Abstract
We have developed two loop-mediated isothermal amplification (LAMP) assays for specific detection of Trypanosoma cruzi and Trypanosoma rangeli based on the 18S ribosomal RNA (rRNA) and the small nucleolar RNA (snoRNA) genes, respectively. The detection limit of the assays is 100 fg and 1 pg for T. cruzi and T. rangeli, respectively, with reactions conducted in 60 minutes. The two LAMP assays were used in detection of T. cruzi and T. rangeli infections in comparison with polymerase chain reaction (PCR) for DNA samples extracted from Rhodnius pallescens bugs collected from palm trees in Panama. Out of a total of 52 DNA samples from R. pallescens bugs 17 (33%) and 14 (27%) were T. cruzi-positive by LAMP and PCR, respectively, while, 7 (13%) and 4 (8%) were T. rangeli-positive by LAMP and PCR, respectively. Further evaluation of these LAMP assays is needed, especially with specimens collected from human patients as well as blood kept for transfusion purposes.
Introduction
Trypanosoma cruzi infects human beings and causes Chagas disease/American trypanosomiasis in Central and South America. It is transmitted by Rhodnius and Triatoma bugs through contaminated feces,1,2 and can also be transmitted vertically by blood transfusion or during organ transplantation.3 The non-pathogenic Trypanosoma rangeli shares the same geographical location and same insect vectors with T. cruzi,2,4,5 hence, necessitating accurate differential diagnosis. Trypanosoma rangeli is capable of transmission both through feces and through the salivary glands.6 Diagnosis of American trypanosomiasis relies on serological techniques, primarily using an indirect immunofluorescence assay (IFA) with T. cruzi epimastigote forms.4 Xenodiagnosis is mainly used for direct detection of T. cruzi parasites, although sometimes it is not effective in patients with very low parasitaemia,7 and furthermore, it is time consuming. Polymerase chain reaction (PCR) allows precise identification of the infecting trypanosome species from blood or tissue samples, and detection of trypanosomes in the vectors.2,5,7 Despite its high specificity and sensitivity, the use of PCR is still not widespread in diagnostic laboratories of endemic areas,4 mainly because of high costs and the requirement for specialized equipment.8,9
The loop-mediated isothermal amplification (LAMP) method is a gene amplification technique that uses 4 or 6 primers that detect DNA of pathogenic organisms with high sensitivity and specificity.10,11 The major advantages of LAMP include: 1) the reaction is isothermal and detection can be conducted within 60 minutes, 2) it requires simple heating devices such as a water bath or laboratory heat block, and 3) the detection of reaction results can be seen immediately after incubation by the naked eye caused by turbidity occurring in positive amplification reactions12 or by addition of fluorescent dyes after incubation such as SYBR green, ethidium bromide, or evagreen, which enable detection under UV light.13 A more convenient loopamp fluorescent detection reagent (FD) (Eiken Chemical Co. Ltd., Tokyo, Japan) has been specifically developed for detection of LAMP products, whereby its addition to the reaction mixture before incubation enables detection of results immediately after incubation by the naked eye under UV without opening the reaction tube.14 We have previously reported on LAMP primers developed for detection of T. cruzi infections,15 although they were never tested with field samples from infected hosts or vectors. In this study, we have developed LAMP assays for detection of T. cruzi and T. rangeli by targeting the 18S ribosomal RNA (rRNA) and small nucleolar RNA (snoRNA) genes, respectively, and further used these assays for detection of T. cruzi and T. rangeli infections in triatomine bugs collected from royal palm trees (Attalea butyracea) in Viento Fronco, Chilibre district, Panama.
Materials and Methods
Insect collection and DNA extraction.
Fifty-two triatomine bugs were collected from royal palm trees (Attalea butyracea) growing nearby households in the community of Viento Fronco, Chilibre district, Colón province, Republic of Panamá. The bugs were transported to the laboratory with the permission of the Autoridad Nacional del Medio Ambiente (ANAM) from April to May 2006. In the laboratory the bugs were identified taxonomically and then dissected aseptically, and DNA was extracted from insect extracts (all internal organs of the bug) using the Puregene DNA Purification kit (Gentra Systems Inc., Minneapolis MN) according to the manufacturer's instructions. Briefly, cell lysis solution and proteinase K (100 μg/mL) were added to whole insect extracts. The mixture was pipetted up and down to lyse the cells and then incubated at 55°C overnight. Samples were cooled at room temperature, protein precipitation solution was added, and the mixture was then centrifuged. The DNA was finally precipitated with 100% isopropanol, washed with 70% ethanol, and then hydrated with 50 μL of DNA hydration solution.
