29,38,53
The results of our study provide useful information on the epidemiology of leptospirosis in the Philippines, which until now was not well studied. However, studies with larger sample sizes on leptospirosis among rats, humans, and other animals in other areas of the Philippines would be beneficial in determining the transmission cycle of leptospirosis and the status of this zoonosis. The observations provided in our study may also be useful in formulating leptospirosis prevention and control measures and guidelines in the Philippines and other countries with similar conditions.
Acknowledgments
We thank all persons who helped made this study feasible, particularly those who have helped in the trapping and collection of rats (market vendors, market managers, hotel staff, Department of Medical Microbiology, College of Public Health, University of the Philippines Manila laboratory aides, farmers); Dr. Arlene G. Bertuso (Department of Parasitology, College of Public Health, University of the Philippines Manila) for arranging the trapping of rats in Laguna; and Ms. Maria Theresa Redaniel for advice on statistical analysis. Part of this paper was presented at the College of Public Health–University of the Philippines Manila/World Health Organization–Western Pacific Regional Office/Kyushu University Joint Scientific Meeting on Leptospirosis in the Asia Pacific Region in the Philippines on November 6–7, 2008 and at the 6th International Leptospirosis Society Meeting in Cochin, India, on September 21–24, 2009.
Disclaimer: The opinions in this article were not influenced by any of the funding agencies.
Footnotes
Financial support: This study was supported by a grant from the Special Coordination Funds on Science and Technology Agency of the Ministry of Education, Sports, Culture, Science and Technology of Japan. Sharon Y. A. M. Villanueva was supported by a scholarship from the Ministry of Education, Culture, Sports, Science and Technology of the Government of Japan.
Authors' addresses: Sharon Y. A. M. Villanueva, Yasutake Yanagihara, and Shin-ichi Yoshida, Department of Bacteriology, Graduate School of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka, 812-8582 Japan, E-mails: svillan@bact.med.kyushu-u.ac.jp, yanagihara@uv.tnc.ne.jp, and shin-ichi@bact.med.kyushu-u.ac.jp. Hirokazu Ezoe, International Research Center for Infectious Diseases, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan, E-mail: ezoe@biken.osaka-u.ac.jp. Rubelia A. Baterna, Lolita L. Cavinta, and Nina G. Gloriani, Department of Medical Microbiology, College of Public Health, University of the Philippines Manila, Ermita, Manila, 1000 the Philippines, E-mails: rabaterna@yahoo.com, cavinta@yahoo.com, and ninagloriani@yahoo.com. Maki Muto and Nobuo Koizumi, Department of Bacteriology, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku, Tokyo, 162-8640 Japan, E-mails: mutomaki@nih.go.jp and nkoizumi@nih.go.jp. Takashi Fukui, Yoshihiro Okamoto, and Toshiyuki Masuzawa, Laboratory of Microbiology and Immunology, Faculty of Pharmaceutical Sciences, Chiba Institute of Science, Choshi, Chiba, 288-0025 Japan, E-mails: tfukui@cis.ac.jp, yokamoto@cis.ac.jp, and masuzawat@cis.ac.jp.
Reprint requests: Sharon Y. A. M. Villanueva, Department of Bacteriology, Graduate School of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582 Japan, E-mail: svillan@bact.med.kyushu-u.ac.jp.
