Abstract
We examined the extent to which different Trypanosoma cruzi strains induce transcriptomic changes in cultured L6E9 myoblasts 72 hours after infection with Brazil (TC I), Y (TC II), CL (TC II), and Tulahuen (TC II) strains. Expression of 6,289 distinct, fully annotated unigenes was quantified with 27,000 rat oligonucleotide arrays in each of the four replicas of all control and infected RNA samples. Considering changes greater than 1.5-fold and P values < 0.05, the Tulahuen strain was the most disruptive to host transcriptome (17% significantly altered genes), whereas the Y strain altered only 6% of the genes. The significantly altered genes in the infected cells were largely different among the strains, and only 21 genes were similarly changed by all four strains. However, myoblasts infected with different strains showed proportional overall gene-expression alterations. These results indicate that infection with different parasite strains modulates similar but not identical pathways in the host cells.
Introduction
Chagas disease, caused by infection with the flagellate protozoan parasite Trypanosoma cruzi, is a widespread disease in Latin America affecting millions of people.1 Infective trypomastigotes invade peripheral cells and transform into multiplicative amastigote forms. The initial (acute) phase of the disease is characterized by intense tissue parasitism involving the heart, skeletal and smooth muscle cells, liver, fat, and brain that is accompanied by intense focal inflammation and necrosis.2 Some patients can evolve to a chronic phase of the disease that can include cardiac and/or digestive forms. The severity of the chronic phase may be related to the efficiency of the host immune response in resolving the infection during the acute phase,3 but this has never been proven. Moreover, there are several reports of differences of tropism of T. cruzi to host tissue, which is also associated with the pathogenesis of chronic Chagas disease.4,5
Differences in the pathogenesis of the disease among patients may vary according to differences in both hosts and parasite strain.6 Among differences in T. cruzi strains are their resistance to chemotherapy, oxidative stress, and infectivity in the mouse.7–9 Although previous in vivo and in vitro microarray analyses using cultured cells10,11 and hearts of mice12–14 infected with T. cruzi showed that this infection results in profound alterations in the host cells, the degree to which these results are applicable to all T. cruzi strains found in infected individuals has not been explored previously.
Since host immune response, tissue parasitism, and parasite strains may be important factors in the pathogenesis of chronic Chagas disease, we have used gene-array analysis to compare the alterations in host cells caused by four different stocks of T. cruzi. The present study characterizes the transcriptomic changes in cultured rat myoblasts that result from infection with each strain and highlights common genes that were similarly or differentially modulated by each strain. Analysis reveals host cell changes that might lead to an understanding of previously observed differences in pathogenesis in vivo.
Methods
Cells and parasites.
The L6E9 rat myoblast cell line was maintained in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin at 37°C with 5% CO2 atmosphere.15 Cells were dissociated with trypsin/ ethylenediaminetetracetic acid (EDTA) solution (Gibco), and 106 cells were plated in 100-mm2 cell-culture dishes. After 24 hours of plating, cells were washed with Phosphate Buffered Saline (PBS) containing Ca2+ and Mg2+ (Gibco) and infected with 2 × 106 trypomastigote forms of T. cruzi in DMEM. Parasites of the Y, CL Brener,16 Tulahuen, and Brazil strains were obtained from supernatants of infected L6E9 cultures. Forty-eight hours post-infection, cells were washed twice with Ca2+/Mg2+ PBS to remove free trypomastigotes in the supernatant, and they were re-fed with fresh supplemented DMEM. Total RNA was harvested at 72 hours post-infection using guanidinium thiocyanate-phenol-chloroform extraction (TRIZOL) reagent (Invitrogen, Carlsbad, CA), following the protocol indicated by the manufacturer, when at least 25% of the cultured cells were infected, presented only intracellular amastigotes, and had no release of trypomastigotes, which would lead to re-infection of culture.
T. cruzi genotyping.
The different isolates of T. cruzi used in this work were identified using the method described by Fernandes and others17 according to their phylogenetic lineage. Briefly, genomic DNA from 5 × 108 epimastigote forms of the Y, CL, Brazil, and Tulahuen strains was extracted using the DNeasy kit (Qiagen, Hilden, Germany). Multiplex polymerase chain reaction (PCR) was performed using 150 ng of DNA, and the primers were designed to recognize the mini-exon gene of the parasites using a pool of five nucleotides: three were derived from a hypervariable region of the T. cruzi mini-exon repeat (T. cruzi 1 [TC1], 5¢ ACA CTT TCT GTG GCG CTG ATC G; TC 2, 5¢ TTG CTC GCA CAC TCG GCT GCA T; TC 3, 5¢ CCG CGW ACA ACC CCT MAT AAA AAT G) and an oligonucleotide from a specific region of the Trypanosoma rangelii non-transcribed spacer (TR; 5¢ CCT ATT GTG ATC CCC ATC TTC G). A common downstream oligonucleotide corresponds to sequences present in the most conserved region of the mini-exon gene (ME; 5¢ TAC CAA TAT AGT ACA GAA ACT G). PCR reaction was performed using the Multiplex PCR kit (Qiagen) with the initial denaturing cycle of 95°C (15 seconds) followed by 30 cycles of 94°C (30 seconds, denaturing), 60°C (30 seconds, annealing), and 72°C (30 seconds, extension); a final extension cycle of 72°C lasts for 10 minutes and is followed by a soak cycle (4°C) using a PTC-100 Thermocycler (M.J. Research Inc., Massachusetts, (USA)). PCR fragments were loaded into a 2% agarose/ Tris base, boric acid, EDTA (TBE) gel with 0.008% ethidium bromide, and images were acquired using Kodak 1D Scientific Imaging Systems.
Light microscopy.
Cells (6 × 104) were plated into glass cover slips in 24-well plates. After 24 hours, 106 trypomastigotes were added to cultures in fresh supplemented DMEM, and infection was followed as described above. After 72 hours of infection, cells were washed three times and fixed with glutaraldehyde for Giemsa staining.18 Cover slips were digitally photographed using a Zeiss Axioplan microscope.
Microarray analysis.