For optimization, specificity, and sensitivity of the reactions, the phenol-chloroform method, as described by Sambrook and Russel,16 was used for DNA extraction of T. cruzi (Tulahuan strain) epimastigotes from in vitro cultures and Triatoma infestans and Rhodnius prolixus from pathogen-free colonies of the National Research Center for Protozoan Diseases, Obihiro University and uninfected human blood DNA. The T. rangeli (Panama strain) DNA was kindly provided by Azael Saldaña, Parasitology Unit, College of Medicine, University of Panamá.
LAMP.
Table 1 shows LAMP primer sets for T. cruzi and T. rangeli parasites targeting the 18S rRNA and the snoRNA-c11 genes, respectively, designed using the LAMP primer explorer software version 4 (http://primerexplorer.jp/e/). The LAMP reactions were performed as previously described by Notomi and others.11 Briefly, a total volume of 25 μL containing 12.5 μL of 2× LAMP buffer (40 mM Tris-HCl [pH 8.8], 20 mM KCl, 16 mM MgSO4, 20 mM [NH4]2SO4, 0.2% Tween 20, 1.6 M Betaine, 2.8 mM of each deoxyribonucleotide triphosphates (dNTPs)), 1.3 μL primer mix (5 pmol each of F3 and B3, 40 pmol each of FIP and BIP and 20 pmol each of LF and LB), 8.2 μL distilled water, 1 μL (8 units) Bst DNA polymerase (New England Biolabs, Tokyo, Japan), and 2 μL of template DNA. In reactions whereby 1 μL FD was added to enable the detection by the naked eye, the volume of distilled water was adjusted appropriately. The reaction mixture was incubated at 63°C for 60 min using a Loopamp real-time turbidimeter (LA200, Teramecs, Tokyo, Japan).
Table 1.
LAMP primer sets used in this study
| Species | Accession no. | Size of target | Gene | Primer sequence |
|---|---|---|---|---|
| Trypanosoma cruzi | AF301912 | 187 bp | FIP: | 5′- CGTGAGTTGAGGGAAGGCATGAGTTGTTGGCAGACTTCGGT-3′ |
| BIP: | 5′-GCATCCAGGAATGAAGGAGGGTTCGTCTTGGTGCGGTCTA-3′ | |||
| F3: | 5′-CCGTGTGGCACTGTTTGT-3′ | |||
| B3: | 5′-TGAAGAATGCCTTCGCTGT-3′ | |||
| LF: | 5′-CATGTGAGATGCGAAGGG-3′ | |||
| LB: | 5′-CATGTGAGATGCGAAGGG-3′ | |||
| Trypanosoma rangeli | AY028385 | 172 bp | FIP: | 5′-TCATGCGTCGCAGCCGTACGCGAGAACGGGAGCA-3′ |
| BIP: | 5′-TTGCAGTTTCCTGTCAGCCTGACGTTTCAGTGTGAGCTGAGT-3′ | |||
| F3: | 5′-CGAGGACGGGCGAGAA-3′ | |||
| B3: | 5′-AAAAGGGGGGAAAGCAAGT-3′ | |||
| LF: | 5′-CCCGCCTTCTTCGCTCT-3′ | |||
| LB: | 5′-GCGCGTGACGACACAAC-3′ |
PCR.
The PCR reactions were performed with specific T. cruzi primers and T. rangeli primers reported previously.17 The primer sequences are as follows:
For T. cruzi, Tcru1: 5′-AAA TAA TGT ACG GGK GAG ATG CAT GA-3′ and Tcru2: 5′-GGT TCG ATT GGG GTT GGT GTA ATA TA-3′ and for T. rangeli, TrINT1: 5′-CGC CCA TTC GTT TGT CC-3′ and TrINT2: 5′-TCC AGC GCC ATC ACT GAT C-3′. The PCR mixture (25 μL total volume) contained PCR Buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2), 2 mM each of the dNTPs, 5 pmol of each primer, and 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Japan). The reaction mixtures were incubated in a PCR thermocycler (Applied Biosystems, Singapore) at 94°C for 10 min (initial denaturation step), and then subjected to 35 cycles consisting of 45 s at 94°C (denaturation step), 1 min at 58°C (annealing step), and 1 min at 72°C (extension), followed by terminal elongation for 7 min at 72°C. The PCR products were electrophoresed in a 1.5% agarose gel and stained with ethidium bromide solution for visualization under UV light.