References
- 1.Famatiga EG, Topacio TM, Suva MH, Oliveros FM. Studies on leptospirosis in animals and man in the Philippines V. Serological survey of leptospirosis among occupationally exposed Filipinos. Southeast Asian J Trop Med Public Health. 1972;3:482–487. [Google Scholar]
- 2.World Health Organization . Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control. Geneva: World Health Organization and International Leptospirosis Society; 2003. [Google Scholar]
- 3.Faine S, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- 4.Wisseman CL, Jr, Traub R, Gochenour WS, Jr, Smadel JE, Lancaster WE. Leptospirosis of man and animals in urban, rural and jungle areas of southeast Asia. Am J Trop Med Hyg. 1955;4:29–40. doi: 10.4269/ajtmh.1955.4.29. [DOI] [PubMed] [Google Scholar]
- 5.Levett PN. Leptospirosis: a forgotten zoonosis? Clin Appl Immunol Rev. 2004;4:435–448. [Google Scholar]
- 6.Cerquiera GM, Picardeau M. A century of Leptospira strain typing. Infect Genet Evol. 2009;9:760–768. doi: 10.1016/j.meegid.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann JL, Bellenger E, Perolat P, Baranton G, Saint Girons I. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J Clin Microbiol. 1992;30:1696–1702. doi: 10.1128/jcm.30.7.1696-1702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlos ER, Kundin WD, Tsai CC, Watten RH, Irving GS, Villanueva C. Leptospirosis in the Philippines. VII. Serologic and isolation studies on horses. Southeast Asian J Trop Med Public Health. 1971;2:151–152. [PubMed] [Google Scholar]
- 9.Famatiga EG. Leptospirosis in Philippine monkeys. Southeast Asian J Trop Med Public Health. 1973;4:316–318. [PubMed] [Google Scholar]
- 10.Padre LP, Watt G, Tuazon ML, Gray MR, Laughlin LW. A serologic survey of rice-field leptospirosis in Central Luzon, Philippines. Southeast Asian J Trop Med Public Health. 1988;19:197–199. [PubMed] [Google Scholar]
- 11.Masuzawa T, Dancel LA, Miyake M, Yanagihara Y. Serological analysis of human leptospirosis in the Philippines. Microbiol Immunol. 2001;45:93–95. doi: 10.1111/j.1348-0421.2001.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 12.Yanagihara Y, Villanueva SY, Yoshida S, Okamoto Y, Masuzawa T. Current status of leptospirosis in Japan and Philippines. Comp Immunol Microbiol Infect Dis. 2007;30:399–413. doi: 10.1016/j.cimid.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Famatiga EG. Leptospira study on fevers of unknown origin in the Philippines. Southeast Asian J Trop Med Public Health. 1971;2:153–163. [PubMed] [Google Scholar]
- 14.Basaca-Sevilla V, Cross JH, Alcantara A, Barrito B, Pastrana E, Sevilla J, Balagot R, Dungca S. An outbreak of leptospirosis in an agricultural penal colony in the Philippines. Phil J Microbiol Infect Dis. 1981;10:73–82. [Google Scholar]
- 15.Carlos ER, Kundin WD, Watten RH, Tsai CC, Irving GS, Carlos ET, Directo AC, Castillo LP, Gaitmaitan O. Leptospirosis in the Philippines VI. Serologic and isolation studies on carabaos. Southeast Asian J Trop Med Public Health. 1970;1:481–482. [PubMed] [Google Scholar]
- 16.Carlos ER, Kundin WD, Watten RH, Tsai CC, Irving GS, Carlos ET, Directo AC. Leptospirosis in the Philippines: canine studies. Am J Vet Res. 1971;32:1451–1454. [PubMed] [Google Scholar]
- 17.Agudelo-Florez P, Londono AF, Quiroz VH, Angel JC, Moreno N, Loaiza ET, Munoz LF, Rodas JD. Prevalence of Leptospira spp. in urban rodents from a groceries trade center of Medellin, Colombia. Am J Trop Med Hyg. 2009;81:906–910. doi: 10.4269/ajtmh.2009.09-0195. [DOI] [PubMed] [Google Scholar]
- 18.Andrade J, Brandao AP. Epidemiology of human leptospirosis, with special reference to greater Rio de Janeiro, Brazil, from 1970 to 1982. Mem Inst Oswaldo Cruz. 1987;82:91–100. doi: 10.1590/s0074-02761987000100016. [DOI] [PubMed] [Google Scholar]
- 19.Aragon PR, Famatiga EG. Studies on leptospirosis. II. Laboratory evidence of human infection. Acta Med Philipp. 1965;1:196–198. [PubMed] [Google Scholar]