We used the protocol optimized in our laboratory19 according to the standards of the Microarray Gene Expression Data Society. Briefly, 20 μg Trizol extracted total RNA from each culture dish was reverse transcribed in the presence of fluorescent Alexa Fluor 555-aha-dUTP (green fluorescent emission) or Alexa 647-aha-dUTP (red emission; Invitrogen) to label cDNAs. Differently labeled RNA samples from biological replicas of control (uninfected cells cultured for the same duration) or infected with one strain at a time were co-hybridized (“multiple yellow” strategy20) overnight at 50°C with rat 27k oligonucleotide arrays printed by Duke University (full technical information available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18175). All spots affected by local corruption, with saturated pixels, or with foreground fluorescence less than twice the background fluorescence (where noise may obscure the quantity) were removed from the analysis. The background subtracted signals were normalized iteratively,19 alternating red/green, interblock, lowess and scale intraslide and interslide normalization until the fluctuation of the ratio between the spot median and the corresponding block median of valid spots became less than 5% between successive iteration steps. Normalized expression levels were organized into redundancy groups (each group composed of all spots probing the same gene) and were represented by the weighted average of the values of individual spots. The abundance of host cell transcripts was considered as significantly altered after infection if the absolute fold-change was greater than 1.5-fold and the P value of the heteroscedastic t test (two-sample, unequal variance), applied to the means of the background-subtracted normalized fluorescence values in the four biological replicas of the compared transcriptomes, was greater than 0.05. This composite criterion to identify the significantly altered gene expression minimizes the number of false hits without eliminating too many true hits. The 50% change cut-off (1.5-fold) was selected to be significantly larger than the overall less than 10% interslide technical noise determined for the bacterial controls.
Gene categories.
GenMapp and MappFinder programs (www.genmapp.org; Gladstone Institute, University of California, San Francisco, CA) were used to determine whether or not altered gene expressions differed significantly from chance for the overlapping functional and structural Gene Ontology (GO) categories.
Results
Multiplex PCR was performed to confirm correspondence of the strains of T. cruzi used in this work to the phylogenetic classification as described in the literature. We verified that the Y, CL, and Tulahuen parasites displayed a 250-bp PCR product, indicating that they belong to the T. cruzi II (TC II) group. The Brazil strain, whose effects in host cells and animals have been characterized previously by our group,15,21 belongs to the TC I group, because it displays a 200-bp PCR product (Figure 1E).
Figure 1.
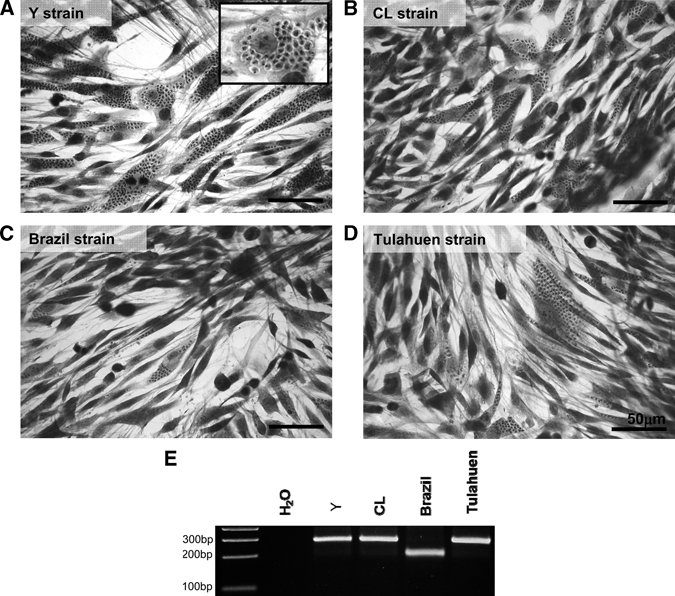
Characterization of L6E9 cell infection by four T. cruzi strains. Giemsa staining of L6E9 cells after 72 hours of infection with (A) Y, (B) CL, (C) Brazil, and (D) Tulahuen strains of T. cruzi was performed. All strains studied successfully infected the myoblasts, and at 72 hours post infection (hpi), it was possible to observe amastigote forms in the host cell's cytoplasm as shown in details in A inset. (Bars = 50 μm.) To confirm genetic background of the parasite stocks used for infection, genomic DNA from epimastigote forms was isolated and amplified with Multiplex PCR with primers derived from a hypervariable region of the T. cruzi mini exon. (E) The Y, CL, and Tulahuen strains had a PCR product of 250 bp, indicating that they belong to the T. cruzi (TC) II family, and the Brazil strain had a 200-bp product, indicating that it belongs to TC I family.
Data complying with the Minimum Information about Microarray Experiments (MIAME) were deposited in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18175) as series GSE18175. In this experiment, we quantified 6,289 distinct, well-annotated unigenes in all samples.
The impact of infection with each of the four strains of T. cruzi on the transcriptome changes of the same immortalized cell line was strikingly different (Figure 2). Thus, as shown in Table 5, there were only two (0.03%) genes significantly decreased, and 19 (0.3%) increased by all four T. cruzi strains. However, 4,340 (69%) genes were not significantly altered by any of the strains. The Venn diagrams in Figure 2 illustrate this observation by showing the number of genes equally increased (Figure 2B) or decreased (Figure 2C) by two, three, or all four strains of T. cruzi.
Figure 2.
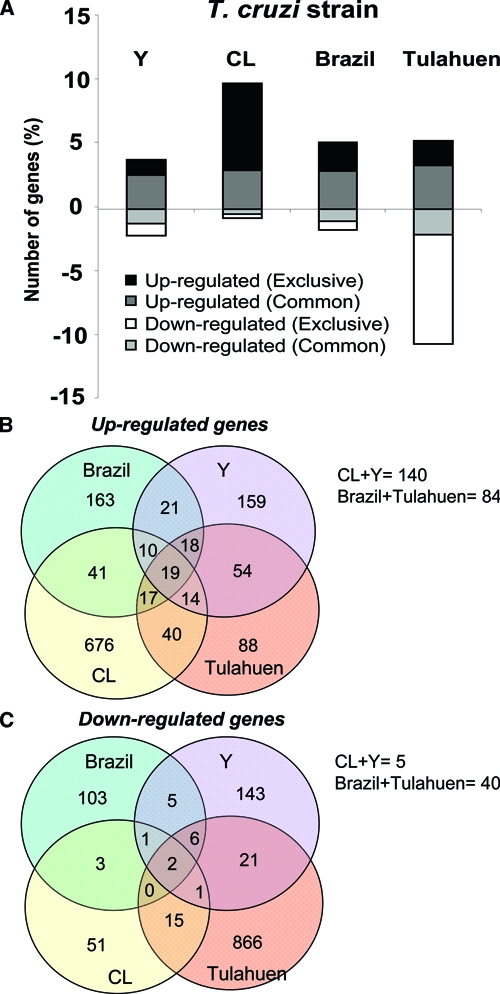
Profiles of transcriptomic changes caused by each strain of T. cruzi in host cells. The percentage of genes modulated in L6E9 cells by each T. cruzi strain is shown in A. Genes with fold-change value ³ 1.5 and P value ≤ 0.05 were quantified and plotted in this histogram, in which bars represent a percentage of the valid genes. Bars in white and black represent genes whose expression was decreased or increased exclusively by each strain, respectively. Bars in gray and light gray represent genes that were increased and decreased, respectively, and shared this modulation by at least two strains. The Venn diagrams represent the number of genes significantly (fold change ³ 1.5 and P value < 0.05) (B) increased and (C) decreased in common by all four strains, by three strains, by pairs of strains, and by individual strains of the parasite. The number of genes equally altered by CL + Y and Brazil + Tulahuen strains is indicated below the diagrams. Note the low number of genes altered in the same direction by all strains studied (19 increased and 2 decreased).