Results and Discussion
The loopamp real-time turbidimetry device enables observation of primer kinetics. Therefore, at the commencement of the study, LAMP reactions were conducted at 60, 63, 65, and 67°C to determine the optimal reaction temperature for both the T. cruzi and T. rangeli primer sets. The temperature at which the reaction would reach and cross the positive reaction threshold, which is 0.1 of released turbidity18 in a short period of time or faster than others, was selected as the optimal reaction temperature (these reactions were done in five repetitions). As a result, the 63°C temperature was chosen as the optimal reaction temperature because the reaction threshold time for positive reactions was achieved faster at this temperature (data not shown). The T. cruzi 18S LAMP assay specifically amplified T. cruzi DNA without amplifying negative control DNA of T. rangeli, vector insects, and human host (Figure 1A). The detection limit of the assay was 100 fg of serially diluted T. cruzi DNA (Figure 1B and C). The T. rangeli snoRNA LAMP assay also specifically amplified T. rangeli DNA without amplifying the negative control DNA (Figure 2A) with a detection limit of 1 pg as determined from serially diluted DNA (Figure 2B and C). In this study, six LAMP primers were used for amplification of each target trypanosome DNA. In this way eight distinct regions were recognized on the target gene, thereby ensuring specificity, high sensitivity, and rapid reaction whereby amplification is achieved within 60 minutes.10
Figure 1.
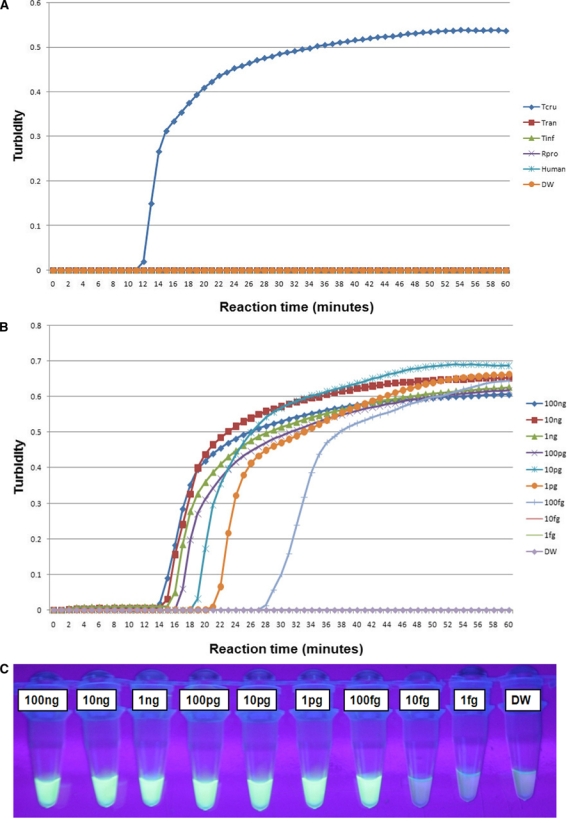
Loop-mediated isothermal amplification (LAMP) reactions with the 18S rRNA primer set for amplification of Trypanosoma cruzi DNA. Standard positive reaction threshold is 0.1 of the value of the turbidity released. (A) Specificity test of the LAMP assay using the real-time turbidimetry device. Tcru – T. cruzi; Tran – Trypanosoma rangeli; Tinf – Triatoma infestans; Rpro – Rhodnius prolixus; Human – DNA extracted from uninfected human blood; and DW – distilled water used as non-DNA negative control. Sensitivity test on serially diluted T. cruzi DNA from 100 ng down to 1 fg: (B) detection using the real-time turbidimetry device, (C) detection under UV light by the naked eye using FD reagent. The green/bright fluorescence indicates a positive reaction and the dark/less fluorescent color indicates a negative reaction. This figure appears in color at www.ajtmh.org.
Figure 2.