- 20.Arambulo PV III, Topacio TM, Jr, Famatiga EG, Sarmiento RV, Lopez S. Leptospirosis among abattoir employees, dog pound workers, and fish inspectors in the city of Manila. Southeast Asian J Trop Med Public Health. 1972;3:212–220. [PubMed] [Google Scholar]
- 21.Basaca-Sevilla V, Cross JH, Pastrana E. Leptospirosis in the Philippines. Southeast Asian J Trop Med Public Health. 1986;17:71–74. [PubMed] [Google Scholar]
- 22.Ciceroni L, Ciarrocchi S, Ciervo A, Petrucca A, Pinto A, Calderaro A, Viani I, Galati L, Dettori G, Chezzi C. Differentiation of leptospires of the serogroup Pomona by monoclonal antibodies, pulsed-field gel electrophoresis and arbitrarily primed polymerase chain reaction. Res Microbiol. 2002;153:37–44. doi: 10.1016/s0923-2508(01)01284-0. [DOI] [PubMed] [Google Scholar]
- 23.Ciceroni L, Lombardo D, Pinto A, Ciarrocchi S, Simeoni J. Prevalence of antibodies to Leptospira serovars in sheep and goats in Alto Adige-South Tyrol. J Vet Med B Infect Dis Vet Public Health. 2000;47:217–223. doi: 10.1046/j.1439-0450.2000.00333.x. [DOI] [PubMed] [Google Scholar]
- 24.Ciceroni L, Pinto A, Benedetti E, Pizzocaro P, Lupidi R, Cinco M, Gelosa L, Grillo R, Rondinella V, Maruccio L, Mansuet S, Ioli A, Franzin L, Giannico F, Cacciapuoti B. Human leptospirosis in Italy, 1986–1993. Eur J Epidemiol. 1995;11:707–710. doi: 10.1007/BF01720308. [DOI] [PubMed] [Google Scholar]
- 25.Fresh JW, Tsai CC, Lai CH, Chang CT. Leptospirosis in man and rodents in Taiwan. Am J Trop Med Hyg. 1968;17:760–768. doi: 10.4269/ajtmh.1968.17.760. [DOI] [PubMed] [Google Scholar]
- 26.Kawabata H, Sakakibara S, Imai Y, Masuzawa T, Fujita H, Tsurumi M, Sato F, Takano A, Nogami S, Kaneda K, Watanabe H. First record of Leptospira borgpetersenii isolation in the Amami Islands, Japan. Microbiol Immunol. 2006;50:429–434. doi: 10.1111/j.1348-0421.2006.tb03811.x. [DOI] [PubMed] [Google Scholar]
- 27.Koizumi N, Muto M, Yamada A, Watanabe H. Prevalence of Leptospira spp. in the kidneys of wild boars and deer in Japan. J Vet Med Sci. 2009;71:797–799. doi: 10.1292/jvms.71.797. [DOI] [PubMed] [Google Scholar]
- 28.Koizumi N, Uchida M, Makino T, Taguri T, Kuroki T, Muto M, Kato Y, Watanabe H. Isolation and characterization of Leptospira spp. from raccoons in Japan. J Vet Med Sci. 2009;71:425–429. doi: 10.1292/jvms.71.425. [DOI] [PubMed] [Google Scholar]
- 29.Masuzawa T, Okamoto Y, Une Y, Takeuchi T, Tsukagoshi K, Koizumi N, Kawabata H, Ohta S, Yoshikawa Y. Leptospirosis in squirrels imported from United States to Japan. Emerg Infect Dis. 2006;12:1153–1155. doi: 10.3201/eid1207.060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthias MA, Ricaldi JN, Cespedes M, Diaz MM, Galloway RL, Saito M, Steigerwalt AG, Patra KP, Ore CV, Gotuzzo E, Gilman RH, Levett PN, Vinetz JM. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl Trop Dis. 2008;2:e213. doi: 10.1371/journal.pntd.0000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topacio TM, Gavino LG, Famatiga E, Suva M. Leptospirosis in animals and man in the Philippines. VIII. Serological incidence in native dogs. Int J Zoonoses. 1974;1:32–42. [PubMed] [Google Scholar]
- 32.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levett PN. Sequence-based typing of Leptospira: epidemiology in the genomic era. PLoS Negl Trop Dis. 2007;1:e120. doi: 10.1371/journal.pntd.0000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, Apiwatanaporn A, Slack AT, Suputtamongkol Y, White NJ, Feil EJ, Day NP, Peacock SJ. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1:e56. doi: 10.1371/journal.pntd.0000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed N, Devi SM, Valverde Mde L, Vijayachari P, Machang'u RS, Ellis WA, Hartskeerl RA. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob. 2006;5:28. doi: 10.1186/1476-0711-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawabata H, Dancel LA, Villanueva SY, Yanagihara Y, Koizumi N, Watanabe H. flaB-polymerase chain reaction (flaB-PCR) and its restriction fragment length polymorphism (RFLP) analysis are an efficient tool for detection and identification of Leptospira spp. Microbiol Immunol. 2001;45:491–496. doi: 10.1111/j.1348-0421.2001.tb02649.x. [DOI] [PubMed] [Google Scholar]