Table 5.
Similar results
| Gene name | Gene symbol | Fold change (strains) | |||
|---|---|---|---|---|---|
| Brazil | CL | Tulahuen | Y | ||
| C1q and tumor necrosis factor-related protein 1 | C1qTNF1 | –1.90 | –1.80 | –2.70 | –1.80 |
| Matrix metallopeptidase 14 | MMP14 | –2.00 | –1.90 | –2.00 | –1.70 |
| Cardiotrophin-like cytokine factor 1 | Clcf1 | 2.20 | 2.30 | 1.80 | 2.30 |
| Cut-like 1 (Drosophila) | Cutl1 | 7.72 | 2.58 | 5.36 | 1.59 |
| DNA-damage inducible transcript 3 | Ddit3 | 9.30 | 3.08 | 2.41 | 2.64 |
| Excision repair cross-complementing rodent repair deficiency, complementation group 3 | Ercc3 | 2.19 | 2.67 | 1.59 | 2.36 |
| G protein-coupled receptor, family C, group 5, member A | Gprc5a | 1.94 | 2.53 | 1.87 | 1.99 |
| GrpE-like 1, mitochondrial | Grpel1 | 1.60 | 1.95 | 1.66 | 1.91 |
| Pericentriolar material 1 | Pcm1 | 2.70 | 4.25 | 2.23 | 3.70 |
| Proprotein convertase subtilisin/kexin type 7 | Pcsk7 | 6.93 | 2.87 | 2.38 | 2.15 |
| Serologically defined colon cancer antigen 3 | Sdccag3 | 3.36 | 2.71 | 2.67 | 2.50 |
| Solute carrier family 1 (glutamate/neutral amino acid transporter) | Slc1a4 | 1.59 | 2.50 | 1.82 | 1.68 |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | Ywhaq | 2.84 | 2.85 | 1.77 | 2.56 |
Analysis of functional classes of genes modulated by the infection with different T. cruzi strains was performed with GenMapp software, and it revealed a very small number of gene functional categories with an abundance that was similarly affected by each pair of parasite strains (Table 7). Below, we describe in more detail the differences that were observed.
Table 7.
Gene categories
| Gene Ontology (GO) ID | GO Name | No. changed | No. measured | No. in GO | Percent changed | P value |
|---|---|---|---|---|---|---|
| Y strain | ||||||
| 5634 | Nucleus | 22 | 557 | 1,643 | 3.95000 | 0.004 |
| 45449 | Regulation of transcription | 12 | 264 | 884 | 4.55000 | 0.010 |
| 6355 | Regulation of transcription, DNA-dependent | 11 | 237 | 790 | 4.64000 | 0.012 |
| 5509 | Calcium-ion binding | 8 | 149 | 538 | 5.37000 | 0.013 |
| 6350 | Transcription | 12 | 278 | 920 | 4.32000 | 0.014 |
| 46872 | Metal-ion binding | 20 | 593 | 1,913 | 3.37000 | 0.049 |
| 5743 | Mitochondrial inner membrane | 4 | 22 | 63 | 18.18000 | 0.006 |
| 19866 | Organelle inner membrane | 4 | 29 | 79 | 13.79000 | 0.026 |
| 16757 | Transferase activity, transferring glycosyl groups | 5 | 48 | 144 | 10.42000 | 0.042 |
| 43234 | Protein complex | 9 | 396 | 1,280 | 2.27000 | 0.052 |
| 16853 | Isomerase activity | 4 | 34 | 74 | 11.77000 | 0.052 |
| CL strain | ||||||
| 5829 | Cytosol | 4 | 68 | 121 | 5.88000 | 0.002 |
| 5622 | Intracellular | 18 | 1,331 | 3,766 | 1.35000 | 0.004 |
| 15031 | Protein transport | 5 | 154 | 348 | 3.25000 | 0.011 |
| 6512 | Ubiquitin cycle | 3 | 102 | 281 | 2.94000 | 0.049 |
| 3824 | Catalytic activity | 8 | 1,611 | 5,073 | 0.50000 | 0.054 |
| 6886 | Intracellular protein transport | 3 | 103 | 237 | 2.91000 | 0.054 |
| 16757 | Transferase activity, transferring glycosyl groups | 10 | 48 | 144 | 20.83000 | 0.011 |
| 15992 | Proton transport | 6 | 24 | 64 | 25.00000 | 0.023 |
| 4197 | Cysteine-type endopeptidase activity | 7 | 34 | 91 | 20.59000 | 0.032 |
| 6811 | Ion transport | 18 | 118 | 555 | 15.25000 | 0.038 |
| 3924 | Gtpase activity | 5 | 22 | 45 | 22.73000 | 0.040 |
| 16491 | Oxidoreductase activity | 25 | 185 | 539 | 13.51000 | 0.045 |
| 6812 | Cation transport | 14 | 86 | 375 | 16.28000 | 0.046 |
| Brazil strain | ||||||
| 8237 | Metallopeptidase activity | 3 | 38 | 133 | 7.89000 | 0.020 |
| 4984 | Olfactory receptor activity | 9 | 24 | 1,034 | 37.50000 | 0.000 |
| 1584 | Rhodopsin-like receptor activity | 9 | 46 | 1,391 | 19.57000 | 0.001 |
| 4872 | Receptor activity | 20 | 195 | 2,013 | 10.26000 | 0.002 |
| 7186 | G-protein–coupled receptor protein-signaling pathway | 11 | 82 | 1,581 | 13.41000 | 0.005 |
| 43234 | Protein complex | 12 | 396 | 1,280 | 3.03000 | 0.022 |
| 7264 | Small gtpase mediated signal transduction | 8 | 66 | 179 | 12.12000 | 0.027 |
| 5615 | Extracellular space | 4 | 23 | 136 | 17.39000 | 0.029 |
| 6512 | Ubiquitin cycle | 1 | 102 | 281 | 0.98000 | 0.046 |
| 5525 | GTP binding | 10 | 99 | 237 | 10.10000 | 0.050 |
| Tulahuen strain | ||||||
| 6457 | Protein folding | 16 | 68 | 138 | 23.53000 | 0.001 |
| 5488 | Binding | 219 | 1,786 | 5,708 | 12.26000 | 0.010 |
| 3743 | Translation initiation factor activity | 7 | 25 | 37 | 28.00000 | 0.019 |
| 6695 | Cholesterol biosynthesis | 4 | 11 | 15 | 36.36000 | 0.021 |
| 6955 | Immune response | 10 | 46 | 258 | 21.73913 | 0.033 |
| 6629 | Lipid metabolism | 20 | 120 | 304 | 16.67000 | 0.053 |
| 4984 | Olfactory receptor activity | 7 | 24 | 1,034 | 29.17000 | 0.000 |
| 1584 | Rhodopsin-like receptor activity | 9 | 46 | 1,391 | 19.57000 | 0.001 |
| 16020 | Membrane | 63 | 740 | 3,798 | 8.51000 | 0.003 |
| 19866 | Organelle inner membrane | 7 | 29 | 79 | 24.14000 | 0.003 |
| 5743 | Mitochondrial inner membrane | 5 | 22 | 63 | 22.73000 | 0.012 |
| 4872 | Receptor activity | 20 | 195 | 2,013 | 10.26000 | 0.014 |
| 16021 | Integral to membrane | 41 | 494 | 2,888 | 8.30000 | 0.018 |
| 5856 | Cytoskeleton | 13 | 114 | 337 | 11.40000 | 0.021 |
| 7186 | G-protein–coupled receptor protein-signaling pathway | 10 | 82 | 1,581 | 12.20000 | 0.029 |
| 9058 | Biosynthesis | 9 | 284 | 923 | 3.17000 | 0.036 |
| 16874 | Ligase activity | 2 | 124 | 329 | 1.61000 | 0.046 |
| 5739 | Mitochondrion | 15 | 149 | 290 | 10.07000 | 0.048 |
Y strain.