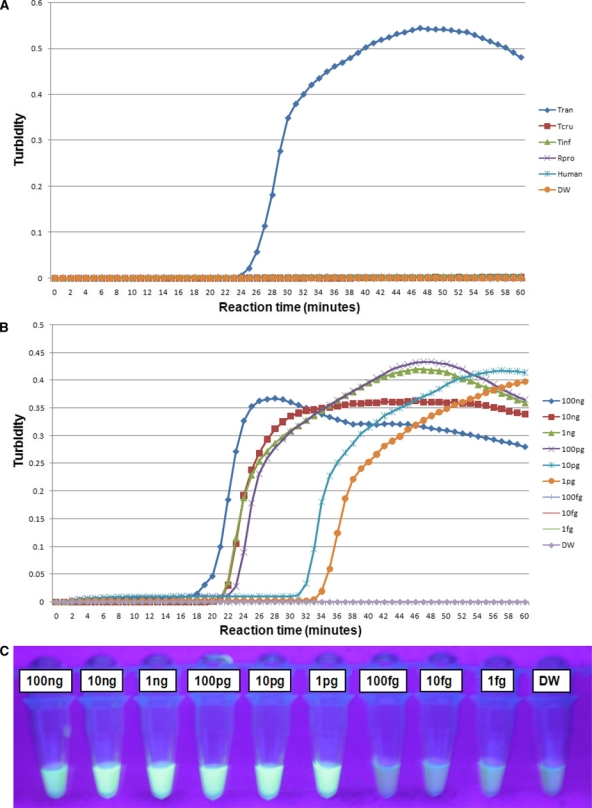
Loop-mediated isothermal amplification (LAMP) reactions with the snoRNA primer set for amplification of Trypanosoma rangeli DNA. Standard positive reaction threshold is 0.1 of the value of the turbidity released. (A) Specificity test of LAMP assay using the real-time turbidimetry device. Tran – T. rangeli; Tcru – Trypanosoma cruzi; Tinf – Triatoma infestans; Rpro – Rhodnius prolixus; Human – DNA extracted from uninfected human blood; and DW – distilled water used as non-DNA negative control. Sensitivity test on serially diluted T. rangeli DNA from 100 ng down to 1 fg: (B) detection using the real-time turbidimetry device, (C) detection under UV light by the naked eye using FD reagent. The green/bright fluorescence indicates a positive reaction and the dark/less fluorescent color indicates a negative reaction. This figure appears in color at www.ajtmh.org.
Rhodnius pallescens is considered to be the most important and widespread vector of T. cruzi and T. rangeli in Panama.19 In the current study, we therefore evaluated detection performance of the newly developed T. cruzi and T. rangeli LAMP assays on DNA extracted from triatomine bugs (37 nymphae and 15 adults) collected from palm trees in Panama. Of the 37 DNA samples extracted from R. pallescens nymphae, 10 (27%) and 7 (19%) were T. cruzi-positive by LAMP and PCR, respectively. Of the 15 adult R. pallescens DNA samples, 7 (47%) were positive for T. cruzi infections by both LAMP and PCR assays (Table 2). Therefore, out of a total of 52 DNA samples from R. pallescens, 17 (33%) and 14 (27%) were T. cruzi-positive by LAMP and PCR, respectively.
Table 2.
Detection of Trypanosoma rangeli and Trypanosoma cruzi infections from triatomine bugs
| Insect stage | Total no. of samples | T. rangeli | T. cruzi | ||
|---|---|---|---|---|---|
| PCR | LAMP | PCR | LAMP | ||
| +ve* | +ve | +ve | +ve | ||
| Nymph | 37 | 0 (0%) | 3 (8%) | 7† (19%) | 10† (27%) |
| Adult | 15 | 4‡ (27%) | 4‡ (27%) | 7‡ (47%) | 7‡ (47%) |
| Total | 52 | 4 (8%) | 7 (13%) | 14 (27%) | 17 (33%) |
Positive detection.
Both loop-mediated isothermal amplification (LAMP) and polymerase chain reaction (PCR) positively detected T. cruzi from same seven samples, whereas 3 samples were positive by LAMP only.
Four samples were detected as T. cruzi and T. rangeli mixed infections by both LAMP and PCR.
However, for T. rangeli, 3/37 (8%) were positively detected by LAMP from R. pallescens nymphae DNA, while none were positive by PCR. Of the 15 adult R. pallescens DNA samples, 4/15 (27%) were positively detected for T. rangeli infections by both LAMP and PCR (Table 2). Therefore, out of a total of 52 DNA samples from R. pallescens, 7 (13%) and 4 (8%) were T. rangeli-positive by LAMP and PCR, respectively. The LAMP assays developed in this study have shown a slightly higher detection performance than PCR. This is in agreement with previous studies where LAMP and PCR were compared for detection of salivarian trypanosome infections.8,20–22 Mixed infections were detected from four adult triatomine bug samples by both LAMP and PCR. This highlighted the importance of a species-specific assay for each trypanosome species.