- 37.Ribeiro RL, Machry L, Brazil JM, Ramos TM, Avelar KE, Pereira MM. Technical improvement to prevent DNA degradation of Leptospira spp. in pulsed field gel electrophoresis. Lett Appl Microbiol. 2009;49:289–291. doi: 10.1111/j.1472-765X.2009.02641.x. [DOI] [PubMed] [Google Scholar]
- 38.Slack AT, Symonds ML, Dohnt MF, Smythe LD. Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiol. 2006;6:95. doi: 10.1186/1471-2180-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slack AT, Khyairani-Bejo S, Symonds ML, Dohnt MF, Galloway RL, Steigerwalt AG, Bahaman AR, Craig S, Harrower BJ, Smythe LD. Leptospira kmetyi sp. nov., isolated from an environmental source in Malaysia. Int J Syst Evol Microbiol. 2009;59:705–708. doi: 10.1099/ijs.0.002766-0. [DOI] [PubMed] [Google Scholar]
- 40.Slack At, Galloway RL, Symonds ML, Dohnt MF, Smythe LD. Reclassfication of Leptospira meyeri serovar Perameles to Leptospira interrogans serovar Perameles through serological and molecular analysis: evidence of a need for changes to current procedures in Leptospira taxonomy. Int J Syst Evol Microbiol. 2009;59:705–708. doi: 10.1099/ijs.0.000992-0. [DOI] [PubMed] [Google Scholar]
- 41.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 42.Department of Health . Field Health Information System Annual Report, 2001. Manila: National Epidemiology Center of the Department of Health, Philippines; [Google Scholar]
- 43.Carlos ER, Kundin WD, Watten RH, Tsai CC, Irving GS, Carlos ET, Directo AC. Leptospirosis in the Philippines: feline studies. Am J Vet Res. 1971;32:1455–1456. [PubMed] [Google Scholar]
- 44.Kmety E, Dikken H. Classification of the Species Leptospira interrogans and History of its Serovars. Groningen, The Netherlands: University Press; 1993. [Google Scholar]
- 45.Geisen V, Stengel C, Brem S, Muller W, Greene C, Hartmann K. Canine leptospirosis infections: clinical signs and outcome with different suspected Leptospira serogroups (42 cases) J Small Anim Pract. 2007;48:324–328. doi: 10.1111/j.1748-5827.2007.00324.x. [DOI] [PubMed] [Google Scholar]
- 46.O'Keefe JS, Jenner JA, Sandifer NC, Antony A, Williamson NB. A serosurvey for antibodies to Leptospira in dogs in the lower North Island of New Zealand. N Z Vet J. 2002;50:23–25. doi: 10.1080/00480169.2002.36245. [DOI] [PubMed] [Google Scholar]
- 47.Thiermann AB. The Norway rat as a selective chronic carrier of Leptospira icterohaemorrhagiae. J Wildl Dis. 1981;17:39–43. doi: 10.7589/0090-3558-17.1.39. [DOI] [PubMed] [Google Scholar]
- 48.Haake DA. Hamster model of leptospirosis. Curr Protoc Microbiol. 2006;12 doi: 10.1002/9780471729259.mc12e02s02. Unit 12E 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009;7:736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto S, Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol. 1995;61:1104–1109. doi: 10.1128/aem.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto S, Harayama S. Phylogenetic analysis of Acetinobacter strains based on the nucleotide sequences of gyrB genes and on the amino acid sequences of their products. Int J Syst Bacteriol. 1996;46:506–511. doi: 10.1099/00207713-46-2-506. [DOI] [PubMed] [Google Scholar]
- 52.Kasai H, Ezaki T, Harayama S. Differentiation of phylogenetically related slowly growing Mycobacteria by their gyrB sequences. J Clin Microbiol. 2000;38:301–308. doi: 10.1128/jcm.38.1.301-308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galloway RL, Levett PN. Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars. Am J Trop Med Hyg. 2008;78:628–632. [PubMed] [Google Scholar]
- 54.Herrmann JL, Baris C, Bellenger E, Perolat P, Baranton G, Saint Girons I. Genome conservation in isolates of Leptospira interrogans. J Bacteriol. 1991;173:7582–7588. doi: 10.1128/jb.173.23.7582-7588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