We observed that the Y strain altered expression of 426 (6%) of the 6,996 quantified host cell genes. Among these genes, 150 were decreased, and 276 were increased after 72 hours of infection. When we identified individual genes whose expression was altered exclusively by the Y strain, we found that 87 genes were increased, such as α-1-integrin (1.76-fold), intercellular adhesion molecule (1.79-fold), presenilin 2 (3.81-fold), transforming growth-factor beta regulated gene (1.73-fold), and vascular cell-adhesion molecule 1 (1.91-fold). Sixty-five genes were decreased, such as cardiac Ca2+ ATPase (−1.6-fold), cadherin (−2.88-fold), and Cyp26b1 (cytochrome P450, family 26, subfamily b, polypeptide 1; −2.83-fold) (Table 1). The Y strain affected the expression of 11 gene categories including calcium and metal binding, regulation of transcription, and complexes of protein (Table 7).
Table 1.
Y Strain
| Gene name | Gene symbol | Fold change |
|---|---|---|
| Alanyl-tRNA synthetase | Aars | 1.85 |
| Actin, gamma, cytoplasmic 1 | Actg1 | 1.55 |
| Chloride intracellular channel 2 | Clic2 | 1.55 |
| Cyclin-dependent kinase inhibitor 2B (p15; inhibits CDK4) | Cdkn2b | 1.65 |
| Glycogen synthase kinase 3 beta | Gsk3b | 1.52 |
| Integrin alpha 1 | Itga1 | 1.76 |
| Intercellular adhesion molecule 1 | Icam1 | 1.79 |
| Lectin, galactose binding, soluble 3 | Lgals3 | 1.52 |
| Presenilin 2 | Psen2 | 3.814 |
| Protein C receptor, endothelial | Procr | 2.04 |
| Pyruvate kinase, muscle | Pkm2 | 1.70 |
| Syndecan -protein | Sdcbp | 1.854 |
| Transforming growth-factor beta-regulated gene 1 | Tbrg1 | 1.73 |
| Vascular cell-adhesion molecule 1 | Vcam1 | 1.91 |
| X-linked myotubular myopathy gene 1 | Mtm1 | 1.53 |
| Actin, alpha 1, skeletal muscle | Acta1 | –4.84 |
| Acyl-CoA synthetase long-chain family member 3 | Acsl3 | –2.63 |
| ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | Atp2a2 | –1.60 |
| Cadherin 15 | Cdh15 | –2.88 |
| Calsenilin, presenilin-binding protein, EF hand transcription factor | Csen | –1.62 |
| Cytochrome P450, family 26, subfamily b, polypeptide 1 | Cyp26b1 | –2.83 |
| Guanine monphosphate synthetase | Gmps | –1.94 |
| Myosin light chain, phosphorylatable, fast skeletal muscle | Mylpf | –1.68 |
| Rho family GTPase 2 | Rnd2 | –1.62 |
CL strain.
When the rat myoblasts were infected with the CL strain, we observed that 763 of 7,133 (11%) genes were significantly altered (53 decreased and 710 increased). The CL strain uniquely modulated the expression of 517 genes (494 were increased and 23 decreased), which corresponded to alterations in 14 gene categories including protein, intracellular protein, proton and ion transport, and ubiquitin cycle (Table 7). Some of the genes that the CL strain modulated were adenylate cyclase 2 (1.56-fold), α-spectrin (1.7-fold), annexin A1 (1.7-fold), caspase 7 (1.76-fold), epsin 2 (1.97), glutamate receptor (3.38), matrix metallopeptidase (MMP) 3 (2.51-fold), and syntaxin (−1.56-fold) (Table 2).
Table 2.
CL strain
| Gene name | Gene symbol | Fold change |
|---|---|---|
| Adenylate cyclase 2 | Adcy3 | 1.56 |
| Alpha-spectrin 2 | Spna2 | 1.70 |
| Angiopoietin-like 2 | AF159049 | 1.56 |
| Annexin A1 | Anxa1 | 1.56 |
| Casein kinase 1, gamma 1 | Csnk1g1 | 1.95 |
| Caspase 7 | Casp7 | 1.76 |
| Chemokine-like factor | Cklf | 1.76 |
| Chondroitin sulfate proteoglycan 6 | Cspg6 | 1.64 |
| Desmuslin | Dmn | 1.60 |
| Epsin 2 | Epn2 | 1.97 |
| Glutamate receptor, ionotropic, N-methyl D-aspartate 2B | Grin2b | 3.38 |
| Integrin alpha 5 | Itga5 | 1.84 |
| Interleukin 1 receptor, type I | Il1r1 | 2.27 |
| Matrix metallopeptidase 10 | Mmp10 | 2.40 |
| Matrix metallopeptidase 3 | Mmp3 | 2.52 |
| Phosphatidylethanolamine N-methyltransferase | Pemt | 2.03 |
| Phospholipase D1 | Pld1 | 1.58 |
| Plasminogen activator, urokinase receptor | Plaur | 1.76 |
| Proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | Psmd2 | 1.99 |
| Chaperonin subunit 4 (delta) | Cct4 | –1.57 |
| Syntaxin 8 | Stx8 | –1.56 |
| T-cell immunomodulatory protein | Cda08 | –2.61 |
Brazil strain.