In this study, we present LAMP assays based on 18S rRNA and snoRNA genes for detecting and differentiation of T. cruzi and T. rangeli infections. The Bst DNA polymerase used in the LAMP reaction is not affected by blood and tissue-derived components such as myoglobin, heme-blood protein complexes, and immunoglobin G.8,23,24 This gives LAMP an advantage of greater detection efficiency in comparison to PCR for field-derived samples. Furthermore, the FD reagent that is added to the reaction tube before incubation enables detection of LAMP results by the naked eye immediately after the reaction without opening the reaction tube, thereby reducing the risk of contamination. This study brings LAMP to the fore as a possible alternative molecular diagnostic tool for confirmation of the presence of T. cruzi and T. rangeli infections in vectors, clinical samples, transfusion blood samples, and during organ transplantation.
Acknowledgments
The cooperation of Viento Fronco residents in Panama during sampling is greatly acknowledged, with special thanks to Roberto Rojas for assistance during bug capturing. We also thank Azael Saldaña for kindly revising this manuscript.
Footnotes
Financial support: The first author was supported by a research grant fellowship for young scientists from the Japanese Society for the Promotion of Science (JSPS). FR and AMC-S received training on protozoan diseases at NRCPD as Japan International Cooperation Agency (JICA) participants. This study was also made possible by a Grant-in-aid for scientific research from JSPS to NI.
Authors' addresses: Oriel M. M. Thekisoe, Andrea M. Coronel-Servian, Shinya Fukumoto, Shin-Ichiro Kawazu, and Noboru Inoue, National Research Center for Protozoan Diseases, Obihiro University of Agriculture and Veterinary Medicine, Obihiro, Hokkaido, Japan, E-mails: thekisoe@yahoo.com, andycoronel@yahoo.com, fukumoto@obihiro.ac.jp, skawazu@obihiro.ac.jp, and ircpmi@obihiro.ac.jp. Carol V. Rodriguez and Francisco Rivas, College of Medicine, Department of Microbiology, University of Panamá, Panama, E-mails: carolrodriguez_24@hotmail.com and frivast@hotmail.com. Chihiro Sugimoto, Research Center for Zoonosis Control, Hokkaido University, Sapporo, Hokkaido, Japan, E-mail: sugimoto@czc.hokudai.ac.jp.
References
- 1.Pavia PX, Vallejo GA, Montilla M, Nicholls RS, Puerta CJ. Detection of Trypanosoma cruzi and Trypanosoma rangeli infections in triatomine vectors by amplification of the histone H2A/SIRE and the sno-RNA-C11 genes. Rev Inst Med Trop Sao Paulo. 2007;49:23–30. doi: 10.1590/s0036-46652007000100005. [DOI] [PubMed] [Google Scholar]
- 2.Vallejo GA, Guhl F, Chiari E, Macedo AM. Species specific detection of Trypanosoma cruzi and Trypanosoma rangeli in vector and mammalian hosts by polymerase chain reaction amplification of kinetoplast minicircle DNA. Acta Trop. 1999;72:203–212. doi: 10.1016/s0001-706x(98)00085-0. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Guillen MC, Barnabe C, Guegan JF, Tibayrenc M, Velasquez-Rojas M, Martinez-Munguia J, Salgado-Rosas H, Torres-Rasgado E, Rosas-Ramirez MI, Perez-Fuentes R. High prevalence anti-Trypanosoma cruzi antibodies, among blood donors in the state of Puebla, a non-endemic area of Mexico. Mem Inst Oswaldo Cruz. 2002;97:947–952. doi: 10.1590/s0074-02762002000700004. [DOI] [PubMed] [Google Scholar]
- 4.de Moraes MH, Guarneri AA, Girardi FP, Rodriques JB, Eger I, Tyler KM, Steindel M, Grisard EC. Different serological cross-reactivity of Trypanosoma rangeli forms in Trypanosoma cruzi infected patients sera. Parasit Vectors. 2008;1:20. doi: 10.1186/1756-3305-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morales L, Romero I, Diez H, Del Portillo P, Montilla M, Nicholls S, Puerta C. Characterization of a candidate Trypanosoma rangeli small nucleolar RNA gene and its application in a PCR-based parasite detection. Exp Parasitol. 2002;102:72–80. doi: 10.1016/s0014-4894(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 6.Stevens JR, Brisse S. In: The Trypanosomiases. Maudlin I, Holmes PH, Miles MA, editors. Oxfordshire, UK: CABI Publishing; 2004. pp. 1–23. (Systematics of trypanosomes of medical and veterinary importance). [Google Scholar]