The Brazil strain induced significant alteration of 7.31% genes of L6E9 by more than 1.5-fold. Among these 494 modulated genes, 117 were decreased, and 377 were increased. Some interesting genes that had their transcription altered by infection with the Brazil strain were cardiomyopathy associated gene (3.37-fold), MMP 1B (2.44-fold), mitogen-activated protein kinase 9 (2.12-fold), fibroblast growth factor (1.94-fold), desmin (1.63-fold), and tropomyosin 4 (−2.45-fold) (Table 3). Ten functional categories of genes were highly modulated in L6E9 by Brazil strain, including metallopeptidase activity, small GTPase-mediated signal transduction, and ubiquitin cycle (Table 7).
Table 3.
Brazil strain
| Gene name | Gene symbol | Fold change |
|---|---|---|
| H2A histone family, member Y | H2afy | 3.70 |
| Cardiomyopathy associated 3 | Cmya3 | 3.37 |
| Protein kinase C, gamma | Prkcc | 2.70 |
| Myocyte enhancer factor 2D | Mef2d | 2.43 |
| Mitogen-activated protein kinase 9 | Mapk9 | 2.12 |
| Fibroblast growth factor 21 | Fgf21 | 1.95 |
| ATPase, H+/K+ exchanging, beta polypeptide | Atp4b | 1.90 |
| Phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide | Pik3c2g | 1.78 |
| Paladin | Pald | 1.67 |
| Desmin | Des | 1.63 |
| Phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 | Pik3r3 | –1.53 |
| Coronin, actin-binding protein, 1B | Coro1b | –1.91 |
| Synaptotagmin XI | Syt11 | –1.91 |
| Proteasome (prosome, macropain) 28 subunit, beta | Psme2 | –2.35 |
| Keratin 25D | Krt25d | –2.38 |
| Tropomyosin 4 | Tpm4 | –2.46 |
Tulahuen strain.
The Tulahuen strain of T. cruzi induced the highest percentage of altered gene expression in the myoblasts (17.35%) with 761 decreased genes and 383 increased genes, of which, 617 and 139, respectively, were uniquely observed in Tulahuen-infected dishes. Additionally, infection with this strain significantly modulated the expression of 18 gene categories, including cholesterol biosynthesis, immune response, lipid metabolism, and receptor activity (Table 7). Some relevant examples of host cell genes significantly modulated during infection were p-cadherin (4.64-fold), cardiac ankyrin repeat kinase (2.54-fold), chymotrypsinogen B (2.64-fold), H2A histone family member Z (3.18-fold), interleukin 11 (1.68-fold), myotrophin (1.51-fold), caspase 9 (−1.75-fold), cytochrome c oxidase, subunit Va (−1.61-fold), heavy and light chain dynein (−1.86- and −1.81-fold, respectively), farnesyltransferase (−1.65-fold), hypoxia-inducible factor 1α subunit (−1.51-fold), janus kinase 3 (−1.64-fold), matrix metallopeptidase 11 (−3.46-fold), and cardiac troponin T2 (−2.89-fold) (Table 4).
Table 4.
Tulahuen strain
| Gene name | Gene symbol | Fold change |
|---|---|---|
| Activin A receptor type II-like 1 | Acvrl1 | 7.60 |
| Cadherin 3, type 1, P-cadherin (placental) | Cdh3 | 4.65 |
| Cardiac ankyrin repeat kinase | Cark | 2.54 |
| CDK5 regulatory subunit-associated protein 1 | Cdk5rap1 | 2.40 |
| Chymotrypsinogen B | Ctrb | 2.64 |
| Glucose 6 phosphatase, catalytic, 3 | G6pc3 | 1.65 |
| Inositol 1,4,5-trisphosphate 3-kinase C | Itpkc | 2.37 |
| Interleukin 11 | Il11 | 1.68 |
| Kinesin family member C1 | Kifc1 | 2.04 |
| Myotrophin | Mtpn | 1.51 |
| ATPase, Ca++ transporting, plasma membrane 1 | Atp2b1 | –2.76 |
| Bcl2 modifying factor | Bmf | –2.93 |
| Calcium channel, voltage-dependent, beta 3 subunit | Cacnb3 | –1.88 |
| Calreticulin | Calr | –2.04 |
| Caspase 9 | Casp9 | –1.75 |
| Cytochrome c oxidase, subunit Va | Cox5a | –1.62 |
| Cytokine-induced apoptosis inhibitor 1 | Ciapin1 | –1.67 |
| Dynein cytoplasmic 1 heavy chain 1 | Dync1h1 | –1.86 |
| Ectonucleoside triphosphate diphosphohydrolase 1 | Entpd1 | –2.90 |
| Farnesyltransferase, CAAX box, alpha | Fnta | –1.66 |
| Hypoxia inducible factor 1, alpha subunit | Hif1a | –1.52 |
| Inositol 1,4,5-triphosphate receptor 3 | Itpr3 | –3.70 |
| Janus kinase 3 | Jak3 | –1.64 |
| Junctional adhesion molecule 3 | Jam3 | –2.25 |
| Laminin, beta 2 | Lamb2 | –2.21 |
| Matrix metallopeptidase 11 | Mmp11 | –3.46 |
| Muscle, skeletal, receptor tyrosine kinase | Musk | –2.73 |
| Phospholipase D2 | Pld2 | –2.88 |
| Protein kinase C, nu | Prkcn | –2.57 |
| Synaptojanin 2 | Synj2 | –2.35 |
| Troponin T2, cardiac | Tnnt2 | –2.89 |
| Tumor protein p53 | Tp53 | –1.92 |
Genes modulated equally by all T. cruzi strains as possible disease biomarkers.
We observed that 13 (0.18%) host cell genes had the same pattern of significant modulation by all four strains of T. cruzi studied. These genes, thus, may represent disease biomarkers that could be useful in detecting the disease independent of the parasite strain. Only two (0.027%) were decreased, MMP-14 (−1.9-fold) and C1q and tumor necrosis factor-related protein 1 (−2.0-fold), and 11 (0.15%) increased, such as solute carrier family 1 (glutamate/neutral amino acid transporter; 1.9-fold), G protein-coupled receptor (2.1-fold), cardiotrophin-like cytokine factor 1 (2.13-fold), pericentriolar material 1 (3.22-fold), and DNA damage-inducible transcript 3 (4.37-fold). Table 5 shows the list of these thirteen genes and their modulation by each strain of the parasite. GenMapp analysis revealed that only nine gene categories were equally expressed by some pairs of strains such as receptor activity (Brazil and Tulahuen) and ubiquitin cycle (Brazil and CL) (Table 7).
Oppositely modulated genes.