- 7.Avila HA, Pereira JB, Thieman O, de Paiva E, Degrave W, Morel CM, Simpson L. Detection of Trypanosoma cruzi in blood specimens of chronic Chagas patients by polymerase chain reaction amplification of kinetoplast minicircle DNA: comparison with serology and xenodiagnosis. J Clin Microbiol. 1993;31:2421–2426. doi: 10.1128/jcm.31.9.2421-2426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, Sugimoto C, Igarashi I. Loop-mediated isothermal amplification (LAMP) for detection of African trypanosomes. J Clin Microbiol. 2003;38:2778–2780. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poon LL, Wong BW, Ma EH, Chan KH, Chow LM, Abeyewickreme W, Tangpukdee N, Yuen KW, Guan Y, Looareesuwan S, Peiris JS. Sensitive and inexpensive molecular test for falciparum malaria: Detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem. 2006;52:303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 10.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 11.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 13.Qiao Y-M, Guo Y-C, Zhang X-E, Zhou Y-F, Zhang Z-P, Wei H-P, Yang R-F, Wang D-B. Loop-mediated isothermal amplification for rapid detection of Bacillus anthracis spores. Biotechnol Lett. 2007;29:1939–1946. doi: 10.1007/s10529-007-9472-9. [DOI] [PubMed] [Google Scholar]
- 14.Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 15.Thekisoe OM, Kuboki N, Nambota A, Fujisaki K, Sugimoto C, Igarashi I, Yasuda J, Inoue N. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop. 2007;102:182–189. doi: 10.1016/j.actatropica.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. In: Molecular Cloning. Third edition. Sambrook J, Russell DW, editors. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. pp. 6.1–6.30. (Preparation and analysis of eukaryotic genomic DNA). [Google Scholar]
- 17.Desquesnes M, Dávila AM. Applications of PCR-based tools for detection and identification of animal trypanosomes: a review and perspectives. Vet Parasitol. 2002;109:213–231. doi: 10.1016/s0304-4017(02)00270-4. [DOI] [PubMed] [Google Scholar]
- 18.Mori Y, Kitao M, Tomita N, Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004;59:145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Pineda V, Montalvo E, Alvarez D, Santamaria AM, Calzada JE, Saldana A. Feeding sources and trypanosome infection index of Rhodnius pallescens in Chagas disease endemic area of Amador County, Panama. Rev Inst Med Trop Sao Paulo. 2008;50:113–116. doi: 10.1590/s0036-46652008000200009. [DOI] [PubMed] [Google Scholar]
- 20.Njiru ZK, Mikosza AS, Matovu E, Enyaru JC, Ouma JO, Kibona SN, Thompson RC, Ndung'u JM. African trypanosomiasis: sensitive and rapid detection of subgenus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol. 2008;38:589–599. doi: 10.1016/j.ijpara.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Njiru ZK, Mikosza AS, Armstrong T, Enyaru JC, Ndung'u JM, Thompson AR. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl Trop Dis. 2008;2:e147. doi: 10.1371/journal.pntd.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thekisoe OM, Omolo JD, Swai ES, Hayashida K, Zhang J, Sugimoto C, Inoue N. Preliminary application and evaluation of loop-mediated isothermal amplification (LAMP) for detection of bovine theileriosis and trypanosomosis in Tanzania. Onderstepoort J Vet Res. 2007;74:339–342. doi: 10.4102/ojvr.v74i4.119. [DOI] [PubMed] [Google Scholar]
- 23.Grab DJ, Lonsdale-Eccles J, Inoue N. LAMP for tadpoles. Nat Methods. 2005;2:635. doi: 10.1038/nmeth0905-635a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thekisoe OM, Bazie RS, Coronel-Servian AM, Sugimoto C, Kawazu S, Inoue N. Stability of loop-mediated isothermal amplification (LAMP) reagents and its amplification efficiency on crude trypanosome DNA templates. J Vet Med Sci. 2009;71:471–475. doi: 10.1292/jvms.71.471. [DOI] [PubMed] [Google Scholar]