Surprisingly, our arrays revealed that some transcripts increased by one specific strain were decreased by another strain. Table 6 contains all the 24 genes that behaved in this manner. Some genes of interest were cytochrome P450, family 2, subfamily d, polypeptide 22 (3-fold by Brazil strain and −2.9-fold by Y strain), neuropathy target esterase-like 1 (2.1-fold by Y strain and −1.8-fold by CL strain), platelet-derived growth-factor receptor, β polypeptide (1.7-fold by CL strain and −1.6-fold by Tulahuen strain), protein kinase D2 (1.5-fold by Brazil strain and −1.6-fold by Y strain), and RNA polymerase 1-1 (1.6-fold by CL strain and −1.6-fold by Tulahuen strain).
Table 6.
Opposite results
| Gene name | Gene symbol | Fold change (strains) | |||
|---|---|---|---|---|---|
| Brazil | CL | Tulahuen | Y | ||
| ADP-ribosylation factor guanine nucleotide-exchange factor 2 | Arfgef2 | – | 1.56 | –1.54 | – |
| CDC-like kinase 2 | LOC365842 | – | 1.52 | –1.64 | – |
| Cytochrome P450, family 2, subfamily d, polypeptide 22 | Cyp2d22 | 2.97 | – | – | –2.92 |
| NAD synthetase 1 | Nadsyn1 | – | 1.56 | –1.63 | – |
| Neuropathy target esterase-like 1 | Ntel1 | – | –1.78 | – | 2.11 |
| Platelet-derived growth-factor receptor, beta polypeptide | Pdgfrb | – | 1.68 | –1.64 | – |
| Pleckstrin homology, Sec7, and coiled-coil domains 1 | Pscd1 | – | 1.53 | –1.53 | – |
| Preoptic regulatory factor-2 | PORF-2 | – | 1.62 | –1.56 | – |
| Protein kinase D2 | Prkd2 | 1.50 | – | – | –1.62 |
| Regenerating islet-derived 1 | Reg1 | 1.67 | –2.31 | – | – |
| RNA polymerase 1-1 | Rpo1-1 | – | 1.56 | –1.56 | – |
| Son of sevenless homolog 1 | Sos1 | –1.53 | 1.82 | – | – |
| Sulfotransferase family 1A, phenol-preferring, member 1 | Sult1a1 | – | – | 1.90 | –1. 53 |
| Thymoma viral proto-oncogene 2 | Akt2 | – | 1.54 | –1.64 | – |
| Transducer of ErbB-2.1 | Tob1 | – | – | 1.58 | –1.72 |
Correlations between infections with four strains.
Our studies identifying individual genes that were significantly altered by the four strains of T. cruzi revealed a surprising diversity with only a few genes similarly changed by infection with all strains. However, because this analysis selects only individuals, it does not compare subtle global changes throughout the transcriptome. To compare global trancriptomic alteration patterns in L6E9 cells infected by the four strains used in the current investigation, we compared the entire gene-array datasets obtained from the infection with each strain against each of the other strains. With Origin software (OriginLab, Northampton, MA), we plotted results of these pairs as log2 values of their expression ratios. In all six of the comparisons of expression changes induced by infection with separate T. cruzi strains (Figure 3), the regression coefficients (r2 values) for these linear relations were highly significant, and P values in all cases were less than 0.0001. This finding indicates that although infection with each of the parasite strains leads to only partially overlapping alterations in the genes that are most affected, there is an overall similarity in the pattern of gene-expression alterations resulting from infection with all the strains.
Figure 3.
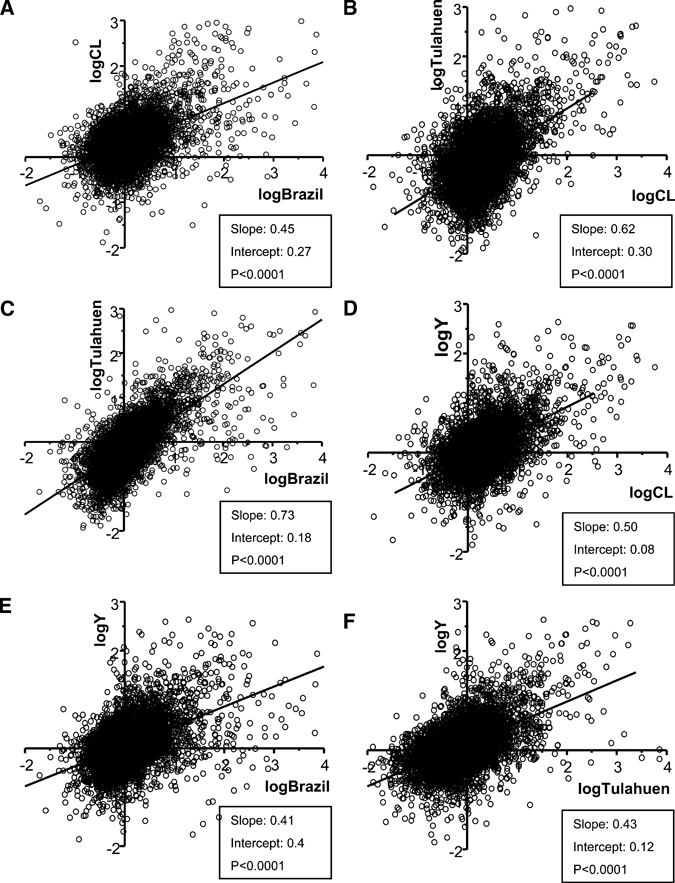
Correlation of datasets obtained by microarray analyses of the infection of L6E9 cell line with Y, CL, Brazil, and Tulahuen strains of T. cruzi. The datasets of each strain were plotted against each other in pairs of parasite strains. The histograms show how the transcriptomic changes induced by all strains have a highly significant correlation (all have P values < 0.0001). R2 values were (A) 0.5, (B) 0.51, (C) 0.67, (D) 0.52, (E) 0.47, and (F) 0.53.
Discussion
Chagas disease represents a spectrum of pathogenesis, varying both in its severity and the organ systems afflicted. Although various host factors, such as competence to launch immune response, may account in part for differences in the pathogenesis of the disease, parasite strain is also an important variable,6 resulting in differences in viability, infectivity, and tissue tropism.7–9 The present study was undertaken to evaluate the extent to which global gene-expression alteration was similarly altered after in vitro infection of a myoblast cell line with four distinct T. cruzi strains.
The microarray analyses described in this study were performed on the myoblast cell line L6E9 during infection with four reference strains of T. cruzi, each with well-characterized rates of in vivo and in vitro infectivity, resistance to chemotherapy, and pathogenesis in vivo. We used the Y and CL strains as representatives of the TC II group of T. cruzi, known to be found in central and eastern Brazil, which is commonly associated with the “mega” syndromes (cardiomegaly, megacolon, and megaesophagous).22 The Tulahuen strain was chosen to represent the TC I group, however, the Multiplex PCR performed with genomic DNA of parasites of this strain revealed that it was actually a strain belonging to the TC II family. The Brazil strain was selected, because it has been shown to cause a dilated cardiomyopathy associated with a reduction in fractional shortening and myocardial wall thinning in mice.13 Genotyping revealed that the Brazil strain belongs to the TC I group, the predominant group in Venezuela and central Brazil, which is usually associated with electrocardiographic (ECG) abnormalities.22
We have compared the datasets obtained from the microarray analyses in two ways, resulting in different but complementary conclusions regarding the pathogenesis of T. cruzi infection in vitro and perhaps, applicable to in vivo infection as well. First, through identification of the genes that had altered expression after infection, we identified a large number of gene expression changes in the infected cells, but only a very small number of these were common to infection with each parasite strain. We concluded from this analysis that each strain induces a particular fingerprint of pathology. One such fingerprint may include genes encoding cell-junction proteins, which was evidenced by our finding that both Tulahuen and Y strains down-regulated several cellular junction genes such as junction plakoglobin, junctional adhesion molecule 3, and adipocyte-specific adhesion molecule as well as cadherin 15 (Y strain). T. cruzi infection was also shown to alter adhesion molecules of the host such as connexin4323 and cadherin-catenin.11,24 These findings highlight an evolving concept that many types of cardiomyopathy target expression or involve mutations in molecular components of the intercalated disk (see reference 25 for review and reference 26 for changes in sepsis). Thus, as pointed out in a recent review,27 cardiomyopathies, including chronic chagasic cardiomyopathy, may be considered to be junctionopathies.
An additional conclusion from the small number of genes found to be commonly regulated in infections with all strains (e.g., cut-like 1, DNA-damage inducible transcript 3, proprotein convertase subtilisin/kexin type 7) may provide a subset of biomarkers that could be potentially useful for diagnosis of acute T. cruzi infection and possibly, also reliably determine both indeterminate phase and chronic disease.
The second type of analysis that we performed on the gene-expression datasets was to compare overall transcriptomic changes in myoblasts infected with each T. cruzi strain through regression analysis of relative expression levels of each gene. For this, we compared each of the six pair-wise combinations of parasite strains, in each case finding a highly significant positive slope. This analysis emphasizes the concept that although Chagas disease shows a spectrum of manifestations, it does indeed represent a syndrome of common phenotypic alterations. The shallow slopes of the regression lines indicate that the changes induced by the parasite strains, although similar, are on average very low in amplitude; this emphasizes again the potential utility of the few commonly altered biomarkers.
The significant alteration in few overlapping genes among myoblasts infected with all strains poses a formidable challenge to the development of disease biomarkers that can be used to detect disease endemic areas where endogenous infective strains differ, although the few genes that we have detected with strong and significant expression changes offer such a possibility. Nevertheless, the overall transcriptomic signature of the disease that is revealed in the strong correlations between subtle expression alteration of all genes may provide a source whereby additional biomarkers may be discovered as common principal components of the acute response.
Acknowledgments
The authors thank Vicki L. Braunstein (AECOM) and Angela Santos (LUC–Fiocruz) for the technical support with maintenance of the T. cruzi strains and Nadia Nehme and Dr. Octavio Fernandes (Fiocruz) and Aisha Cordero (AECOM) for the assistance with T. cruzi genotyping. We also thank Ethan Mackenzie (AECOM) for the help with GenMapp analysis. D.A. was supported in part by a grant from the Fogarty International Center-NIH D43 W007129 (HBT) and grants from The US National Institutes of Health AI-076248(HBT), HL-73732(HBT, DCS) and from CNPq, CNPq, PAPES IV-FIOCRUZ.
Footnotes
Authors' addresses: Daniel Adesse, Luciana Ribeiro Garzoni, and Maria de Nazareth Meirelles, Laboratório de Ultraestrutura Celular, Instituto Oswaldo Cruz, FIOCRUZ, Rio de Janeiro, Brazil. Dumitru A. Iacobas, Sanda Iacobas, and David C. Spray, Dominick P. Purpura Department of Neuroscience, Albert Einstein College of Medicine, Bronx, NY. Herbert B. Tanowitz, Department of Pathology, Albert Einstein College of Medicine, Bronx, NY.
References
- 1.de Souza W. Chagas' disease: facts and reality. Microbes Infect. 2007;9:544–545. doi: 10.1016/j.micinf.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Tanowitz HB, Kirchhoff LV, Simon D, Morris SA, Weiss LM, Wittner M. Chagas' disease. Clin Microbiol Rev. 1992;5:400–419. doi: 10.1128/cmr.5.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higuchi Mde L, Benvenuti LA, Martins Reis M, Metzger M. Pathophysiology of the heart in Chagas' disease: current status and new developments. Cardiovasc Res. 2003;60:96–107. doi: 10.1016/s0008-6363(03)00361-4. [DOI] [PubMed] [Google Scholar]
- 4.Carneiro M, Romanha AJ, Chiari E. Biological characterization of Trypanosoma cruzi strains from different zymodemes and schizodemes. Mem Inst Oswaldo Cruz. 1991;86:387–393. doi: 10.1590/s0074-02761991000400002. [DOI] [PubMed] [Google Scholar]
- 5.Andrade SG. Trypanosoma cruzi: clonal structure of parasite strains and the importance of principal clones. Mem Inst Oswaldo Cruz. 1999;94((Suppl 1)):185–187. doi: 10.1590/s0074-02761999000700026. [DOI] [PubMed] [Google Scholar]
- 6.Andrade SG, Magalhães JB. Biodemes and zymodemes of Trypanosoma cruzi strains: correlations with clinical data and experimental pathology. Rev Soc Bras Med Trop. 1996;30:27–35. doi: 10.1590/s0037-86821997000100006. [DOI] [PubMed] [Google Scholar]
- 7.Veloso VM, Carneiro CM, Toledo MJ, Lana M, Chiari E, Tafuri WL, Bahia MT. Variation in susceptibility to benznidazole in isolates derived from Trypanosoma cruzi parental strains. Mem Inst Oswaldo Cruz. 2001;96:1005–1011. doi: 10.1590/s0074-02762001000700021. [DOI] [PubMed] [Google Scholar]
- 8.Mielniczki-Pereira AA, Chiavegatto CM, López JA, Colli W, Alves MJ, Gadelha FR. Trypanosoma cruzi strains, Tulahuen 2 and Y, besides the difference in resistance to oxidative stress, display differential glucose-6-phosphate and 6-phosphogluconate dehydrogenases activities. Acta Trop. 2007;101:54–60. doi: 10.1016/j.actatropica.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez G, Wallace A, Olivares M, Diaz N, Aguilera X, Apt W, Solari A. Biological characterization of Trypanosoma cruzi zymodemes: in vitro differentiation of epimastigotes and infectivity of culture metacyclic trypomastigotes to mice. Exp Parasitol. 1990;71:125–133. doi: 10.1016/0014-4894(90)90015-5. [DOI] [PubMed] [Google Scholar]
- 10.Shigihara T, Hashimoto M, Shindo N, Aoki T. Transcriptome profile of Trypanosoma cruzi-infected cells: simultaneous up- and down-regulation of proliferation inhibitors and promoters. Parasitol Res. 2008;102:715–722. doi: 10.1007/s00436-007-0819-x. [DOI] [PubMed] [Google Scholar]
- 11.Imai K, Mimori T, Kawai M, Koga H. Microarray analysis of host gene-expression during intracellular nests formation of Trypanosoma cruzi amastigotes. Microbiol Immunol. 2005;49:623–631. doi: 10.1111/j.1348-0421.2005.tb03654.x. [DOI] [PubMed] [Google Scholar]
- 12.Mukherjee S, Belbin TJ, Spray DC, Iacobas DA, Weiss LM, Kitsis RN, Wittner M, Jelicks LA, Scherer PE, Ding A, Tanowitz HB. Microarray analysis of changes in gene expression in a murine model of chronic chagasic cardiomyopathy. Parasitol Res. 2003;91:187–196. doi: 10.1007/s00436-003-0937-z. [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee S, Nagajyothi F, Mukhopadhyay A, Machado FS, Belbin TJ, Campos de Carvalho A, Guan F, Albanese C, Jelicks LA, Lisanti MP, Silva JS, Spray DC, Weiss LM, Tanowitz HB. Alterations in myocardial gene expression associated with experimental Trypanosoma cruzi infection. Genomics. 2008;91:423–432. doi: 10.1016/j.ygeno.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg N, Popov VL, Papaconstantinou J. Profiling gene transcription reveals a deficiency of mitochondrial oxidative phosphorylation in Trypanosoma cruzi-infected murine hearts: implications in chagasic myocarditis development. Biochim Biophys Acta. 2003;1638:106–120. doi: 10.1016/s0925-4439(03)00060-7. [DOI] [PubMed] [Google Scholar]
- 15.Rowin KS, Tanowitz HB, Wittner M, Nguyen HT, Nadal-Ginard B. Inhibition of muscle differentiation by Trypanosoma cruzi. Proc Natl Acad Sci USA. 1983;80:6390–6394. doi: 10.1073/pnas.80.20.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertelli MS, Brener Z. Infection of tissue culture cells with bloodstream trypomastigotes of Trypanosoma cruzi. J Parasitol. 1980;66:992–997. [PubMed] [Google Scholar]
- 17.Fernandes O, Santos SS, Cupolillo E, Mendonça B, Derre R, Junqueira AC, Santos LC, Sturm NR, Naiff RD, Barret TV, Campbell DA, Coura JR. A mini-exon multiplex polymerase chain reaction to distinguish the major groups of Trypanosoma cruzi and T. rangeli in the Brazilian Amazon. Trans R Soc Trop Med Hyg. 2001;95:97–99. doi: 10.1016/s0035-9203(01)90350-5. [DOI] [PubMed] [Google Scholar]
- 18.Garzoni LR, Caldera A, Meirelles Mde N, de Castro SL, Docampo R, Meints GA, Oldfield E, Urbina JA. Selective in vitro effects of the farnesyl pyrophosphate synthase inhibitor risedronate on Trypanosoma cruzi. Int J Antimicrob Agents. 2004;23:273–285. doi: 10.1016/j.ijantimicag.2003.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Iacobas DA, Iacobas S, Li WE, Zoidl G, Dermietzel R, Spray DC. Genes controlling multiple functional pathways are transcriptionally regulated in connexin43 null mouse heart. Physiol Genomics. 2005;20:211–223. doi: 10.1152/physiolgenomics.00229.2003. [DOI] [PubMed] [Google Scholar]
- 20.Iacobas DA, Fan C, Iacobas S, Spray DC, Haddad GG. Transcriptomic changes in developing kidney exposed to chronic hypoxia. Biochem Biophys Res Commun. 2006;13:329–338. doi: 10.1016/j.bbrc.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 21.Combs TP, Nagajyothi F, Mukherjee S, de Almeida CJ, Jelicks LA, Schubert W, Lin Y, Jayabalan DS, Zhao D, Braunstein VL, Landskroner-Eiger S, Cordero A, Factor SM, Weiss LM, Lisanti MP, Tanowitz HB, Scherer PE. The adipocyte as an important target cell for Trypanosoma cruzi infection. J Biol Chem. 2005;24:24085–24094. doi: 10.1074/jbc.M412802200. [DOI] [PubMed] [Google Scholar]
- 22.Miles MA, Cedillos RA, Póvoa MM, de Souza AA, Prata A, Macedo V. Do radically dissimilar Trypanosoma cruzi strains (zymodemes) cause Venezuelan and Brazilian forms of Chagas' disease. Lancet. 1981;20:1338–1340. doi: 10.1016/s0140-6736(81)92518-6. [DOI] [PubMed] [Google Scholar]
- 23.Adesse D, Garzoni LR, Huang H, Tanowitz HB, de Nazareth Meirelles M, Spray DC. Trypanosoma cruzi induces changes in cardiac connexin43 expression. Microbes Infect. 2008;10:21–28. doi: 10.1016/j.micinf.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Melo TG, Meirelles Mde N, Pereira MC. Trypanosoma cruzi alters adherens junctions in cardiomyocytes. Microbes Infect. 2008;10:1405–1410. doi: 10.1016/j.micinf.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 25.Saffitz JE, Hames KY, Kanno S. Remodeling of gap junctions in ischemic and nonischemic forms of heart disease. J Membr Biol. 2007;218:65–71. doi: 10.1007/s00232-007-9031-2. [DOI] [PubMed] [Google Scholar]
- 26.Celes MR, Torres-Dueñas D, Alves-Filho JC, Duarte DB, Cunha FQ, Rossi MA. Reduction of gap and adherens junction proteins and intercalated disc structural remodeling in the hearts of mice submitted to severe cecal ligation and puncture sepsis. Crit Care Med. 2007;35:2176–2185. doi: 10.1097/01.ccm.0000281454.97901.01. [DOI] [PubMed] [Google Scholar]
- 27.Spray DC, Tanowitz HB. Pathology of mechanical and gap junctional co-coupling at the intercalated disc: is sepsis a junctionopathy. Crit Care Med. 2007;35:2231–2232. doi: 10.1097/01.CCM.0000281460.33497.56. [DOI] [PubMed] [Google Scholar]


